Use of SSR Markers for the Exploration of Genetic Diversity and DNA Finger-Printing in Early-Maturing Upland Cotton (Gossypium hirsutum L.) for Future Breeding Program
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Material
2.2. DNA Extraction, PCR Amplification and Electrophoresis Detection
2.3. Band Recording and Data Analysis
3. Results
3.1. Selection of SSR Primers and Polymorphisms in the Amplified Products
3.2. DNA Fingerprinting Analysis of the 79 Cotton Accessions
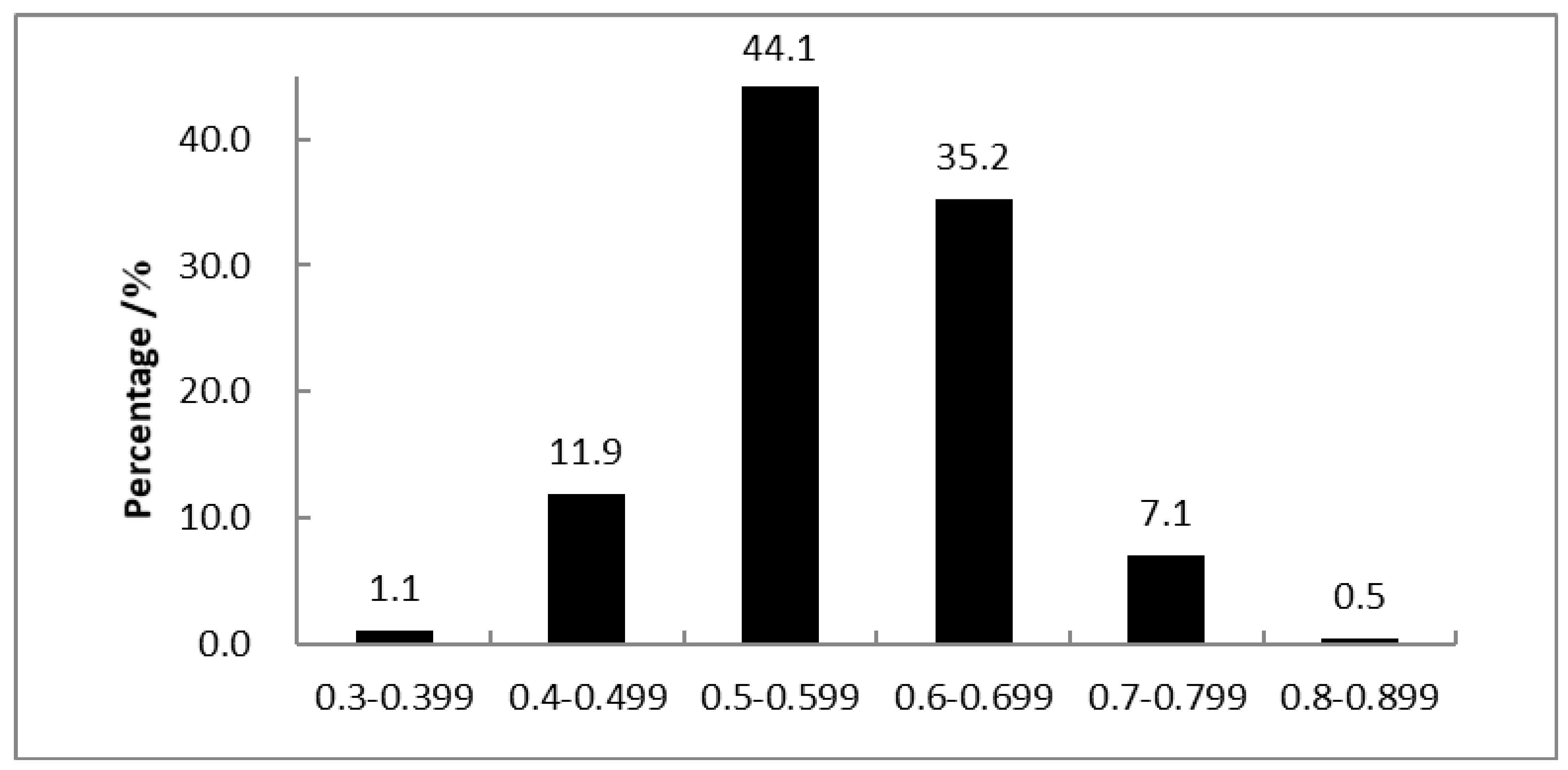
3.3. Genetics Diversity Analysis
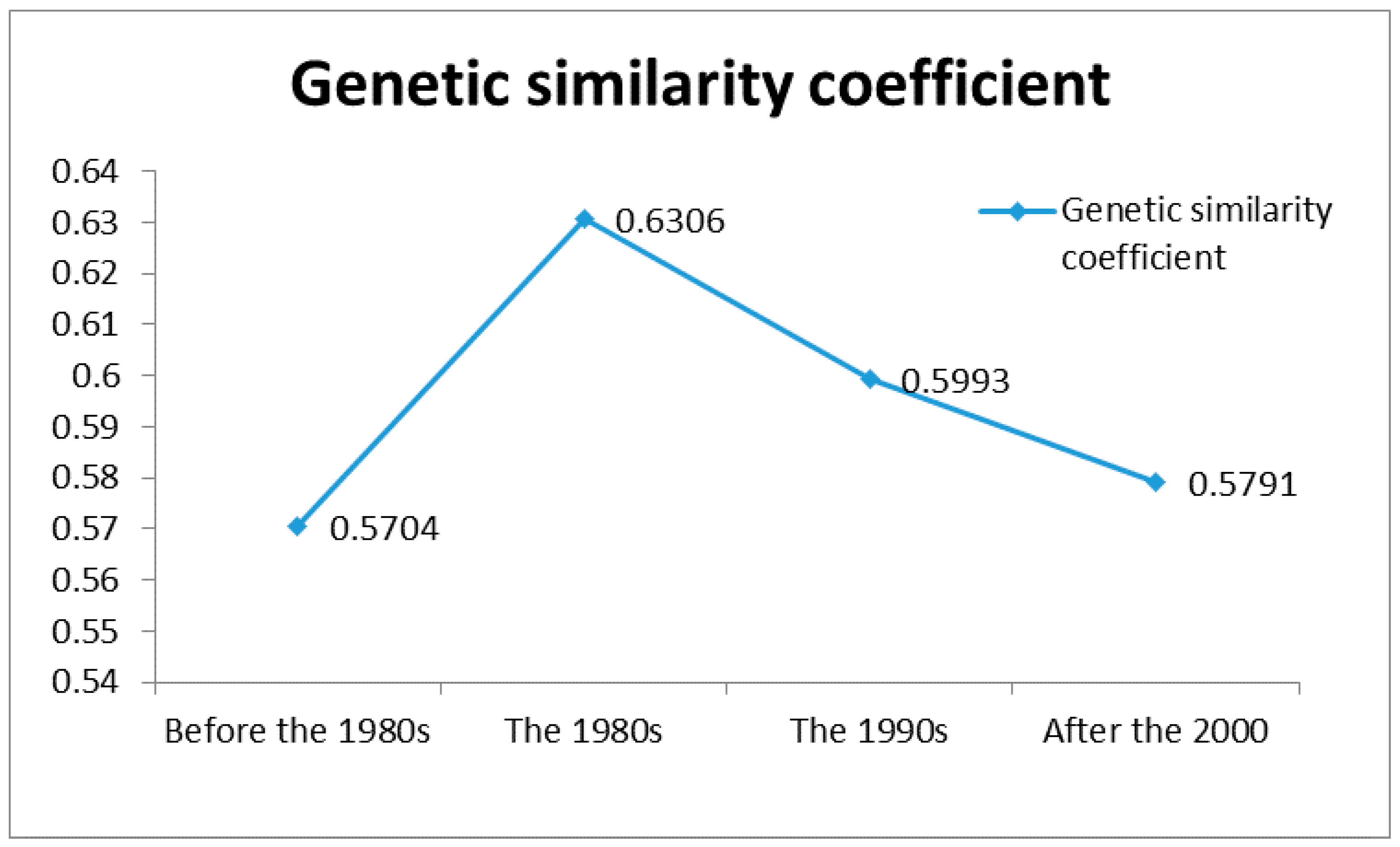
3.4. Region-Wise Genetic Diversity Analysis
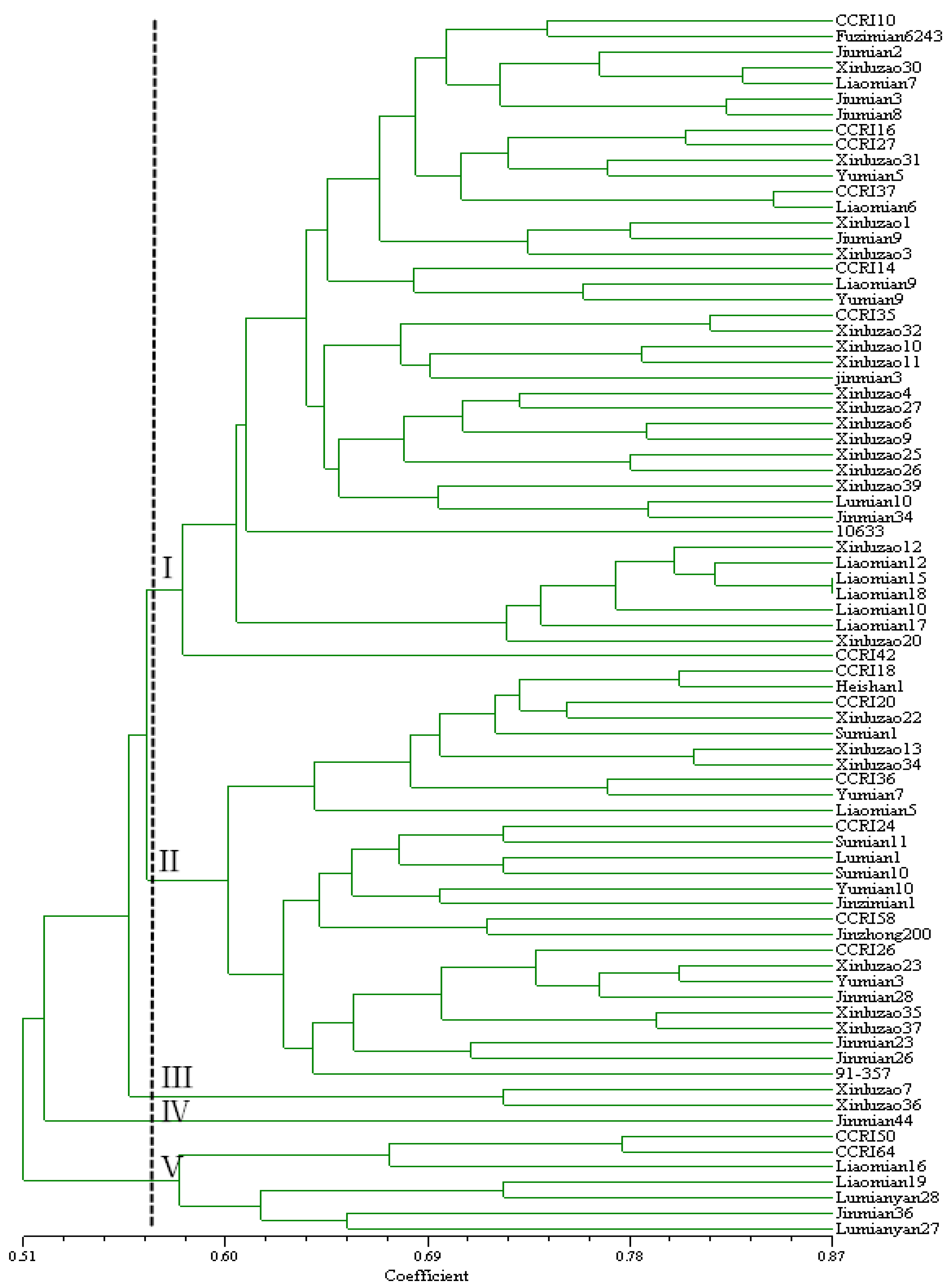
3.5. Hierarchical Cluster Analysis
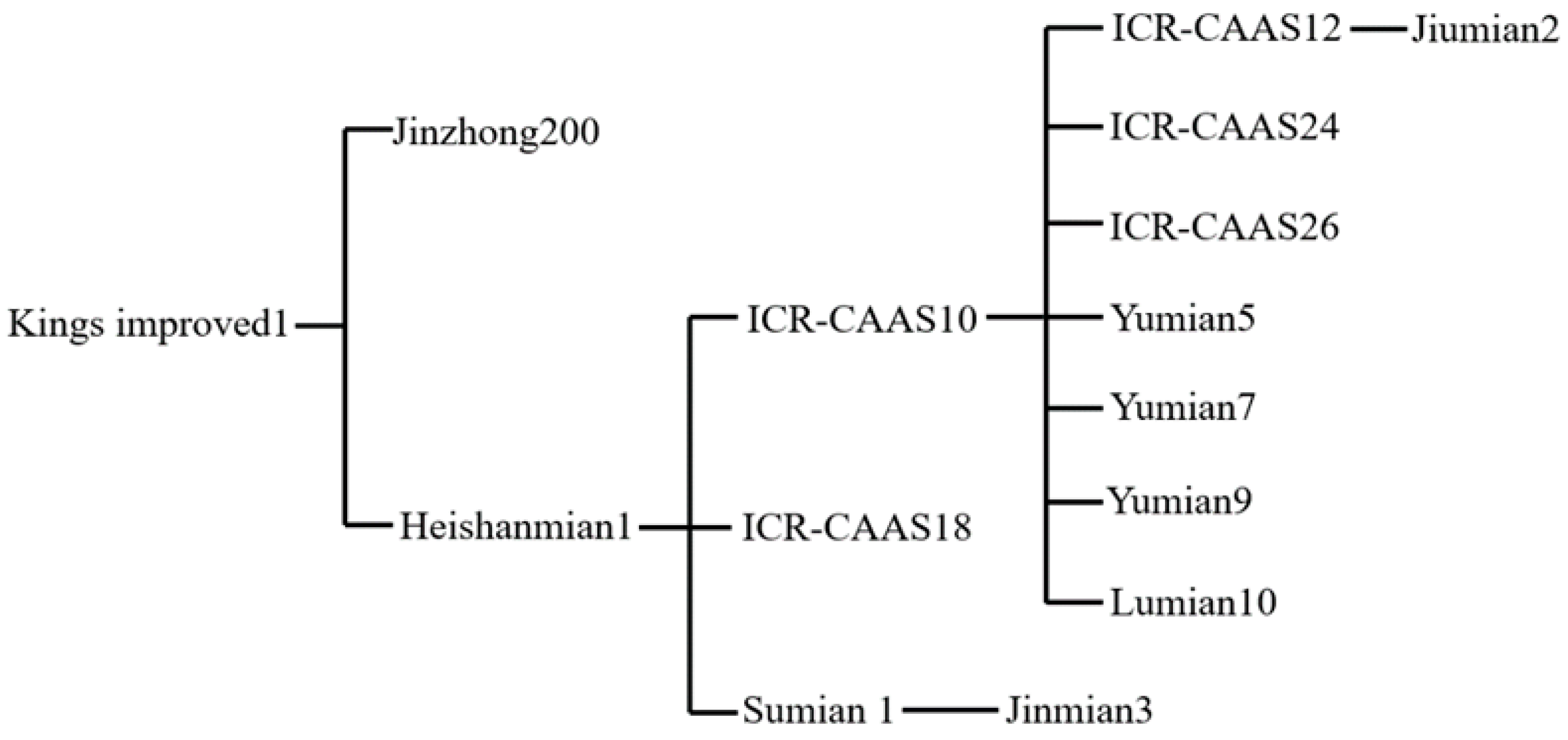
3.6. Pedigree/Parentage Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dai, J.; Kong, X.; Zhang, D.; Li, W.; Dong, H. Technologies and theoretical basis of light and simplified cotton cultivation in China. Field Crops Res. 2017, 214, 142–148. [Google Scholar] [CrossRef]
- Malik, W.; Abid, M.A.; Cheema, H.M.N.; Khan, A.A.; Iqbal, M.Z.; Qayyum, A.; Hanif, M.; Bibi, N.; Yuan, S.N.; Yasmeen, A.; et al. From Qutn to Bt cotton: Development, adoption and prospects. A review. Cytol. Genet. 2015, 49, 408–419. [Google Scholar] [CrossRef]
- Ma, Z.; He, S.; Wang, X.; Sun, J.; Zhang, Y.; Zhang, G.; Wu, L.; Li, Z.; Liu, Z.; Sun, G.; et al. Resequencing a core collection of upland cotton identifies genomic variation and loci influencing fiber quality and yield. Nat. Genet. 2018, 50, 803–813. [Google Scholar] [CrossRef]
- Liu, B.; Liang, M.; Huang, Z.; Tan, X. Duration–severity–area characteristics of drought events in eastern China determined using a three-dimensional clustering method. Int. J. Climatol. 2021, 41, E3065–E3084. [Google Scholar] [CrossRef]
- Feng, L.; Dai, J.; Tian, L.; Zhang, H.; Li, W.; Dong, H. Review of the technology for high-yielding and efficient cotton cultivation in the northwest inland cotton-growing region of China. Field Crops Res. 2017, 208, 18–26. [Google Scholar] [CrossRef]
- Yu, S.X.; Wang, H.T.; Wei, H.L.; Su, J.J. Research progress and application of early maturity in upland cotton. Cott. Sci. 2017, 29, 1–10. [Google Scholar]
- Li, C.; Wang, X.; Dong, N.; Zhao, H.; Xia, Z.; Wang, R.; Converse, R.L.; Wang, Q. QTL analysis for early-maturing traits in cotton using two upland cotton (Gossypium hirsutum L.) crosses. Breed. Sci. 2013, 63, 154–163. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Xiao, C.; Li, X.; Kuang, Z.; Wang, W.L.Y. DNA Fingerprints Construction and Genetic Diversity Analysis of Short-season Cotton by SSR Markers. Acta Agric. Boreali-Sin. 2014, 29, 98–104. [Google Scholar]
- Iqbal, M.J.; Aziz, N.; Saeed, N.A.; Zafar, Y.; Malik, K.A. Genetic diversity evaluation of some elite cotton varieties by RAPD analysis. Theor. Appl. Genet. 1997, 94, 139–144. [Google Scholar] [CrossRef]
- Noormohammadi, Z.; Rahnama, A.; Sheidai, M. EST-SSR and SSR analyses of genetic diversity in diploid cotton genotypes from Iran. Nuclear 2013, 56, 171–178. [Google Scholar] [CrossRef]
- Kumar, P.; Nimbal, S.; Budhlakoti, N.; Singh, V.; Sangwan, R.S. Genetic diversity and population structure analysis for morphological traits in upland cotton (Gossypium hirsutum L.). J. Appl. Genet. 2022, 63, 87–101. [Google Scholar] [CrossRef] [PubMed]
- Qian, Z.; Chen, H.; Cui, Y. Analysis of the SSR loci and development of molecular markers in Spinacia oleracea transcriptome. J. Agric Biotechnol. 2016, 24, 1688–1697. [Google Scholar]
- Xiong, Y.; Lei, X.; Bai, S.; Xiong, Y.; Liu, W.; Wu, W.; Yu, Q.; Dong, Z.; Yang, J.; Ma, X. Genomic survey sequencing, development and characterization of single-and multi-locus genomic SSR markers of Elymus sibiricus L. BMC Plant Biol. 2021, 21, 1–12. [Google Scholar] [CrossRef]
- Kumar, P.; Nimbal, S.; Sangwan, R.S.; Budhlakoti, N.; Singh, V.; Mishra, D.C.; Sagar; Choudhary, R.R. Identification of novel marker–trait associations for lint yield contributing traits in upland cotton (Gossypium hirsutum L.) using SSRs. Front. Plant Sci. 2021, 12, 855. [Google Scholar] [CrossRef] [PubMed]
- Santosh, H.B.; Meshram, M.; Santhy, V.; Waghmare, V.N. Microsatellite marker based diversity analysis and DNA fingerprinting of Asiatic cotton (Gossypium arboreum) varieties of India. J. Plant Biochem. Biotechnol. 2021, 31, 421–428. [Google Scholar] [CrossRef]
- Nie, X.-H.; You, C.-Y.; Li, X.-F.; Qin, J.-H.; Huang, C.; Guo, H.-L.; Wang, X.-Q.; Zhao, W.-X.; Lin, Z.-X. Construction of DNA fingerprinting and analysis of genetic diversity for Xinluzao cotton varieties. Acta Agron. Sin. 2014, 40, 2104–2117. [Google Scholar] [CrossRef]
- Li, X.; Shahzad, K.; Guo, L.; Qi, T.; Zhang, X.; Wang, H.; Tang, H.; Qiao, X.; Zhang, J.; Wu, J.; et al. Using yield quantitative trait locus targeted SSR markers to study the relationship between genetic distance and yield heterosis in upland cotton (Gossypium hirsutum). Plant Breed. 2019, 138, 105–113. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.Y.; Li, X.Y.; Gong, Z.L.; Wang, J.; Fan, L.; Zheng, J.; Liang, Y.; Guo, J.; Mamat, M.; Ai, X. DNA fingerprinting construction and genetic diversity analysis based on SSR markers for upland cotton in Xinjiang. Cotton Sci. 2018, 30, 308–315. [Google Scholar]
- Punitha, D.; Raveendran, T.S. DNA fingerprinting studies in colored cotton genotypes. Plant Breed. 2010, 123, 101–103. [Google Scholar] [CrossRef]
- Li, C.Q.; Li, H.P.; Zhao, H.H.; Dong, N.; Wang, Q.L.; Ai, N.J. SSR core primers reveal DNA fingerprinting of 30 Upland cotton (Gossypium hirsutum L.) varieties in China. Acta Bot. Bor-Occid. Sin. 2014, 34, 2210–2219. [Google Scholar]
- Li, C.; Chen, B.; Xu, X.; Li, D.; Dong, J. Simple sequence repeat markers associated/linked with agronomic traits, as core primers, are eminently suitable for DNA fingerprinting in Upland cotton. Breed. Sci. 2018, 68, 393–403. [Google Scholar] [CrossRef] [Green Version]
- Han, Z.F.; Wang, J.H.; Shen, G.F.; Wang, Z.; Zhao, F.; Li, R.; Zhang, J.; Kong, F.; Zhao, J. Construction of DNA fingerprinting and analysis of genetic diversity with SSR markers for major varieties from the Yellow River Valley. Cotton Sci. 2011, 23, 545–551. [Google Scholar]
- Li, C.Q.; Wang, X.Y.; Zhang, X.F.; Wang, Q.L. SSR fingerprinting establishment of Baimian series cotton inbred varieties (lines). Cott. Sci. 2011, 23, 228–234. [Google Scholar]
- Kuang, M.; Weihua, Y.; Hongxia, X. Construction of DNA fingerprinting and analysis of genetic diversity with SSR markers for cotton major cultivars in China. China Agric. Sci. 2011, 44, 20–27. [Google Scholar]
- Zhang, J.; Stewart, J.M. Economical and rapid method for extracting cotton genomic DNA. J. Cott. Sci. 2000, 4, 193–201. [Google Scholar]
- Yu, S.; Fan, S.; Wang, H.; Wei, H.; Pang, C. Progresses in research on cotton high yield breeding in China. Sci. Agric. Sin. 2016, 49, 3465–3476. [Google Scholar]
- Jasim, A.S.; Rafii, M.Y.; Latif, M.A.; Sakimin, S.Z.; Arolu, I.W.; Miah, G. Genetic Diversity of Aromatic Rice Germplasm Revealed by SSR Markers. Biomed. Res. Int. 2018, 2018, 7658032. [Google Scholar] [CrossRef] [Green Version]
- Alam, M.; Juraimi, A.S.; Rafii, M.Y.; Hamid, A.A.; Arolu, I.W.; Latif, M.A. Application of EST-SSR marker in detection of genetic variation among purslane (Portulaca oleracea L.) accessions. Braz. J. Bot. 2015, 38, 119–129. [Google Scholar] [CrossRef]
- Arolu, I.W.; Rafii, M.Y.; Hanafi, M.M.; Mahmud, T.M.M.; Askani, S. Genetic Divergence and Evaluation of Yield Potential of Jatropha curcas Accessions Collected from Peninsular Malaysia. Pertanika J. Trop. Agric. Sci. 2015, 38, 127–142. [Google Scholar]
- Noor, F.; Alshamrani, R.; Gull, M.; Mehmood, M.A.; Aslam, S. Identification of conserved and novel mature miRNAs in selected crops as future targets for metabolic engineering. Asian J. Agric. Biol. 2021, 2021, 202012551. [Google Scholar] [CrossRef]
- Bligh, H.F.J.; Blackhall, N.W.; Edwards, K.J.; McClung, A.M. Using amplified fragment length polymorphisms and simple sequence length polymorphisms to identify cultivars of brown and white milled rice. Crop Sci. 1999, 39, 1715–1721. [Google Scholar] [CrossRef]
- Jeung, J.U.; Hwang, H.G.; Moon, H.P.; Jena, K.K. Fingerprinting temperate japonica and tropical indica rice genotypes by comparative analysis of DNA markers. Euphytica 2005, 146, 239–251. [Google Scholar] [CrossRef]
- Sheidai, M.; Afshar, F.; Keshavarzi, M.; Talebi, S.M.; Noormohammadi, Z.; Shafaf, T. Genetic diversity and genome size variability in Linum austriacum (Lineaceae) populations. Biochem. Syst. Ecol. 2014, 57, 20–26. [Google Scholar] [CrossRef]
- Al-Mehemdi, A.F.; Elsahookie, M.M.; Al-Issawi, M.H. Analysis of genotype environment interaction in fennel using Sudoku design. Asian J. Agric. Biol. 2020, 8, 61–68. [Google Scholar] [CrossRef]
- Noormohammadi, Z.; Ibrahim-Khalili, N.; Sheidai, M.; Alishah, O. Genetic fingerprinting of diploid and tetraploid cotton cultivars by retrotransposon-based markers. Nuclear 2018, 61, 137–143. [Google Scholar] [CrossRef]
- Ul-Allah, S.; Ahmad, S.; Iqbal, M.; Naeem, M.; Ijaz, M.; Ahmad, M.Q. Creation of new genetic diversity in cotton germplasm through chemically induced mutation. Int. J. Agric. Biol. 2019, 22, 51–56. [Google Scholar]
- Ikram, A.; Aslam, H.M.U.; Atiq, M.; Amrao, L.; Ali, S.; Khan, N.A.; Naveed, K. Screening of resistant germplasm against powdery mildew of pea and its management through nutrients and plant activators. Asian J. Agric. Biol. 2020, 8, 85–91. [Google Scholar] [CrossRef]
- Van-Becelaere, G.; Lubbers, E.L.; Paterson, A.H.; Chee, P.W. Pedigree-vs. DNA marker-based genetic similarity estimates in cotton. Crop Sci. 2005, 45, 2281–2287. [Google Scholar] [CrossRef] [Green Version]
- Khan, A.I.; Awan, F.S.; Sadia, B.; Rana, R.M.; Khan, I.A. Genetic diversity studies among coloured cotton genotypes by using RAPD markers. Pak. J. Bot. 2010, 42, 71–77. [Google Scholar]
- Badigannavar, A.; Myers, G.O.; Jones, D.C. Molecular diversity revealed by AFLP markers in upland cotton genotypes. J. Crop Improv. 2012, 26, 627–640. [Google Scholar] [CrossRef]
- Zhu, L.; Tyagi, P.; Kaur, B.; Kuraparthy, V. Genetic diversity and population structure in elite US and race stock accessions of upland cotton (Gossypium hirsutum). J. Cotton Sci. 2019, 23, 38–47. [Google Scholar]
- Javaid, A.; Awan, F.S.; Azhar, F.M.; Khan, I.A. Assessment of allelic diversity among drought-resistant cotton genotypes using microsatellite markers. Genet. Mol. Res. 2017, 16, 28549206. [Google Scholar] [CrossRef] [PubMed]
- Gurmessa, D. Genetic diversity study of improved cotton (G. hirsutum L.) varieties in Ethiopia using simple sequence repeats markers. J. Biotechnol. 2019, 7, 6–14. [Google Scholar]
- McCarty, J.C.; Deng, D.D.; Jenkins, J.N.; Geng, L. Genetic diversity of day-neutral converted landrace Gossypium hirsutum L. accessions. Euphytica 2018, 214, 173. [Google Scholar] [CrossRef]
- Ali, I.; Khan, N.U.; Gul, S.; Khan, S.U.; Bibi, Z.; Aslam, K.; Shabir, G.; Haq, H.; Aslam, S.; Hussain, I.; et al. Genetic Diversity and Population Structure Analysis in Upland Cotton Germplasm. Int. J. Agric. Biol. 2019, 22, 669–676. [Google Scholar] [CrossRef]
- Lacape, J.-M.; Nguyen, T.-B.; Thibivilliers, S.; Bojinov, B.; Courtois, B.; Cantrell, R.G.; Burr, B.; Hau, B. A combined RFLP SSR AFLP map of tetraploid cotton based on a Gossypium hirsutum × Gossypium barbadense backcross population. Genome 2003, 46, 612–626. [Google Scholar] [CrossRef]
- Qin, J.; Li, Y.H.; Liu, Z.X.; Guan, R.X.; Zhang, M.C.; Chang, R.Z.; Li, M.; Ma, Z. Genetic relationship among parents of elite soybean (Glycine max) cultivars Suinong 14 pedigree revealed by SSR markers. Sci. Agric. Sin. 2008, 41, 3999–4007. [Google Scholar]
- Zhang, L.; Huang, Y.; Shen, X.; Liu, L.; Zhao, W.; Qiang, S. Analysis of genetic relationship of local varieties of Morus alba var. multicaulis from the lower area of Yellow River based on ISSR marker. J. Plant Resour. Environ. 2010, 19, 21–27. [Google Scholar]
- Benchasri, S.; Simla, S.; Harakotr, B. The effect of genotypic variability on the yield and yield components of okra (Abelmoschus esculentus L. Moench) in Thailand. Asian J. Agric. Biol. 2020, 8, 480–490. [Google Scholar] [CrossRef]
- Ahmad, N.; Raziuddin, A.F.; Iqbal, T.; Khan, N.; Nauman, M.; Hameed, F. Genetic analysis of biochemical traits in F3 populations of rapeseed (Brassica napus L.). Asian J. Agric. Biol. 2020, 8, 491–500. [Google Scholar] [CrossRef]
- Çelİk, S. Genetic Diversity Analysis of Some Upland Cotton (Gossypium hirsutum L.) Genotypes Using SSR Markers. Available online: https://dergipark.org.tr/en/download/article-file/1975684 (accessed on 10 June 2022).
- Gondal, M.R.; Saleem, M.Y.; Rizvi, S.A.; Riaz, A.; Naseem, W.; Muhammad, G.; Hayat, S.; Iqbal, M. Assessment of drought tolerance in various cotton genotypes under simulated osmotic settings. Asian J. Agric. Biol. 2021, 2021, 202008437. [Google Scholar] [CrossRef]
- Ali, I.; Khan, N.U.; Rahman, M.; Gul, R.; Bibi, Z.; Gul, S.; Ahmed, S.; Ali, S.; Ali, N.; Afrid, K.; et al. Genotype by environment and biplot analyses for yield and fiber traits in upland cotton. Int. J. Agric. Biol. 2018, 20, 1979–1990. [Google Scholar]
- Buyyarapu, R.; Kantety, R.V.; Yu, J.Z.; Saha, S.; Sharma, G.C. Development of new candidate gene and EST-based molecular markers for Gossypium species. Int. J. Plant Genom. 2011, 2011, 894598. [Google Scholar]
- Ijaz, U.; Pervaiz, T.; Ahmed, T.; Seemab, R.; Muhammad, S.M.; Noman, M.; Nadeem, M.; Azeem, F. Plant Cis-regulatory elements: Methods of identification and applications. Asian J. Agric. Biol. 2020, 8, 207–222. [Google Scholar] [CrossRef]
- Barbara, D.; Margit, C.M. High resolution melting (HRM) analysis of DNA–Its role and potential in food analysis. Food Chem. 2014, 158, 245–254. [Google Scholar]
- Cinzia, M.; Monica, M.M.; Antonella, P.; Valentina, F.; Wilma, S.; Valentina, R. Traceability of PDO Olive Oil “Terra di Bari” Using High Resolution Melting. J. Chem. 2015, 2015, 496986. [Google Scholar]

| SN | SRR Locus | Chromosome | PIC | Genetic Diversity (H′) | Effective Number of Alleles (Ne) |
|---|---|---|---|---|---|
| 1 | NAU4073 | Chr.01 | 0.8168 | 5.7030 | 5.4582 |
| 2 | NAU2457 | Chr.01 | 0.2577 | 1.5311 | 1.3471 |
| 3 | NAU2083 | Chr.01 | 0.4356 | 1.8725 | 1.7717 |
| 4 | NAU4044 | Chr.01 | 0.8822 | 9.1008 | 8.4884 |
| 5 | MON_COT064 | Chr.02 | 0.4980 | 1.9960 | 1.9920 |
| 6 | NAU1190 | Chr.03 | 0.7956 | 5.3171 | 4.8933 |
| 7 | MON_CGR6528 | Chr.03 | 0.7423 | 3.9381 | 3.8800 |
| 8 | MON_CGR6683 | Chr.03 | 0.4576 | 1.9158 | 1.8437 |
| 9 | NAU1269 | Chr.05 | 0.7314 | 3.8504 | 3.7234 |
| 10 | MON_CGR5732 | Chr.05 | 0.7307 | 3.8447 | 3.7138 |
| 11 | NAU1225 | Chr.05 | 0.7307 | 3.8447 | 3.7138 |
| 12 | MON_DC40122 | Chr.05 | 0.6884 | 3.5157 | 3.2096 |
| 13 | NAU1221 | Chr.05 | 0.7307 | 3.8447 | 3.7138 |
| 14 | MON_CGR5651 | Chr.06 | 0.6811 | 3.4515 | 3.1354 |
| 15 | MON_DPL0702 | Chr.06 | 0.7349 | 3.8787 | 3.7721 |
| 16 | MON_COT002 | Chr.06 | 0.7266 | 3.8116 | 3.6572 |
| 17 | BNL1694 | Chr.07 | 0.7442 | 3.9534 | 3.9092 |
| 18 | MUSS095 | Chr.07 | 0.7431 | 3.9447 | 3.8921 |
| 19 | MON_DC30218 | Chr.07 | 0.5000 | 2.0000 | 2.0000 |
| 20 | NAU3859 | Chr.09 | 0.4980 | 1.9960 | 1.9920 |
| 21 | DPL0431 | Chr.10 | 0.6873 | 3.4966 | 3.1981 |
| 22 | NAU3784 | Chr.11 | 0.7071 | 3.6561 | 3.4141 |
| 23 | MON_CER0098 | Chr.11 | 0.7250 | 3.8002 | 3.6363 |
| 24 | NAU3563 | Chr.11 | 0.8579 | 7.4448 | 7.0361 |
| 25 | NAU2671 | Chr.12 | 0.6978 | 3.5853 | 3.3095 |
| 26 | MON_DPL0491 | Chr.12 | 0.2397 | 1.4972 | 1.3153 |
| 27 | NAU3991 | Chr.13 | 0.6801 | 3.4455 | 3.1264 |
| 28 | MON_COT009 | Chr.13 | 0.6886 | 3.5149 | 3.2116 |
| 29 | BNL1421 | Chr.13 | 0.7107 | 3.6867 | 3.4567 |
| 30 | NAU3308 | Chr.14 | 0.6721 | 3.3728 | 3.0500 |
| 31 | GH304 | Chr.15 | 0.7085 | 3.6704 | 3.4305 |
| 32 | MUSS440 | Chr.15 | 0.8784 | 8.9176 | 8.2218 |
| 33 | NAU2343 | Chr.15 | 0.7354 | 3.8824 | 3.7790 |
| 34 | BNL2646 | Chr.15 | 0.6360 | 3.1112 | 2.7475 |
| 35 | NAU2742 | Chr.17 | 0.6635 | 3.3170 | 2.9714 |
| 36 | MON_DPL0308 | Chr.18 | 0.5896 | 2.7536 | 2.4365 |
| 37 | NAU3011 | Chr.18 | 0.4980 | 1.9960 | 1.9920 |
| 38 | NAU5262 | Chr.18 | 0.7407 | 3.9252 | 3.8560 |
| 39 | MON_CGR6151 | Chr.19 | 0.4768 | 1.9539 | 1.9115 |
| 40 | NAU1187 | Chr.19 | 0.7307 | 3.8447 | 3.7138 |
| 41 | NAU1042 | Chr.19 | 0.7307 | 3.8447 | 3.7138 |
| 42 | MON_CGR5590 | Chr.19 | 0.7193 | 3.7542 | 3.5631 |
| 43 | TMB1791 | Chr.19 | 0.7387 | 3.9101 | 3.8277 |
| 44 | MON_CGR6439 | Chr.20 | 0.2604 | 1.5362 | 1.3520 |
| 45 | DPL0442 | Chr.20 | 0.7317 | 3.8542 | 3.7271 |
| 46 | MON_SHIN1421 | Chr.20 | 0.7418 | 3.9344 | 3.8728 |
| 47 | MON_CGR5565 | Chr.20 | 0.9043 | 11.0904 | 10.4502 |
| 48 | BNL1551 | Chr.21 | 0.7006 | 3.6046 | 3.3401 |
| 49 | GH222 | Chr.22 | 0.8094 | 5.5713 | 5.2460 |
| 50 | MON_CGR6410 | Chr.22 | 0.7378 | 3.9022 | 3.8136 |
| 51 | CIR253 | Chr.22 | 0.6158 | 2.9431 | 2.6031 |
| 52 | MUSS139 | Chr.23 | 0.7426 | 3.9408 | 3.8848 |
| 53 | MON_CGR5202 | Chr.24 | 0.3230 | 1.6552 | 1.4772 |
| 54 | MON_CGR6932 | Chr.25 | 0.7423 | 3.9381 | 3.8800 |
| 55 | CRI151 | 0.7312 | 3.8485 | 3.7200 | |
| 56 | MON_CGR6389 | 0.7430 | 3.9442 | 3.8913 | |
| 57 | NAU3181 | 0.1841 | 1.3919 | 1.2256 | |
| 58 | MON_C2-0118 | 0.7397 | 3.9179 | 3.8418 | |
| 59 | CRI002 | 0.6296 | 3.0356 | 2.7000 | |
| 60 | MUCS375 | 0.3519 | 1.7103 | 1.5429 | |
| 61 | MON_CGR6784 | 0.6384 | 3.1230 | 2.7658 | |
| 62 | CIR096 | 0.7210 | 3.7659 | 3.5837 | |
| 63 | NAU3254 | 0.8289 | 6.4664 | 5.8436 | |
| 64 | MON_DPL0133 | 0.5479 | 2.4396 | 2.2119 | |
| 65 | MON_CER0168 | 0.7397 | 3.9179 | 3.8421 | |
| 66 | MUSB0175 | 0.4734 | 1.9470 | 1.8989 | |
| 67 | MON_DPL0906 | 0.4999 | 1.9998 | 1.9997 | |
| 68 | GH111 | 0.4734 | 1.9470 | 1.8989 | |
| 69 | GH112 | 0.4647 | 1.9297 | 1.8680 | |
| 70 | MON_DC40266 | 0.7037 | 3.6271 | 3.3744 | |
| 71 | MON_DC40286 | 0.6888 | 3.5120 | 3.2131 |
| Cultivar | Specific Primer | Cultivar | Specific Primer |
|---|---|---|---|
| ICR-CAAS64 | NAU1190, MUSS440 | Xinluzao20 | NAU3254 |
| Liaomian17 | NAU3254 | Liaomian19 | NAU3254 |
| Xinluzao25 | NAU4044 | Jiumian9 | NAU4044 |
| Liaomian5 | NAU4044 | Lumianyan28 | NAU4044 |
| Jinmian23 | NAU4073 |
| Region | Genetic Similarity Coefficient | CAAS | YRB | Northwest Inland Region | Liaohe River Basin | United States | Former Soviet Union |
|---|---|---|---|---|---|---|---|
| CAAS | Max | 0.8058 | 0.7817 | 0.8169 | 0.8451 | 0.7447 | 0.6691 |
| Min | 0.3630 | 0.3643 | 0.3704 | 0.3582 | 0.4789 | 0.4296 | |
| Mean | 0.6032 | 0.5796 | 0.5943 | 0.5824 | 0.6143 | 0.5672 | |
| YRB | Max | 0.7899 | 0.8028 | 0.7606 | 0.7042 | 0.7042 | |
| Min | 0.4085 | 0.3310 | 0.3475 | 0.3521 | 0.4366 | ||
| Mean | 0.5802 | 0.5636 | 0.5575 | 0.5911 | 0.5661 | ||
| Northwest Inland Region | Max | 0.8239 | 0.8310 | 0.7324 | 0.7254 | ||
| Min | 0.4225 | 0.3582 | 0.4296 | 0.4296 | |||
| Mean | 0.6221 | 0.5936 | 0.5799 | 0.5894 | |||
| Liaohe River Basin | Max | 0.8705 | 0.7465 | 0.6812 | |||
| Min | 0.3944 | 0.4718 | 0.4014 | ||||
| Mean | 0.6134 | 0.5783 | 0.5684 | ||||
| United States | Max | 0.5845 | 0.6549 | ||||
| Min | 0.5845 | 0.5357 | |||||
| Mean | 0.5845 | 0.6027 | |||||
| Former Soviet Union | Max | 0.5286 | |||||
| Min | 0.5286 | ||||||
| Mean | 0.5286 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuang, Z.; Xiao, C.; Ilyas, M.K.; Ibrar, D.; Khan, S.; Guo, L.; Wang, W.; Wang, B.; Huang, H.; Li, Y.; et al. Use of SSR Markers for the Exploration of Genetic Diversity and DNA Finger-Printing in Early-Maturing Upland Cotton (Gossypium hirsutum L.) for Future Breeding Program. Agronomy 2022, 12, 1513. https://doi.org/10.3390/agronomy12071513
Kuang Z, Xiao C, Ilyas MK, Ibrar D, Khan S, Guo L, Wang W, Wang B, Huang H, Li Y, et al. Use of SSR Markers for the Exploration of Genetic Diversity and DNA Finger-Printing in Early-Maturing Upland Cotton (Gossypium hirsutum L.) for Future Breeding Program. Agronomy. 2022; 12(7):1513. https://doi.org/10.3390/agronomy12071513
Chicago/Turabian StyleKuang, Zhengcheng, Caisheng Xiao, Muhammad Kashif Ilyas, Danish Ibrar, Shahbaz Khan, Lishuang Guo, Wei Wang, Baohua Wang, Hui Huang, Yujun Li, and et al. 2022. "Use of SSR Markers for the Exploration of Genetic Diversity and DNA Finger-Printing in Early-Maturing Upland Cotton (Gossypium hirsutum L.) for Future Breeding Program" Agronomy 12, no. 7: 1513. https://doi.org/10.3390/agronomy12071513
APA StyleKuang, Z., Xiao, C., Ilyas, M. K., Ibrar, D., Khan, S., Guo, L., Wang, W., Wang, B., Huang, H., Li, Y., Li, Y., Zheng, J., Saleem, S., Tahir, A., Ghafoor, A., & Chen, H. (2022). Use of SSR Markers for the Exploration of Genetic Diversity and DNA Finger-Printing in Early-Maturing Upland Cotton (Gossypium hirsutum L.) for Future Breeding Program. Agronomy, 12(7), 1513. https://doi.org/10.3390/agronomy12071513







