Integrating Satellite and UAV Data to Predict Peanut Maturity upon Artificial Neural Networks
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Field Evaluations
2.3. UAV Image Acquisition
2.4. Acquisition of PlanetScope Images
2.5. Vegetation Indices (VIs)
2.6. Statistical Analysis
2.7. Description of ANN Models for Prediction of PMI
2.8. Multilayer Perceptron Neural Networks (MLP)
2.9. Radial Basis Activation Function (RBF)
- the input layer,
- a hidden layer that applies a non-linear transformation from the input space to the hidden space, and
- the linear output layer seeks to classify patterns received from the hidden layer.
2.10. Training, Validation, and Performance
3. Results
3.1. Exploratory Analysis of PMI
3.2. Comparation between Remote Sensing Platforms
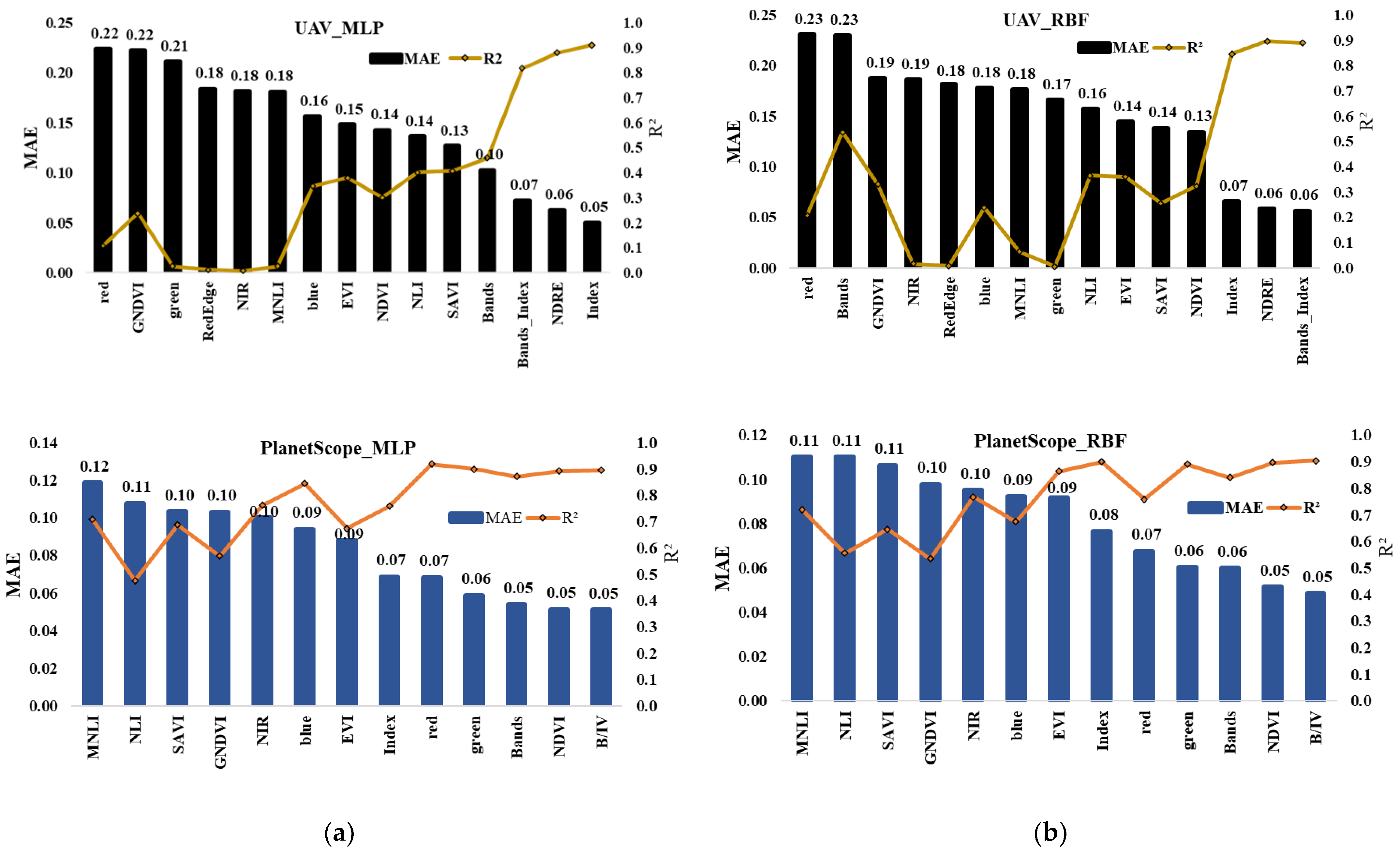
3.3. ANN Performance
3.4. Validation Models
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- West, P.C.; Gerber, J.S.; Engstrom, P.M.; Mueller, N.D.; Brauman, K.A.; Carlson, K.M.; Cassidy, E.S.; Johnston, M.; MacDonald, G.K.; Ray, D.K.; et al. Leverage points for improving global food security and the environment. Science 2014, 345, 325–328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radoglou-Grammatikis, P.; Sarigiannidis, P.; Lagkas, T.; Moscholios, I. A Compilation of UAV Applications for Precision Agriculture. Comput. Netw. 2020, 172, 107148. [Google Scholar] [CrossRef]
- Jung, J.; Maeda, M.; Chang, A.; Bhandari, M.; Ashapure, A.; Landivar-Bowles, J. The Potential of Remote Sensing and Artificial Intelligence as Tools to Improve the Resilience of Agriculture Production Systems. Curr. Opin. Biotechnol. 2021, 70, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Virnodkar, S.S.; Pachghare, V.K.; Patil, V.C.; Jha, S.K. Remote Sensing and Machine Learning for Crop Water Stress Determination in Various Crops: A Critical Review. Precis. Agric. 2020, 21, 1121–1155. [Google Scholar] [CrossRef]
- Colvin, B.C.; Tseng, Y.-C.; Tillman, B.L.; Rowland, D.L.; Erickson, J.E.; Culbreath, A.K.; Ferrell, J.A. Consideration of peg strength and disease severity in the decision to harvest peanut in southeastern USA. J. Crop Improv. 2018, 32, 287–304. [Google Scholar] [CrossRef]
- Williams, E.J.; Drexler, J.S. A Non-Destructive Method for Determining Peanut Pod Maturity. Peanut Sci. 1981, 8, 134–141. [Google Scholar] [CrossRef]
- Ashapure, A.; Jung, J.; Chang, A.; Oh, S.; Maeda, M.; Landivar, J. A Comparative Study of RGB and Multispectral Sensor-Based Cotton Canopy Cover Modelling Using Multi-Temporal UAS Data. Remote Sens. 2019, 11, 2757. [Google Scholar] [CrossRef] [Green Version]
- Rowland, D.L.; Sorensen, R.B.; Butts, C.L.; Faircloth, W.H.; Sullivan, D.G. Canopy Characteristics and Their Ability to Predict Peanut Maturity. Peanut Sci. 2008, 35, 43–54. [Google Scholar] [CrossRef] [Green Version]
- dos Santos, A.F.; Corrêa, L.N.; Lacerda, L.N.; Tedesco-Oliveira, D.; Pilon, C.; Vellidis, G.; da Silva, R.P. High-resolution satellite image to predict peanut maturity variability in commercial fields. Precis. Agric. 2021, 22, 1464–1478. [Google Scholar] [CrossRef]
- Santos, A.F.; Lacerda, L.N.; Rossi, C.; Moreno, L.d.A.; Oliveira, M.F.; Pilon, C.; Silva, R.P.; Vellidis, G. Using UAV and Multispectral Images to Estimate Peanut Maturity Variability on Irrigated and Rainfed Fields Applying Linear Models and Artificial Neural Networks. Remote Sens. 2022, 14, 93. [Google Scholar] [CrossRef]
- Li, R.; Zhao, Z.; Monfort, W.S.; Johnsen, K.; Tse, Z.T.H.; Leo, D.J. Development of a smartphone-based peanut data logging system. Precis. Agric. 2021, 22, 1006–1018. [Google Scholar] [CrossRef]
- Tsouros, D.C.; Bibi, S.; Sarigiannidis, P.G. A Review on UAV-Based Applications for Precision Agriculture. Information 2019, 10, 349. [Google Scholar] [CrossRef] [Green Version]
- de Oliveira, M.F.; dos Santos, A.F.; Kazama, E.H.; Rolim, G.D.S.; da Silva, R.P. Determination of application volume for coffee plantations using artificial neural networks and remote sensing. Comput. Electron. Agric. 2021, 184, 106096. [Google Scholar] [CrossRef]
- Khan, I.; Iqbal, M.; Hashim, M.M. Impact of Sowing Dates on the Yield and Quality of Sugar Beet (Beta Vulgaris l.) Cv. California-Kws. Proc. Pak. Acad. Sci. Part B 2020, 57, 51–60. [Google Scholar]
- Xie, B.; Zhang, H.K.; Xue, J. Deep Convolutional Neural Network for Mapping Smallholder Agriculture Using High Spatial Resolution Satellite Image. Sensors 2019, 19, 2398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hasan, A.S.M.M.; Sohel, F.; Diepeveen, D.; Laga, H.; Jones, M.G. A survey of deep learning techniques for weed detection from images. Comput. Electron. Agric. 2021, 184, 106067. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, Z.; Kang, Y.; Özdoğan, M. Corn Yield Prediction and Uncertainty Analysis Based on Remotely Sensed Variables Using a Bayesian Neural Network Approach. Remote Sens. Environ. 2021, 259, 112408. [Google Scholar] [CrossRef]
- Alvares, C.A.; Stape, J.L.; Sentelhas, P.C.; Gonçalves, J.D.M.; Sparovek, G. Köppen’s Climate Classification Map for Brazil. Meteorol. Z. 2013, 22, 711–728. [Google Scholar] [CrossRef]
- Planet. Planet Imagery Product Specification. 2020. Available online: https://assets.planet.com/marketing/PDF/Planet_Surface_Reflectance_Technical_White_Paper.pdf (accessed on 26 November 2021).
- Rouse, J.W.; Haas, R.H.; Schell, J.A.; Deering, D.W. Monitoring Vegetation Systems in the Great Plains with Erts. NASA Spec. Publ. 1974, 351, 309. [Google Scholar]
- Gitelson, A.A.; Merzlyak, M.N. Signature Analysis of Leaf Reflectance Spectra: Algorithm Development for Remote Sensing of Chlorophyll. J. Plant Physiol. 1996, 148, 494–500. [Google Scholar] [CrossRef]
- Gong, P.; Pu, R.; Biging, G.S.; Larrieu, M.R. Estimation of Forest Leaf Area Index Using Vegetation Indices Derived from Hyperion Hyperspectral Data. IEEE Trans. Geosci. Remote Sens. 2003, 41, 1355–1362. [Google Scholar] [CrossRef] [Green Version]
- Justice, C.O.; Vermote, E.; Townshend, J.R.G.; Defries, R.; Roy, D.P.; Hall, D.K.; Salomonson, V.V.; Privette, J.L.; Riggs, G.; Strahler, A.; et al. The Moderate Resolution Imaging Spectroradiometer (MODIS): Land Remote Sensing for Global Change Research. IEEE Trans. Geosci. Remote Sens. 1998, 36, 1228–1249. [Google Scholar] [CrossRef] [Green Version]
- Goel, N.S.; Qin, W. Influences of Canopy Architecture on Relationships between Various Vegetation Indices and LAI and Fpar: A Computer Simulation. Remote Sens. Rev. 1994, 10, 309–347. [Google Scholar] [CrossRef]
- Huete, A.R. A Soil-Adjusted Vegetation Index (SAVI). Remote Sens. Environ. 1988, 25, 295–309. [Google Scholar] [CrossRef]
- Fitzgerald, G.; Rodriguez, D.; O’Leary, G. Measuring and Predicting Canopy Nitrogen Nutrition in Wheat Using a Spectral Index—The Canopy Chlorophyll Content Index (CCCI). Field Crops Res. 2010, 116, 318–324. [Google Scholar] [CrossRef]
- Jiang, D.; Yang, X.; Clinton, N.; Wang, N. An Artificial Neural Network Model for Estimating Crop Yields Using Remotely Sensed Information. Int. J. Remote Sens. 2004, 25, 1723–1732. [Google Scholar] [CrossRef]
- Savegnago, R.; Nunes, B.; Caetano, S.; Ferraudo, A.; Schmidt, G.; Ledur, M.; Munari, D. Comparison of logistic and neural network models to fit to the egg production curve of White Leghorn hens. Poult. Sci. 2011, 90, 705–711. [Google Scholar] [CrossRef] [PubMed]
- Soares, P.; da Silva, J.; Santos, M. Artificial Neural Networks Applied to Reduce the Noise Type of Ground Roll. J. Seism. Explor. 2015, 24, 1–14. [Google Scholar]
- Jung, Y.H.; Hong, S.K.; Wang, H.S.; Han, J.H.; Pham, T.X.; Park, H.; Kim, J.; Kang, S.; Yoo, C.D.; Lee, K.J. Flexible Piezoelectric Acoustic Sensors and Machine Learning for Speech Processing. Adv. Mater 2020, 32, e1904020. [Google Scholar] [CrossRef]
- Miao, Y.; Mulla, D.J.; Robert, P.C. Identifying important factors influencing corn yield and grain quality variability using artificial neural networks. Precis. Agric. 2006, 7, 117–135. [Google Scholar] [CrossRef]
- Haykin, S.; Lippmann, R. Neural networks, a comprehensive foundation. Int. J. Neural Syst. 1994, 5, 363–364. [Google Scholar] [CrossRef]
- Awal, M.A.; Ikeda, T. Effect of Elevated Soil Temperature on Radiation-Use Efficiency in Peanut Stands. Agric. For. Meteorol. 2003, 118, 63–74. [Google Scholar] [CrossRef]
- Growth Stages of Peanut (Arachis Hypogaea L.)1|Peanut Science. Available online: https://meridian.allenpress.com/peanut-science/article/9/1/35/108765/Growth-Stages-of-Peanut-Arachis-hypogaea-L-1 (accessed on 18 April 2022).
- Shiratsuchi, L.S.; Brandão, Z.N.; Vicente, L.E.; de Castro Victoria, D.; Ducati, J.R.; de Oliveira, R.P.; de Fátima Vilela, M. Sensoriamento Remoto: Conceitos básicos e aplicações na Agricultura de Precisão. In Agricultura de Precisão: Rsultados de um Novo Olhar; Embrapa: Brasília, Brazil, 2014. [Google Scholar]
- Gitelson, A.A.; Viña, A.; Arkebauer, T.J.; Rundquist, D.C.; Keydan, G.; Leavitt, B. Remote estimation of leaf area index and green leaf biomass in maize canopies. Geophys. Res. Lett. 2003, 30, 1248. [Google Scholar] [CrossRef] [Green Version]
- Nguy-Robertson, A.L.; Peng, Y.; Gitelson, A.A.; Arkebauer, T.J.; Pimstein, A.; Herrmann, I.; Karnieli, A.; Rundquist, D.C.; Bonfil, D.J. Estimating Green LAI in Four Crops: Potential of Determining Optimal Spectral Bands for a Universal Algorithm. Agric. For. Meteorol. 2014, 192–193, 140–148. [Google Scholar] [CrossRef]
- Viña, A.; Gitelson, A.A.; Nguy-Robertson, A.L.; Peng, Y. Comparison of Different Vegetation Indices for the Remote Assessment of Green Leaf Area Index of Crops. Remote Sens. Environ. 2011, 115, 3468–3478. [Google Scholar] [CrossRef]
- Abd-El Monsef, H.; Smith, S.E.; Rowland, D.L.; Abd El Rasol, N. Using Multispectral Imagery to Extract a Pure Spectral Canopy Signature for Predicting Peanut Maturity. Comput. Electron. Agric. 2019, 162, 561–572. [Google Scholar] [CrossRef]
- Morlin Carneiro, F.; Angeli Furlani, C.E.; Zerbato, C.; Candida de Menezes, P.; da Silva Gírio, L.A.; Freire de Oliveira, M. Comparison between Vegetation Indices for Detecting Spatial and Temporal Variabilities in Soybean Crop Using Canopy Sensors. Precis. Agric. 2020, 21, 979–1007. [Google Scholar] [CrossRef]
- Taubinger, L.; Amaral, L.; Molin, J.P. Vegetation Indices from Active Crop Canopy Sensor and their Potential Interference Factors on Sugarcane. In Proceedings of the 11th International Conference on Precision Agriculture, Indianapolis, IN, USA, 15–18 July 2012. [Google Scholar]
- Amaral, L.R.; Molin, J.P.; Portz, G.; Finazzi, F.B.; Cortinove, L. Comparison of crop canopy reflectance sensors used to identify sugarcane biomass and nitrogen status. Precis. Agric. 2015, 16, 15–28. [Google Scholar] [CrossRef]
- Schuerger, A.C.; Capelle, G.A.; Di Benedetto, J.A.; Mao, C.; Thai, C.N.; Evans, M.D.; Richards, J.T.; Blank, T.A.; Stryjewski, E.C. Comparison of Two Hyperspectral Imaging and Two Laser-Induced Fluorescence Instruments for the Detection of Zinc Stress and Chlorophyll Concentration in Bahia Grass (Paspalum Notatum Flugge.). Remote Sens. Environ. 2003, 84, 572–588. [Google Scholar] [CrossRef]
- Xie, T.; Yu, H.; Wilamowski, B. Comparison between Traditional Neural Networks and Radial Basis Function Networks. In Proceedings of the 2011 IEEE International Symposium on Industrial Electronics, Gdansk, Poland, 27–30 June 2011; pp. 1194–1199. [Google Scholar]
- Hashemi Fath, A.; Madanifar, F.; Abbasi, M. Implementation of Multilayer Perceptron (MLP) and Radial Basis Function (RBF) Neural Networks to Predict Solution Gas-Oil Ratio of Crude Oil Systems. Petroleum 2020, 6, 80–91. [Google Scholar] [CrossRef]






| VI | Equation | References |
|---|---|---|
| NDVI | (NIR − Red)/(NIR + Red) | [20] |
| GNDVI | (NIR − Green)/(NIR + Green) | [21] |
| MNLI | (NIR2 − Red) (1 + L **)/(NIR2 + Red + L **) | [22] |
| EVI | 2.5 (NIR − Red)/(L ** + NIR + C1 ** Red − C2 **Blue) | [23] |
| NLI | (NIR − Red)/(NIR2 + Red) | [24] |
| SAVI | (1 + L **) (NIR − Red)/(L + NIR + Red) | [25] |
| NDRE *** | (NIR − RE)/(NIR + RE) | [26] |
| Training | Validation | |||||
|---|---|---|---|---|---|---|
| Simulation | Model | Model Structure | MAE | R2 | MAE | R2 |
| B/VI_sat * | RBF | 3:20:1 | 0.05 | 0.89 | 0.05 | 0.90 |
| MLP | 4:20–10:1 | 0.06 | 0.90 | 0.05 | 0.90 | |
| green_sat | RBF | 1:14:1 | 0.06 | 0.90 | 0.06 | 0.90 |
| MLP | 1:20–10:1 | 0.06 | 0.89 | 0.06 | 0.89 | |
| NDVI_sat | RBF | 1:13–1:1 | 0.05 | 0.89 | 0.05 | 0.89 |
| MLP | 1:1–6:1 | 0.05 | 0.90 | 0.05 | 0.90 | |
| NDRE_UAV | RBF | 1:1–1:1 | 0.06 | 0.88 | 0.06 | 0.88 |
| MLP | 1:20–10:1 | 0.06 | 0.90 | 0.06 | 0.90 | |
| Bands_sat | BRF | 3:3–20:1 | 0.05 | 0.87 | 0.05 | 0.87 |
| MLP | 4:20–8:1 | 0.06 | 0.86 | 0.06 | 0.84 | |
| Index_UAV | BRF | 3:3–20:1 | 0.05 | 0.91 | 0.05 | 0.91 |
| B/VI_UAV ** | MLP | 5:11–10:1 | 0.06 | 0.89 | 0.06 | 0.89 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Souza, J.B.C.; de Almeida, S.L.H.; Freire de Oliveira, M.; Santos, A.F.d.; Filho, A.L.d.B.; Meneses, M.D.; Silva, R.P.d. Integrating Satellite and UAV Data to Predict Peanut Maturity upon Artificial Neural Networks. Agronomy 2022, 12, 1512. https://doi.org/10.3390/agronomy12071512
Souza JBC, de Almeida SLH, Freire de Oliveira M, Santos AFd, Filho ALdB, Meneses MD, Silva RPd. Integrating Satellite and UAV Data to Predict Peanut Maturity upon Artificial Neural Networks. Agronomy. 2022; 12(7):1512. https://doi.org/10.3390/agronomy12071512
Chicago/Turabian StyleSouza, Jarlyson Brunno Costa, Samira Luns Hatum de Almeida, Mailson Freire de Oliveira, Adão Felipe dos Santos, Armando Lopes de Brito Filho, Mariana Dias Meneses, and Rouverson Pereira da Silva. 2022. "Integrating Satellite and UAV Data to Predict Peanut Maturity upon Artificial Neural Networks" Agronomy 12, no. 7: 1512. https://doi.org/10.3390/agronomy12071512
APA StyleSouza, J. B. C., de Almeida, S. L. H., Freire de Oliveira, M., Santos, A. F. d., Filho, A. L. d. B., Meneses, M. D., & Silva, R. P. d. (2022). Integrating Satellite and UAV Data to Predict Peanut Maturity upon Artificial Neural Networks. Agronomy, 12(7), 1512. https://doi.org/10.3390/agronomy12071512









