Genetic Dissection of Stem Branch Trait and Envisioning of Fixing Heterosis by Vegetative Reproduction in Oryza rufipogon
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Natural Field Experiment Conditions
2.2. Phenotypes Analysis
2.3. DNA Extraction
2.4. Mapping of qSBR1 and qSBR2
2.5. Statistical Analysis
3. Results
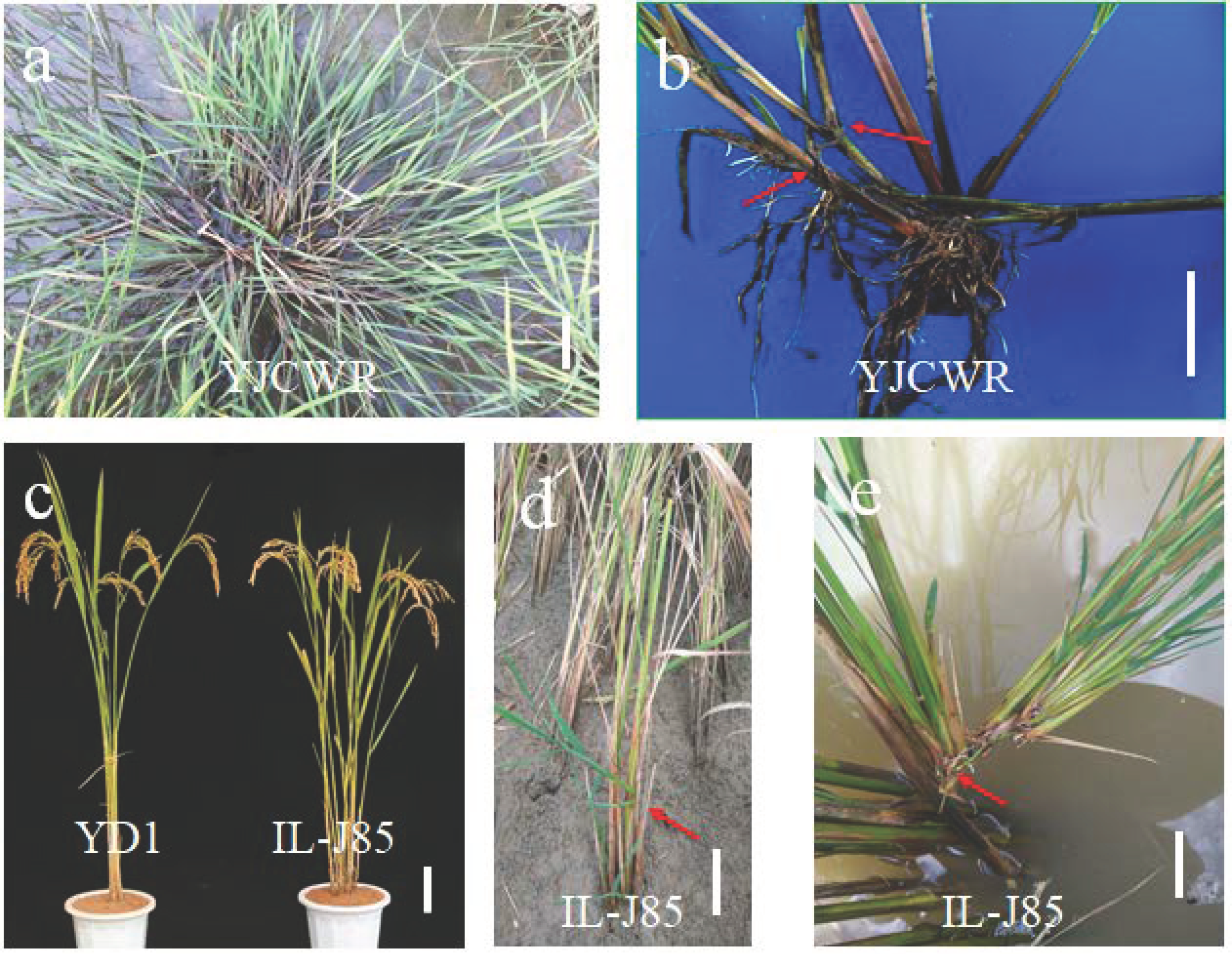
3.1. Phenotypic Analysis of YD1 and IL-J85
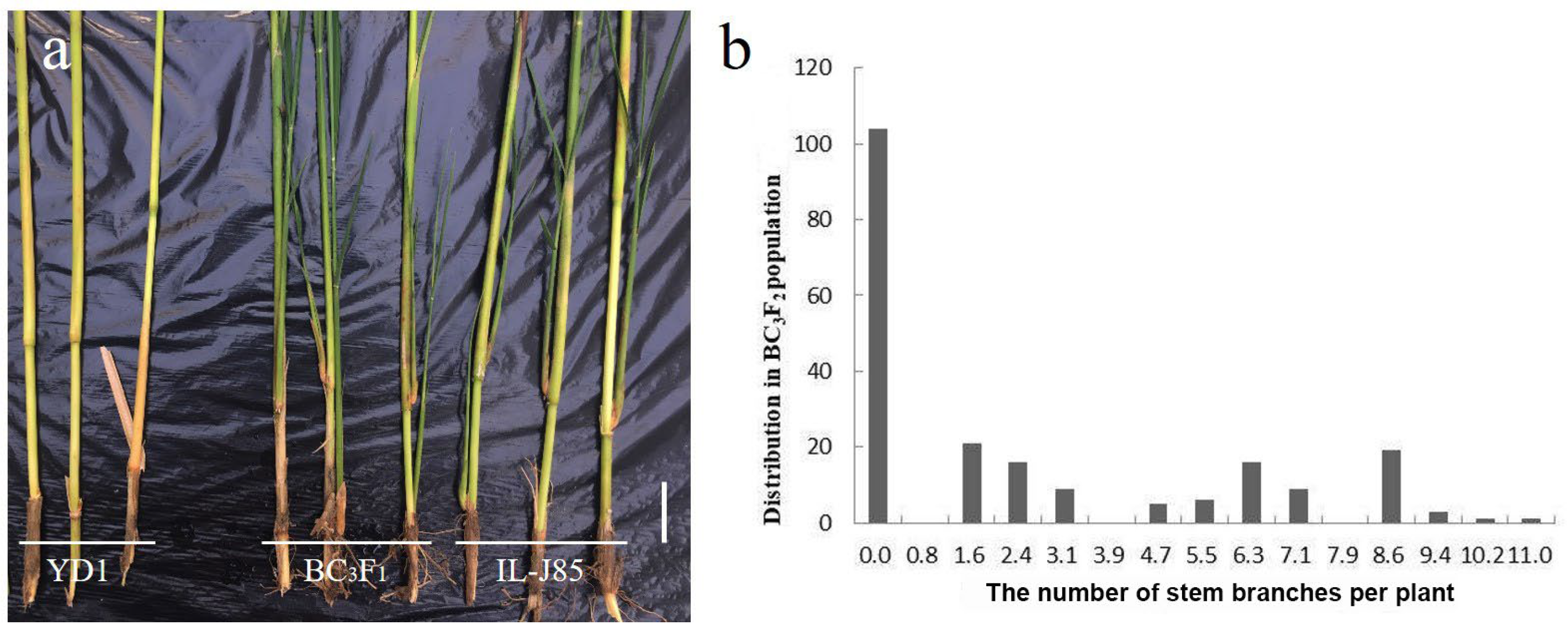
3.2. Genetics Analysis of Stem Branch Trait in IL-J85
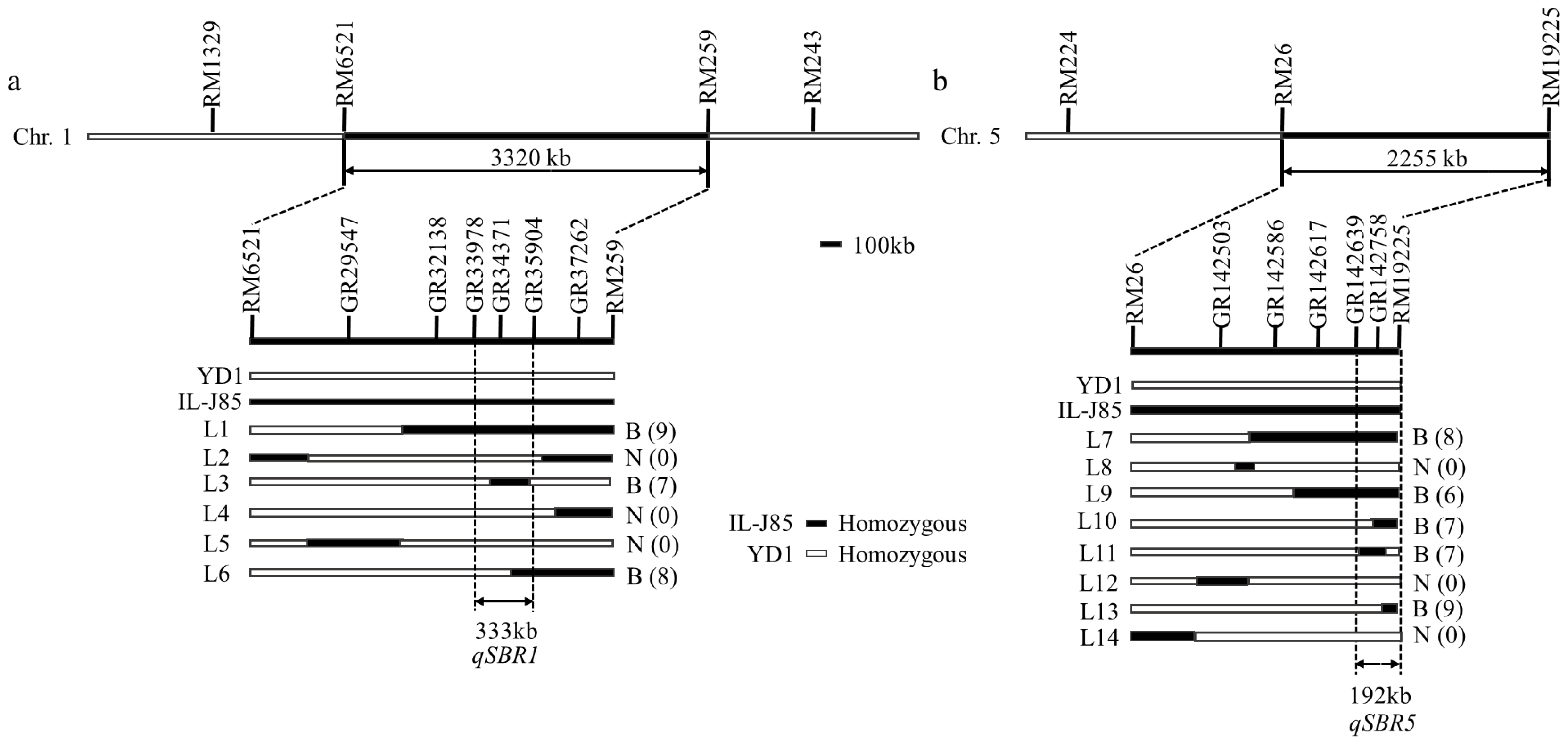
3.3. QTL Mapping for Stem Branch Trait
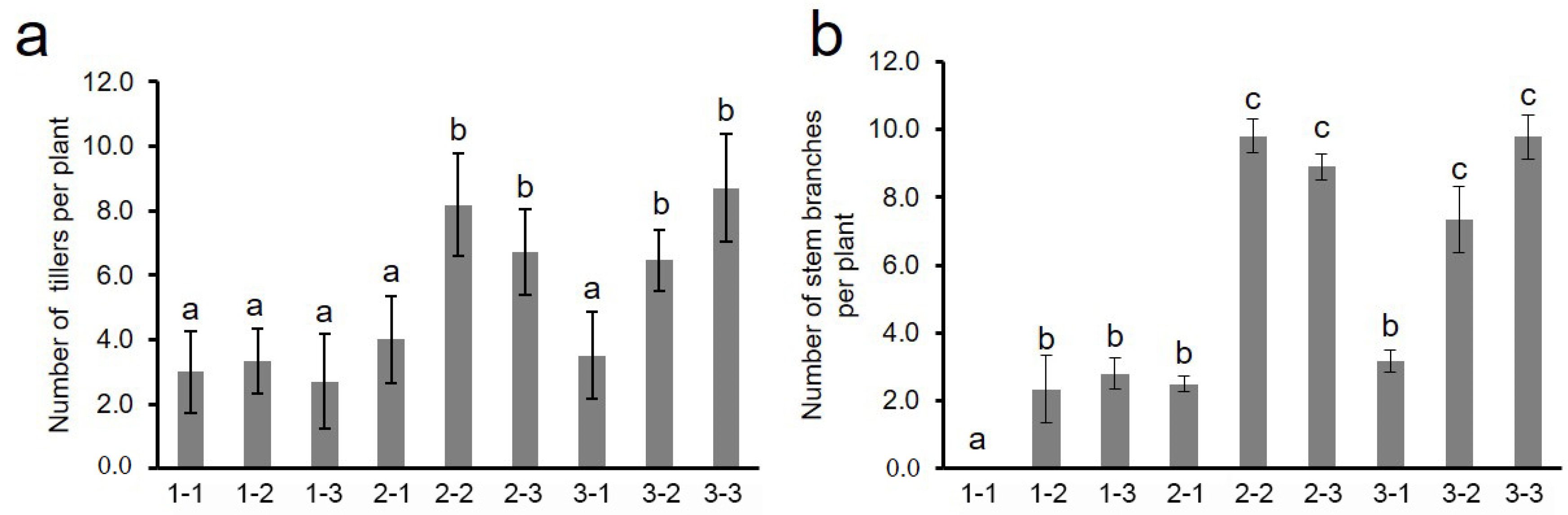
3.4. Effects of Different Genotype Combinations of Two QTLs on Stem Branch and Related Trait
3.5. Breeding Utilization of Stem Branch Trait
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sasaki, T.; Burr, B. International Rice Genome Sequencing Project: The effort to completely sequence the rice genome. Curr. Opin. Plant Biol. 2000, 3, 138–141. [Google Scholar] [CrossRef]
- Elert, E. Rice by the numbers: A good grain. Nature 2014, 514, S50–S51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, R.; Deng, Y.; Ding, Y.; Guo, J.; Qiu, J.; Wang, B.; Wang, C.; Xie, Y.; Zhang, Z.; Chen, J.; et al. Rice functional genomics: Decades’ efforts and roads ahead. Sci. China Life Sci. 2021, 65, 33–92. [Google Scholar] [CrossRef] [PubMed]
- Glover, J.D.; Reganold, J.P.; Bell, L.W.; Borevitz, J.; Brummer, E.C.; Buckler, E.S.; Cox, C.M.; Cox, T.S.; Crews, T.E.; Culman, S.W.; et al. Increased Food and Ecosystem Security via Perennial Grains. Science 2010, 328, 1638–1639. [Google Scholar] [CrossRef] [Green Version]
- Hu, F.Y.; Tao, D.Y.; Sacks, E.; Fu, B.Y.; Xu, P.; Li, J.; Yang, Y.; McNally, K.; Khush, G.S.; Paterson, A.H.; et al. Convergent evolution of perenniality in rice and sorghum. Proc. Natl. Acad. Sci. USA 2003, 100, 4050–4054. [Google Scholar] [CrossRef] [Green Version]
- Tao, D.; Xu, P.; Hu, F.; Yang, Y.; Li, J.; Zhou, J. Hybrid sterility among near-isogenic lines derived from interspecific hybrid between cultivated rice species and Oraza glaberrima. Chin. J. Rice Sci. 2002, 16, 106–110. [Google Scholar]
- Hu, F.; Wang, D.; Zhao, X.; Zhang, T.; Sun, H.; Zhu, L.; Zhang, F.; Li, L.; Li, Q.; Tao, D.; et al. Identification of rhizome-specific genes by genome-wide differential expression Analysis in Oryza longistaminata. BMC Plant Biol. 2011, 11, 18. [Google Scholar] [CrossRef] [Green Version]
- Khush, G. S. Origin, dispersal, cultivation and variation of rice. Plant Mol. Biol. 1997, 35, 25–34. [Google Scholar] [CrossRef]
- Li, C.; Zhou, A.; Sang, T. Genetic analysis of rice domestication syndrome with the wild annual species, Oryza nivara. New Phytol. 2006, 170, 185–194. [Google Scholar] [CrossRef]
- Grillo, M.A.; Li, C.; Fowlkes, A.M.; Briggeman, T.M.; Zhou, A.; Schemske, D.W.; Sang, T. Genetic architecture for the adaptive origin of annual wild rice, Oryza nivara. Evol. Int. J. Org. Evol. 2009, 63, 870–883. [Google Scholar] [CrossRef]
- Chen, E.; Huang, X.; Tian, Z.; Wing, R.A.; Han, B. The Genomics of Oryza Species Provides Insights into Rice Domestication and Heterosis. Annu. Rev. Plant Biol. 2019, 70, 639–665. [Google Scholar] [CrossRef] [PubMed]
- Konishi, S.; Izawa, T.; Lin, S.Y.; Ebana, K.; Fukuta, Y.; Sasaki, T.; Yano, M. An SNP Caused Loss of Seed Shattering During Rice Domestication. Science 2006, 312, 1392–1396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Zhou, A.; Sang, T. Rice Domestication by Reducing Shattering. Science 2006, 311, 1936–1939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bessho-Uehara, K.; Wang, D.R.; Furuta, T.; Minami, A.; Nagai, K.; Gamuyao, R.; Asano, K.; Angeles-Shim, R.B.; Shimizu, Y.; Ayano, M.; et al. Loss of function at RAE2, a previously unidentified EPFL, is required for awnlessness in cultivated Asian rice. Proc. Natl. Acad. Sci. USA 2016, 113, 8969–8974. [Google Scholar] [CrossRef] [Green Version]
- Hua, L.; Wang, D.R.; Tan, L.; Fu, Y.; Liu, F.; Xiao, L.; Zhu, Z.; Fu, Q.; Sun, X.; Gu, P.; et al. LABA1, a Domestication Gene Associated with Long, Barbed Awns in Wild Rice. Plant Cell 2015, 27, 1875–1888. [Google Scholar] [CrossRef] [Green Version]
- Luo, J.; Liu, H.; Zhou, T.; Gu, B.; Huang, X.; Shangguan, Y.; Zhu, J.; Li, Y.; Zhao, Y.; Wang, Y.; et al. n-1 encodes a basic helix-loop-helix protein that regulates awn development, grain size, and grain number in rice. Plant Cell 2013, 25, 3360–3376. [Google Scholar] [CrossRef] [Green Version]
- Yano, K.; Yamamoto, E.; Aya, K.; Takeuchi, H.; Lo, P.-C.; Hu, L.; Yamasaki, M.; Yoshida, S.; Kitano, H.; Hirano, K.; et al. Genome-wide association study using whole-genome sequencing rapidly identifies new genes influencing agronomic traits in rice. Nat. Genet. 2016, 48, 927–934. [Google Scholar] [CrossRef]
- Zhu, B.-F.; Si, L.; Wang, Z.; Zhu, Y.Z.J.; Shangguan, Y.; Lu, D.; Fan, D.; Li, C.; Lin, H.; Qian, Q.; et al. Genetic Control of a Transition from Black to Straw-White Seed Hull in Rice Domestication. Plant Physiol. 2011, 155, 1301–1311. [Google Scholar] [CrossRef] [Green Version]
- Gross, B.L.; Steffen, F.T.; Olsen, K.M. The molecular basis of white pericarps in African domesticated rice: Novel mutations at the Rc gene. J. Evol. Biol. 2010, 23, 2747–2753. [Google Scholar] [CrossRef] [Green Version]
- Ishii, T.; Numaguchi, K.; Miura, K.; Yoshida, K.; Thanh, P.T.; Htun, T.M.; Yamasaki, M.; Komeda, N.; Matsumoto, T.; Terauchi, R.; et al. OsLG1 regulates a closed panicle trait in domesticated rice. Nat. Genet. 2013, 45, 462–465. [Google Scholar] [CrossRef]
- Liu, J.; Chen, J.; Zheng, X.; Wu, F.; Lin, Q.; Heng, Y.; Tian, P.; Cheng, Z.; Yu, X.; Zhou, K.; et al. GW5 acts in the brassinosteroid signalling pathway to regulate grain width and weight in rice. Nat. Plants 2017, 3, 17043. [Google Scholar] [CrossRef] [PubMed]
- Shomura, A.; Izawa, T.; Ebana, K.; Ebitani, T.; Kanegae, H.; Konishi, S.; Yano, M. Deletion in a gene associated with grain size increased yields during rice domestication. Nat. Genet. 2008, 40, 1023–1028. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Li, X.; Liu, F.; Sun, X.; Li, C.; Zhu, Z.; Fu, Y.; Cai, H.; Wang, X.; Xie, D.; et al. Control of a key transition from prostrate to erect growth in rice domestication. Nat. Genet. 2008, 40, 1360–1364. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Huang, W.; Gao, J.P.; Yang, J.; Shi, M.; Zhu, M.Z.; Luo, D.; Lin, H.X. Genetic control of rice plant architecture under domestication. Nat. Genet. 2008, 40, 1365–1369. [Google Scholar] [CrossRef]
- Sun, C.; Wang, X.; Yoshimura, A.; Doi, K. Genetic differentiation for nuclear, mitochondrial and chloroplast genomes in common wild rice (Oryza rufipogon Griff.) and cultivated rice (Oryza sativa L.). Theor. Appl. Genet. 2002, 104, 1335–1345. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.-H.; Shang, F.; Lin, Q.-T.; Lou, C.; Zhang, J. Tillering and panicle branching genes in rice. Gene 2014, 537, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.; Wang, J.; Zhu, X.; Hao, W.; Wang, L.; Li, Q.; Zhang, L.; He, W.; Lu, B.; Lin, H.; et al. Control of rice grain-filling and yield by a gene with a potential signature of domestication. Nat. Genet. 2008, 40, 1370–1374. [Google Scholar] [CrossRef]
- Li, X.; Qian, Q.; Fu, Z.; Wang, Y.; Xiong, G.; Zeng, D.; Wang, X.; Liu, X.; Teng, S.; Hiroshi, F.; et al. Control of tillering in rice. Nature 2003, 422, 618–621. [Google Scholar] [CrossRef]
- Lu, Z.; Shao, G.; Xiong, J.; Jiao, Y.; Wang, J.; Liu, G.; Meng, X.; Liang, Y.; Xiong, G.; Wang, Y.; et al. MONOCULM 3, an ortholog of WUSCHEL in rice, is required for tiller bud formation. J. Genet. Genom. 2015, 42, 71–78. [Google Scholar] [CrossRef]
- Tanaka, W.; Ohmori, Y.; Ushijima, T.; Matsusaka, H.; Matsushita, T.; Kumamaru, T.; Kawano, S.; Hirano, H.-Y. Axillary Meristem Formation in Rice Requires the WUSCHEL Ortholog TILLERS ABSENT1. Plant Cell 2015, 27, 1173–1184. [Google Scholar] [CrossRef] [Green Version]
- Xu, C.; Wang, Y.; Yu, Y.; Duan, J.; Liao, Z.; Xiong, G.; Meng, X.; Liu, G.; Qian, Q.; Li, J. Degradation of MONOCULM 1 by APC/CTAD1 regulates rice tillering. Nat. Commun. 2012, 3, 750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Q.; Wang, D.; Dong, H.; Gu, S.; Cheng, Z.; Gong, J.; Qin, R.; Jiang, L.; Li, G.; Wang, J.L.; et al. Rice APC/C (TE) controls tillering by mediating the degradation of MONOCULM 1. Nat. Commun. 2012, 3, 752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komatsua, M.; Maekawab, M.; Shimamotoa, K.; Kyozuka, J. The LAX1 and FRIZZY PANICLE 2 Genes Determine the Inflorescence Architecture of Rice by Controlling Rachis-Branch and Spikelet Development. Dev. Biol. 2001, 231, 364–373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tabuchi, H.; Zhang, Y.; Hattori, S.; Omae, M.; Shimizu-Sato, S.; Oikawa, T.; Qian, Q.; Nishimura, M.; Kitano, H.; Xie, H.; et al. LAX PANICLE2 of Rice Encodes a Novel Nuclear Protein and Regulates the Formation of Axillary Meristems. Plant Cell 2011, 23, 3276–3287. [Google Scholar] [CrossRef] [Green Version]
- Alder, A.; Jamil, M.; Marzorati, M.; Bruno, M.; Vermathen, M.; Bigler, P.; Ghisla, S.; Bouwmeester, H.; Beyer, P.; Al-Babili, S. The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science 2012, 335, 1348–1351. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.M.; Ai-Ping, Z.; Ping, L.I.; Wang, S.Q.; Liang, Y.Y.; Ling, L.I. Identification and cloning of tillering-related genes osmax1 in rice. Rice Sci. 2015, 22, 255–263. [Google Scholar] [CrossRef] [Green Version]
- Guo, S.; Xu, Y.; Liu, H.; Mao, Z.; Zhang, C.; Ma, Y.; Zhang, Q.; Meng, Z.; Chong, K. The interaction between OsMADS57 and OsTB1 modulates rice tillering via DWARF14. Nat. Commun. 2013, 4, 1566. [Google Scholar] [CrossRef] [Green Version]
- Jiang, L.; Liu, X.; Xiong, G.; Liu, H.; Chen, F.; Wang, L.; Meng, X.; Liu, G.; Yu, H.; Yuan, Y.; et al. DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 2013, 504, 401–405. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Wu, K.; Qian, L.; Liu, Q.; Li, Q.; Pan, Y.; Ye, Y.; Liu, X.; Wang, J.; Zhang, J.; et al. Non-canonical regulation of SPL transcription factors by a human OTUB1-like deubiquitinase defines a new plant type rice associated with higher grain yield. Cell Res. 2017, 27, 1142–1156. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Yu, H.; Xiong, G.; Lu, Z.; Jiao, Y.; Meng, X.; Liu, G.; Chen, X.; Wang, Y.; Li, J. Tissue-Specific Ubiquitination by IPA1 INTERACTING PROTEIN1 Modulates IPA1 Protein Levels to Regulate Plant Architecture in Rice. Plant Cell 2017, 29, 697–707. [Google Scholar] [CrossRef] [Green Version]
- Yao, R.; Li, J.; Xie, D. Recent advances in molecular basis for strigolactone action. Sci. China Life Sci. 2017, 61, 277–284. [Google Scholar] [CrossRef]
- Jiao, Y.; Wang, Y.; Xue, D.; Wang, J.; Yan, M.; Liu, G.; Dong, G.; Zeng, D.; Lu, Z.; Zhu, X.; et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 2010, 42, 541–544. [Google Scholar] [CrossRef]
- Lu, Z.; Yu, H.; Xiong, G.; Wang, J.; Jiao, Y.; Liu, G.; Jing, Y.; Meng, X.; Hu, X.; Qian, Q.; et al. Genome-Wide Binding Analysis of the Transcription Activator Ideal Plant Architecture1 Reveals a Complex Network Regulating Rice Plant Architecture. Plant Cell 2013, 25, 3743–3759. [Google Scholar] [CrossRef] [Green Version]
- Tong, H.; Jin, Y.; Liu, W.; Li, F.; Fang, J.; Yin, Y.; Qian, Q.; Zhu, L.; Chu, C. DWARF AND LOW-TILLERING, a new member of the GRAS family, plays positive roles in brassinosteroid signaling in rice. Plant J. 2009, 58, 803–816. [Google Scholar] [CrossRef]
- Liu, X.; Hu, Q.; Yan, J.; Sun, K.; Liang, Y.; Jia, M.; Meng, X.; Fang, S.; Wang, Y.; Jing, Y.; et al. ζ-Carotene Isomerase Suppresses Tillering in Rice through the Coordinated Biosynthesis of Strigolactone and Abscisic Acid. Mol. Plant 2020, 13, 1784–1801. [Google Scholar] [CrossRef]
- Abdel-Latif, A.; Osman, G. Comparison of three genomic DNA extraction methods to obtain high DNA quality from maize. Plant Methods 2017, 13, 1. [Google Scholar] [CrossRef] [Green Version]
- Gaikwad, K.B.; Singh, N.; Kaur, P.; Rani, S.; Babu, H.P.; Singh, K. Deployment of wild relatives for genetic improvement in rice (Oryza sativa L.). Plant Breed 2021, 140, 23–52. [Google Scholar] [CrossRef]
- Jin, F.X.; Kim, D.M.; Ju, H.G.; Ahn, S.N. Mapping quantitative traits loci for awnness and yield component traits in isogenic lines derived from an Oryza sativa/O. rufipogon cross. J. Crop Sci. Biotechnol. 2009, 12, 9–15. [Google Scholar] [CrossRef]
- Yuan, P.-R.; Kim, H.-J.; Chen, Q.-H.; Ju, H.-G.; Ji, S.-D.; Ahn, S.-N. Mapping QTLs for grain quality using an introgression line population from a cross between Oryza sativa and O. rufipogon. J. Crop Sci. Biotechnol. 2010, 13, 205–212. [Google Scholar] [CrossRef]
- Luo, X.; Wu, S.; Tian, F.; Xin, X.; Zha, X.; Dong, X.; Fu, Y.; Wang, X.; Yang, J.; Sun, C. Identification of heterotic loci associated with yield-related traits in Chinese common wild rice (Oryza rufipogon Griff.). Plant Sci. Int. J. Exp. Plant Biol. 2011, 181, 14–22. [Google Scholar] [CrossRef]
- Liang, Y.; Yan, C.; Zheng, J.; Nan, W.; Qin, X.; Zhang, H. Locating QTL associated with spike traits of Dongxiang wild rice (Oryza rufipogon Griff.). Euphytica 2019, 215, 26. [Google Scholar] [CrossRef]
- Guttikonda, H.; Thummala, S.R.; Agarwal, S.; Mangrauthia, S.K.; Ramanan, R.; Neelamraju, S. Genome-wide transcriptome profile of rice hybrids with and without Oryza rufipogon introgression reveals candidate genes for yield. Sci. Rep. 2020, 10, 4873. [Google Scholar] [CrossRef] [Green Version]
- Food and Agriculture Organization of the United Nations (FAO). The State of Food Insecurity in the World 2009; Progress Report; FAO: Rome, Italy, 2009. [Google Scholar]
- Godfray, H.C.J.; Beddington, J.R.; Crute, I.R.; Haddad, L.; Lawrence, D.; Muir, J.F.; Pretty, J.; Robinson, S.; Thomas, S.M.; Toulmin, C. Food Security: The Challenge of Feeding 9 Billion People. Science 2010, 327, 812–818. [Google Scholar] [CrossRef] [Green Version]
- Monfreda, C.; Ramankutty, N.; Foley, J.A. Farming the planet: 2. Geographic distribution of crop areas, yields, physiological types, and net primary production in the year 2000. Glob. Biogeochem. Cycles 2008, GB1022. [Google Scholar] [CrossRef]
- Pimm, S. L. The World According to Pimm; McGraw-Hill: New York, NY, USA, 2001. [Google Scholar]
- Tilman, D.; Socolow, R.; Foley, J.A.; Hill, J.; Larson, E.; Lynd, L.; Pacala, S.; Reilly, J.; Searchinger, T.; Somerville, C.; et al. Beneficial Biofuels—The Food, Energy, and Environment Trilemma. Science 2009, 325, 270–271. [Google Scholar] [CrossRef] [Green Version]
- Randall, G.W.; Huggins, D.R.; Russelle, M.P.; Fuchs, D.J.; Nelson, W.W.; Anderson, J.L. Nitrate Losses through Subsurface Tile Drainage in Conservation Reserve Program, Alfalfa, and Row Crop Systems. J. Environ. Qual. 1997, 26, 1240–1247. [Google Scholar] [CrossRef]
- Dohleman, F.G.; Long, S.P. More Productive Than Maize in the Midwest: How Does Miscanthus Do It? Plant Physiol. 2009, 150, 2104–2115. [Google Scholar] [CrossRef] [Green Version]

| Traits | YD1 | IL-J85 |
|---|---|---|
| PH (cm) | 108.51 ± 6.27 | 91.52 ± 5.29 ** |
| PL (cm) | 23.28 ± 1.92 | 19.15 ± 1.84 ** |
| FL (cm) | 29.12 ± 5.48 | 22.04 ± 3.19 ** |
| FW (cm) | 1.63 ± 0.12 | 1.29 ± 0.11 ** |
| FL/FW | 17.78 ± 2.73 | 17.02 ± 1.64 |
| NP | 7.43 ± 2.37 | 10.07 ± 2.85 ** |
| PP | 14.97 ± 1.4 | 8.47 ± 0.94 ** |
| SP | 38.00 ± 5.72 | 11.57 ± 3.11 ** |
| 1000-grain weight (g) | 29.05 ± 0.75 | 28.62 ± 1.57 |
| GY (g) | 22.03 ± 7.31 | 21.34 ± 3.51 |
| INL1 (cm) | 21.74 ± 1.62 | 20.89 ± 1.17 |
| INL2 (cm) | 38.48 ± 2.67 | 33.03 ± 1.21 ** |
| INL3 (cm) | 22.31 ± 1.01 | 18.91 ± 1.37 ** |
| INL4 (cm) | 15.44 ± 1.93 | 13.4 ± 1.3 * |
| INL5 (cm) | 6.36 ± 2.55 | 6.43 ± 2.44 |
| TP | 1.9 ± 3.00 | 7.13 ± 3.84 ** |
| TB | 0.00 ± 0.00 | 5.06 ± 2.81 ** |
| PH (cm) | NP | FL (cm) | FW (cm) | FL/FW | PL (cm) | GY (g) | TP | TB | |
| PH (cm) | 1 | ||||||||
| NP | −0.70 ** | 1 | |||||||
| FL (cm) | 0.73 ** | −0.69 ** | 1 | ||||||
| FW (cm) | 0.91 ** | −0.74 ** | 0.88 ** | 1 | |||||
| FL/FW | 0.25 | −0.43 | 0.81 ** | 0.45 * | 1 | ||||
| PL (cm) | 0.89 ** | −0.71 | 0.89 ** | 0.96 ** | 0.52 * | 1 | |||
| GY (g) | 0.45 | −0.41 * | 0.18 | 0.11 | 0.25 | 0.63 ** | 1 | ||
| TP | 0.09 | 0.00 | −0.28 | −0.04 | −0.48 * | −0.19 | −0.18 | 1 | |
| TB | −0.57 * | 0.66 ** | −0.73 ** | −0.84 ** | −0.38 | −0.58 * | −0.34 | −0.01 | 1 |
| Chromosome | Markers | MS | df Error | MS Error | F | p |
|---|---|---|---|---|---|---|
| 1 | RM259 | 16.11 | 238 | 0.58 | 27.57 | 0.00 |
| 3 | RM6147 | 0.24 | 238 | 0.21 | 1.15 | 0.28 |
| 5 | RM274 | 6.63 | 237 | 0.60 | 11.00 | 0.00 |
| 5 | RM26 | 11.98 | 238 | 0.67 | 17.97 | 0.00 |
| 5 | RM5162 | 7.43 | 236 | 0.67 | 11.05 | 0.00 |
| 5 | RM19151 | 7.37 | 238 | 0.86 | 8.54 | 0.00 |
| 5 | RM5261 | 6.23 | 238 | 0.60 | 10.41 | 0.00 |
| 6 | RM3330 | 1.13 | 236 | 0.42 | 2.67 | 0.10 |
| 7 | RM542 | 0.41 | 238 | 0.57 | 0.72 | 0.40 |
| 8 | RM6976 | 1.43 | 237 | 0.74 | 1.95 | 0.16 |
| 9 | RM5526 | 0.55 | 238 | 0.64 | 0.87 | 0.35 |
| 11 | RM3605 | 0.19 | 238 | 0.67 | 0.28 | 0.60 |
| 12 | RM1246 | 0.01 | 238 | 0.27 | 0.02 | 0.88 |
| Markers | Chromosome | MS Error | df Error | F | p |
|---|---|---|---|---|---|
| RM259 | 1 | 7.509 | 2 | 14.631 | 0.000 |
| RM26 | 5 | 3.976 | 2 | 7.746 | 0.000 |
| RM259* RM26 | 3.716 | 3 | 5.430 | 0.000 |
| Trait | QTL | Chromosome | Markers | Range (cm) | LOD | PVE (%) |
|---|---|---|---|---|---|---|
| Stem branch | qSBR1 | 1 | RM6521-RM259 | 16.50–29.78 | 15.65 | 15.65 |
| qSBR5 | 5 | RM26-RM19225 | 109.37–118.39 | 7.53 | 13.45 |
| Traits | YD1 Background | YG37 Background | DJY1 Background | |||
|---|---|---|---|---|---|---|
| S-J85 | C-J85 | S-Y37 | C-Y37 | S-D1 | C-D1 | |
| HD (d) | 65.30 ± 1.11 | 60.52 ± 0.50 * | 65.50 ± 1.00 | 60.60 ± 0.60 * | 64.90 ± 0.99 | 59.42 ± 0.36 * |
| PH (cm) | 93.35 ± 5.85 | 90.76 ± 4.01 | 102.47 ± 4.39 | 95.87 ± 3.29 ** | 108.10 ± 3.42 | 104.85 ± 4.95 * |
| PL (cm) | 19.87 ± 1.65 | 19.74 ± 1.31 | 21.27 ± 1.22 | 22.36 ± 1.26 * | 20.95 ± 1.69 | 22.92 ± 1.45 ** |
| FL (cm) | 30.73 ± 3.32 | 29.31 ± 3.27 | 41.68 ± 4.50 | 33.00 ± 5.60 ** | 39.90 ± 5.10 | 39.88 ± 3.34 |
| FW (cm) | 1.27 ± 0.09 | 1.21 ± 0.10 | 1.67 ± 0.09 | 1.65 ± 0.11 | 1.78 ± 0.09 | 1.73 ± 0.08 |
| NP | 6.27 ± 1.56 | 7.44 ± 2.58 | 11.337 ± 3.68 | 9.27 ± 1.83 | 6.40 ± 1.40 | 7.27 ± 1.49 |
| PP | 8.82 ± 1.25 | 9.63 ± 1.63 | 12.20 ± 1.57 | 8.73 ± 0.96 ** | 13.40 ± 1.50 | 12.73 ± 2.15 |
| SP | 18.45 ± 5.73 | 14.81 ± 4.55 | 39.20 ± 6.93 | 21.73 ± 4.11 ** | 40.20 ± 9.17 | 39.53 ± 8.06 |
| 1000-grain weight (g) | 31.55 ± 0.47 | 30.79 ± 1.40 | 27.58 ± 0.69 | 27.26 ± 0.21 | 29.99 ± 0.16 | 28.95 ± 1.31 ** |
| GY (g) | 12.73 ± 4.05 | 12.65 ± 3.61 | 29.37 ± 6.32 | 24.33 ± 4.74 * | 22.34 ± 5.28 | 23.10 ± 5.93 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, F.; Lu, C.; Wan, J.; Yang, J.; Liu, L.; Zhang, F.; Wu, Z.; Zhang, X.; Chang, G.; Yu, D.; et al. Genetic Dissection of Stem Branch Trait and Envisioning of Fixing Heterosis by Vegetative Reproduction in Oryza rufipogon. Agronomy 2022, 12, 1503. https://doi.org/10.3390/agronomy12071503
Wang F, Lu C, Wan J, Yang J, Liu L, Zhang F, Wu Z, Zhang X, Chang G, Yu D, et al. Genetic Dissection of Stem Branch Trait and Envisioning of Fixing Heterosis by Vegetative Reproduction in Oryza rufipogon. Agronomy. 2022; 12(7):1503. https://doi.org/10.3390/agronomy12071503
Chicago/Turabian StyleWang, Feijun, Chengkai Lu, Jinpeng Wan, Jun Yang, Lei Liu, Feifei Zhang, Zihao Wu, Xiao Zhang, Guimei Chang, Diqiu Yu, and et al. 2022. "Genetic Dissection of Stem Branch Trait and Envisioning of Fixing Heterosis by Vegetative Reproduction in Oryza rufipogon" Agronomy 12, no. 7: 1503. https://doi.org/10.3390/agronomy12071503
APA StyleWang, F., Lu, C., Wan, J., Yang, J., Liu, L., Zhang, F., Wu, Z., Zhang, X., Chang, G., Yu, D., & Xu, P. (2022). Genetic Dissection of Stem Branch Trait and Envisioning of Fixing Heterosis by Vegetative Reproduction in Oryza rufipogon. Agronomy, 12(7), 1503. https://doi.org/10.3390/agronomy12071503







