Wavelet Decomposition and Machine Learning Technique for Predicting Occurrence of Spiders in Pigeon Pea
Abstract
:1. Introduction
2. Methodology
2.1. Study Locations, Surveillance, and Sampling Plans for Spiders and Weather
2.2. Multiple Linear Regression Model
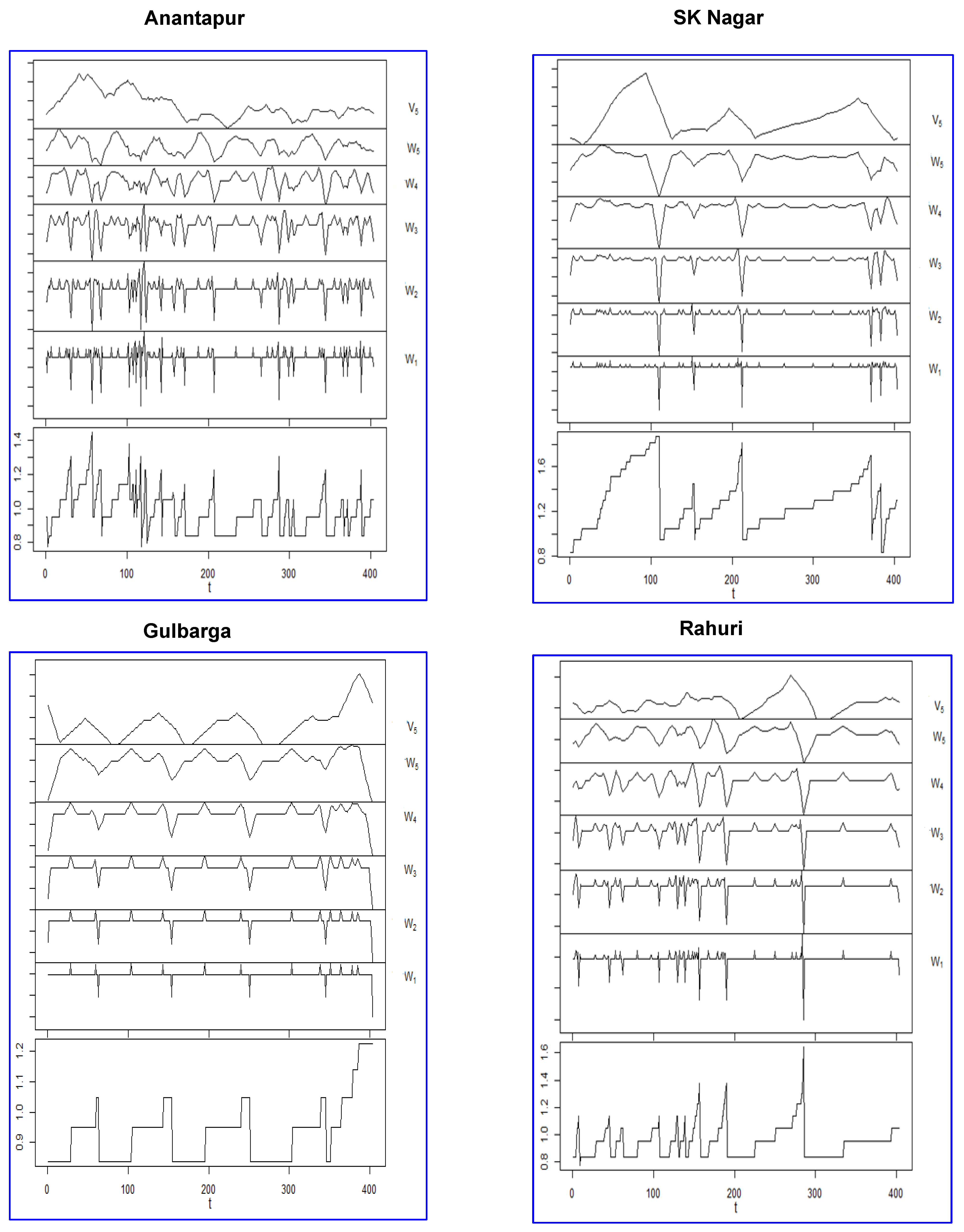
2.3. Wavelets
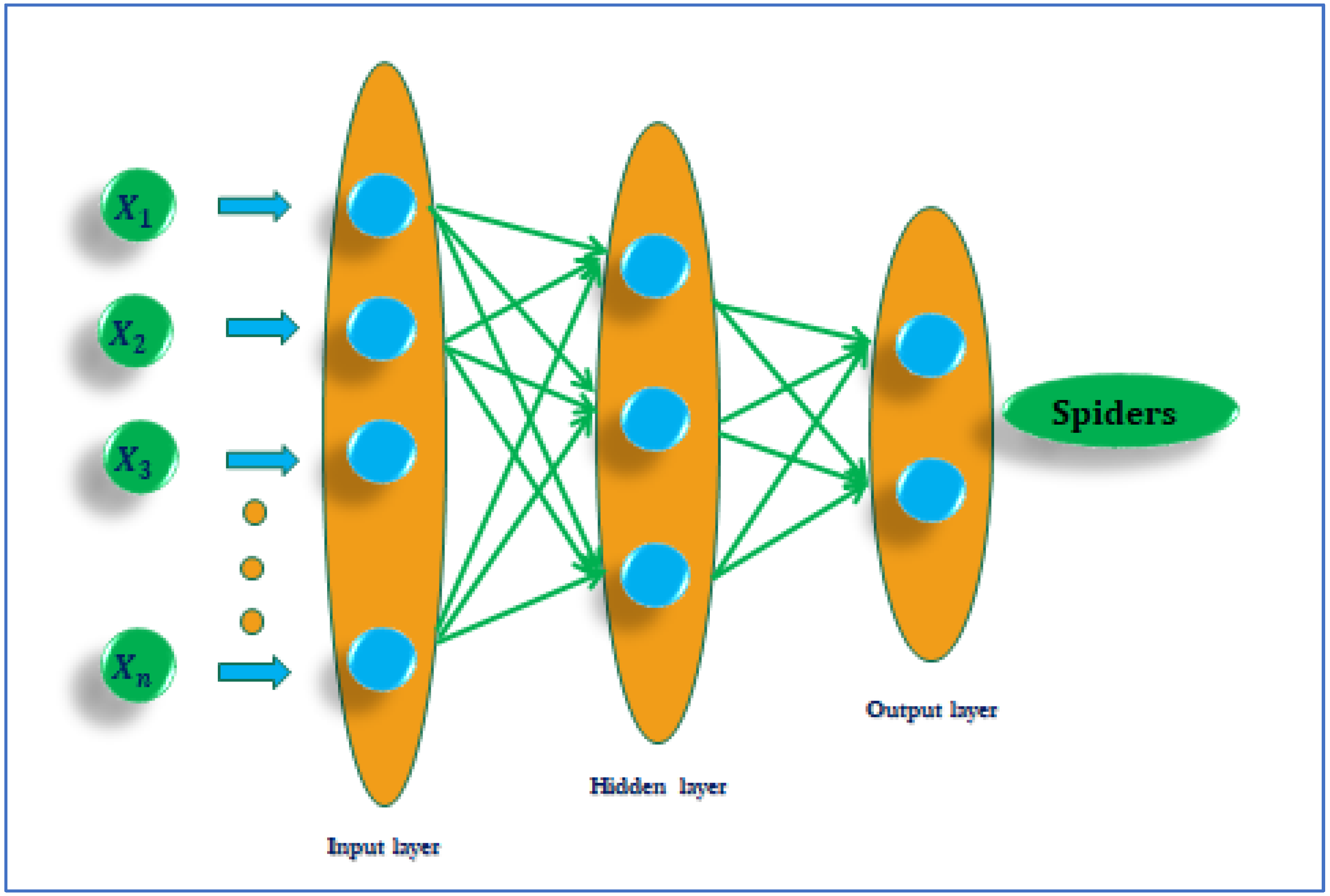
2.4. Artificial Neural Network (ANN)
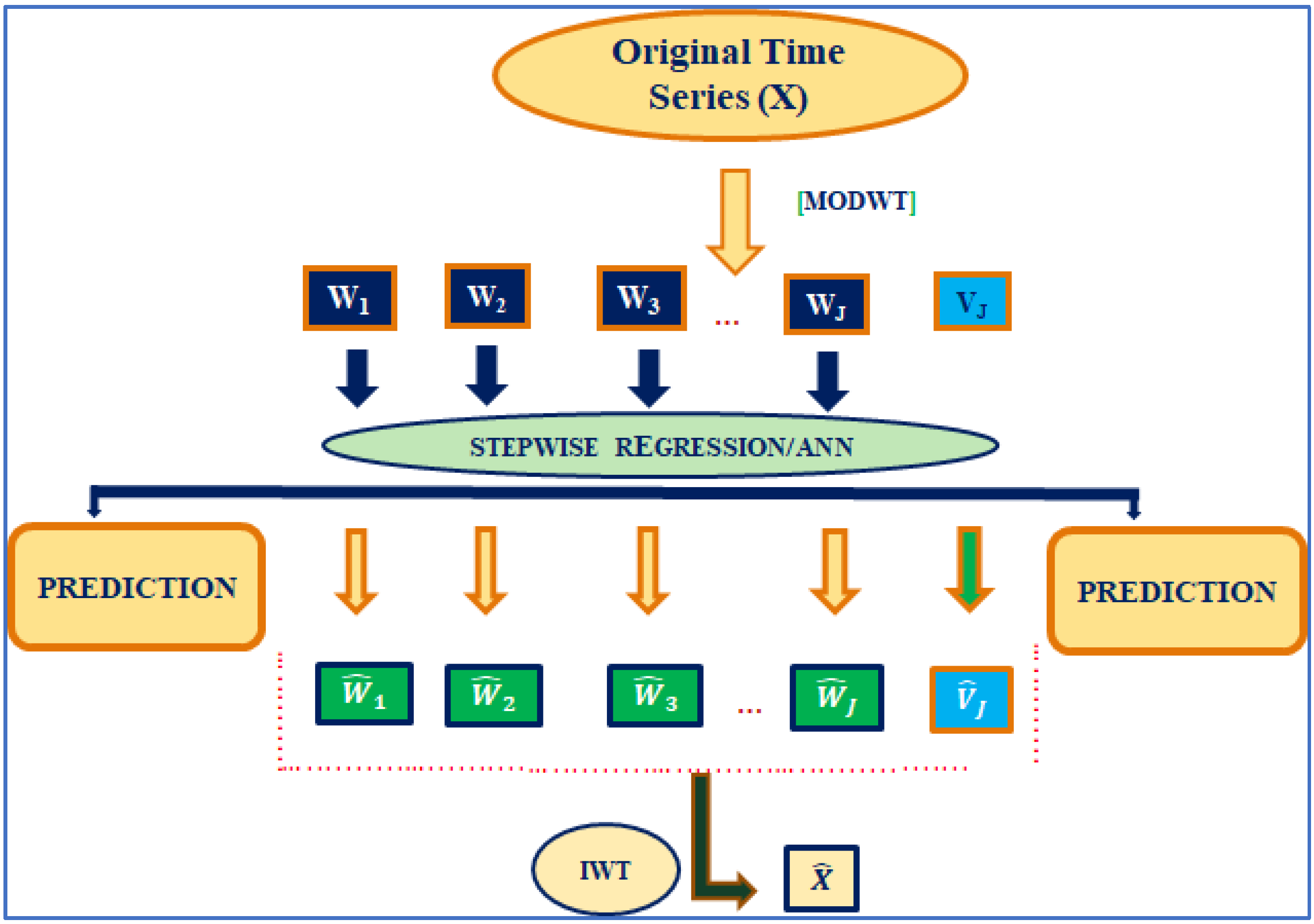
2.5. Wavelet–Linear Regression (W–LR) Approach
2.6. Wavelet–ANN (W–ANN) Approach
2.7. Validation
3. Results and Discussion
3.1. Spiders of Pigeon Pea Ecosystem and Description of Study Locations
3.2. Seasonality of Spiders
3.3. Descriptive Statistics of Spider Occurrence
3.4. Spider–Weather Relations
3.5. Modeling of Spiders
3.6. Validation
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Food and Agriculture Organization, Statistical Database 2003. Available online: https://www.fao.org/documents/card/en/c/19d2a310-cee0-5bfd-bdfe-3b3e64e789ed/ (accessed on 24 January 2022).
- Food and Agriculture Organization. statistical database 2014. Mushrooms and Truffles. Rome: Food and Agriculture Organization of the United Nations. Available online: http://faostat3.fao.org/ (accessed on 1 August 2014).
- Reddy, M.V.; Nene, Y.L. Estimation of yield loss in Pigeon pea due to sterility mosaic. In Proceedings of the International Workshop on Pigeon Pea, Patancheru, AP, India, 15–19 December 1980; The International Crops Research Institute for the Semi-Arid Tropics Center: Patancheru, India; pp. 305–312. [Google Scholar]
- Laxman, S. Production as-pects of Pigeon pea and future prospects. In Uses of Tropical Grain Legumes, Proceedings of the Consultants Meeting, Patancheru, India, 27–30 March 1989; Laxman, S., Silim, S.N., Ariyanayagam, R.P., Reddy, M.V., Eds.; The International Crops Research Institute for the Semi-Arid Tropics Center: Patancheru, India, 1991; pp. 27–121. [Google Scholar]
- Kannaiyan, J.; Nene, Y.L.; Reddy, M.V.; Ryan, J.G.; Raju, T.N. Prevalence of Pigeon pea disease and associated crop losses in Asia, Africa and the Americas. Tropical. Pest Manag. 1984, 30, 62–71. [Google Scholar] [CrossRef] [Green Version]
- Ganapathy, K.N.; Gnanesh, B.N.; Gowda, B.M.; Venkatesha, S.C.; Gomashe, S.S.; Channamallikarjuna, V. AFLP analysis in Pigeon pea (Cajanus cajan (L.) Mill sp.) revealed close relationship of cultivated genotypes with some of its wild relatives. Genet. Resour. Crop Evol. 2011, 58, 837–847. [Google Scholar] [CrossRef]
- Varshney, R.K.; Penmetsa, R.V.; Dutta, S.; Kulwal, P.L.; Saxena, R.K.; Datta, S.; Sharma, T.R.; Rosen, B.N.; Carrasquilla-Garcia, N.; Farmer, A.D.; et al. Pigeon pea genomics initiative (PGI): An international effort to improve crop productivity of Pigeon pea (Cajanus cajan L.). Mol. Breed. 2010, 26, 393. [Google Scholar] [CrossRef] [Green Version]
- Srilaxmi, K.; Paul, R. Diversity of insect pests of Pigeon pea [Cajanus cajan (L.) Millsp.] and their succession in relation to crop phenology in Gulbarga, Karnataka. Ecoscan 2010, 4, 273–276. [Google Scholar]
- Shanower, T.G.; Romeis, J.E.; Minja, M. Insect Pests of Pigeon pea and Their Management. Annu. Rev. Entomol. 1999, 44, 77–96. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, S.K.; Kada, R.; Subbiah, J.; Ahsan, C.R.; Bari, L.; Mai, D.S.; Suong, N.K. Asian Food Safety and Security Association, Dhaka, Bangladesh. In Proceedings of the 2nd AFSSA Conference on Food Safety and Food Security, Dong Nai University of Technology, Bien Hoa, Vietnam, 15–18 August 2014; pp. 66–71. [Google Scholar]
- Patel, M.L.; Patel, K.G.; Pandya, H.V. Navbharath Enterprises, Bangalore, India. Insect Environ. 2005, 11, 23–25. [Google Scholar]
- Box, G.E.P.; Jenkins, G. Time Series Analysis, Forecasting and Control; Holden-Day: San Francisco, CA, USA, 1970. [Google Scholar]
- Paul, R.K.; Das, M.K. Statistical modelling of inland fish production in India. J. Inland Fish. Soc. India 2010, 42, 1–7. [Google Scholar]
- Paul, R.K.; Prajneshu, G.H. Wavelet frequency domain approach for modelling and forecasting of Indian monsoon rainfall time-series data. J. Indian Soc. Agric. Stat. 2013, 67, 319–327. [Google Scholar]
- Paul, R.K.; Alam, W.; Paul, A.K. Prospects of livestock and dairy production in India under time series framework. Indian J. Anim. Sci. 2014, 84, 130–134. [Google Scholar]
- Paul, R.K.; Ghosh, H.; Prajneshu. Development of out-of-sample forecast formulae for ARIMAX-GARCH model and their application. J. Indian Soc. Agric. Stat. 2014, 68, 85–92. [Google Scholar]
- Arya, P.; Paul, R.K.; Kumar, A.; Singh, K.; Sivaramne, N.; Chaudhary, P. Predicting pest population using weather variables: An ARIMAX time series framework. Int. J. Agric. Stat. Sci. 2015, 11, 381–386. [Google Scholar]
- Kim, Y.; Yoo, S.; Gu, Y.; Lim, J.; Han, D.; Baik, S. Crop pests prediction method using Regression and machine learning technology: Survey. IERI Procedia 2014, 6, 52–56. [Google Scholar] [CrossRef] [Green Version]
- Paul, R.K.; Vennila, S.; Yadav, S.K.; Bhat, M.N.; Kumar, M.; Chandra, P.; Paul, A.K.; Prabhakar, M. Weather based Forecasting of Sterility Mosaic Disease in Pigeon pea using Machine Learning Techniques and Hybrid Models. Indian J. Agric. Sci. 2020, 90, 1952–1958. [Google Scholar]
- Paul, R.K.; Vennila, S.; Bhat, M.N.; Yadav, S.K.; Sharma, V.K.; Nisar, S.; Panwar, S. Prediction of early blight severity in tomato (Solanum lycopersicum) by machine learning technique. Indian J. Agric. Sci. 2019, 89, 169–175. [Google Scholar]
- Paul, R.K.; Garai, S. Performance comparison of wavelets-based machine learning technique for forecasting agricultural commodity prices. Soft Comput. 2021, 25, 12857–12873. [Google Scholar] [CrossRef]
- Paul, R.K.; Paul, A.K.; Bhar, L.M. Wavelet-based combination approach for modeling sub-divisional rainfall in India. Theor. Appl. Climatol. 2020, 139, 949–963. [Google Scholar] [CrossRef]
- Calvo, L.; Guzmán, M.; Guzmán, J. Considerations about Application of Machine Learning to the Prediction of Sigatoka Disease. In Proceedings of the World Conference on Computers in Agriculture and Natural Resources, University of Costa Rica, San Jose, Costa Rica, 27–30 July 2014; Available online: http://CIGRProceedings.org (accessed on 10 January 2022).
- Diebold, F.X.; Mariano, R.S. Comparing predictive accuracy. J. Bus. Econ. Stat. 1995, 13, 253–263. [Google Scholar]
- Chatterjee, S.; Hadi, A.S. Sensitivity Analysis in Linear Regression; John Wiley and Sons, Inc.: New York, NY, USA, 1988. [Google Scholar]
- Daubechies, I. Ten Lectures on Wavelets; SIAM: Philadelphia, PA, USA.
- Percival, D.B.; Walden, A.T. Wavelet Methods for Time-Series Analysis; Cambridge University Press: Cambridge, UK, 2000. [Google Scholar]
- Ogden, T. Essential Wavelets for Statistical Applications and Data Analysis; Birkhauser: Boston, MA, USA, 1997. [Google Scholar]
- Paul, R.K. WaveletANN: Wavelet ANN Model. R Package Version 0.1.0. 2019. Available online: https://CRAN.R-project.org/package=WaveletANN (accessed on 2 January 2022).
- Hunter, M.D. Trophic promiscuity, intraguild predation and the problem of omnivores. Agric. For. Entomol. 2009, 11, 125–131. [Google Scholar] [CrossRef]
- CPC. Crop Protection Compendium. CAB International. 2009. Available online: http://www.cabi.org/compendia/cpc/ (accessed on 4 January 2022).
- Takabayashi, J.; Sabelis, W.M.; Janssen, A.; Shiojiri, K.; van Wijk, M. Can plants betray the presence of multiple herbivore species to predators and parasitoids? The role of learning in phytochemical information networks. Ecol. Res. 2006, 21, 3–8. [Google Scholar] [CrossRef] [Green Version]
- Khan, Z.R.; James, D.G.; Midega, C.A.O.; Pickett, J.A. Chemical ecology and conservation biological control. Biol. Control. 2008, 45, 210–224. [Google Scholar] [CrossRef]
- Schnee, C.; Köllner, T.G.; Held, M.; Turlings, T.C.J.; Gershenzon, J.; Degenhardt, J. The products of a single maize sesquiterpene synthase form a volatile defense signal that attracts natural enemies of maize herbivores. Proc. Natl. Acad. Sci. USA 2013, 103, 1129–1134. [Google Scholar] [CrossRef] [Green Version]
- Degenhardt, J. Indirect Defense Responses to Herbivory in Grasses. Plant Physiol. 2009, 149, 96–102. [Google Scholar] [CrossRef] [Green Version]
- Unsicker, S.B.; Kunert, G.; Gershenzon, J. Protective perfumes: The role of vegetative volatiles in plant defense against herbivores. Curr. Opin. Plant Biol. 2009, 12, 479–485. [Google Scholar] [CrossRef]
- Satyagopal, K.; Sushil, S.N.; Jeyakumar, P.; Shankar, G.; Sharma, O.P.; Boina, D.R.; Sain, S.K.; Lavanya, N.; Sunanda, B.S.; Ram, A.; et al. AESA Based IPM Package for Redgram; Directorate of Plant Protection: Faridabad, India, 2014; p. 42.
- Anderson, T.W.; Darling, D.A. Asymptotic theory of certain “goodness of fit” criteria based on stochastic processes. Ann. Math. Stat. 1952, 23, 193–212. [Google Scholar] [CrossRef]
- Hanna, C.J.; Cobb, V.A. Critical Thermal Maximum of the Green Lynx Spider, Peucetia viridans (Araneae, Oxyopidae). J. Arachnol. 2007, 35, 193–196. [Google Scholar] [CrossRef]
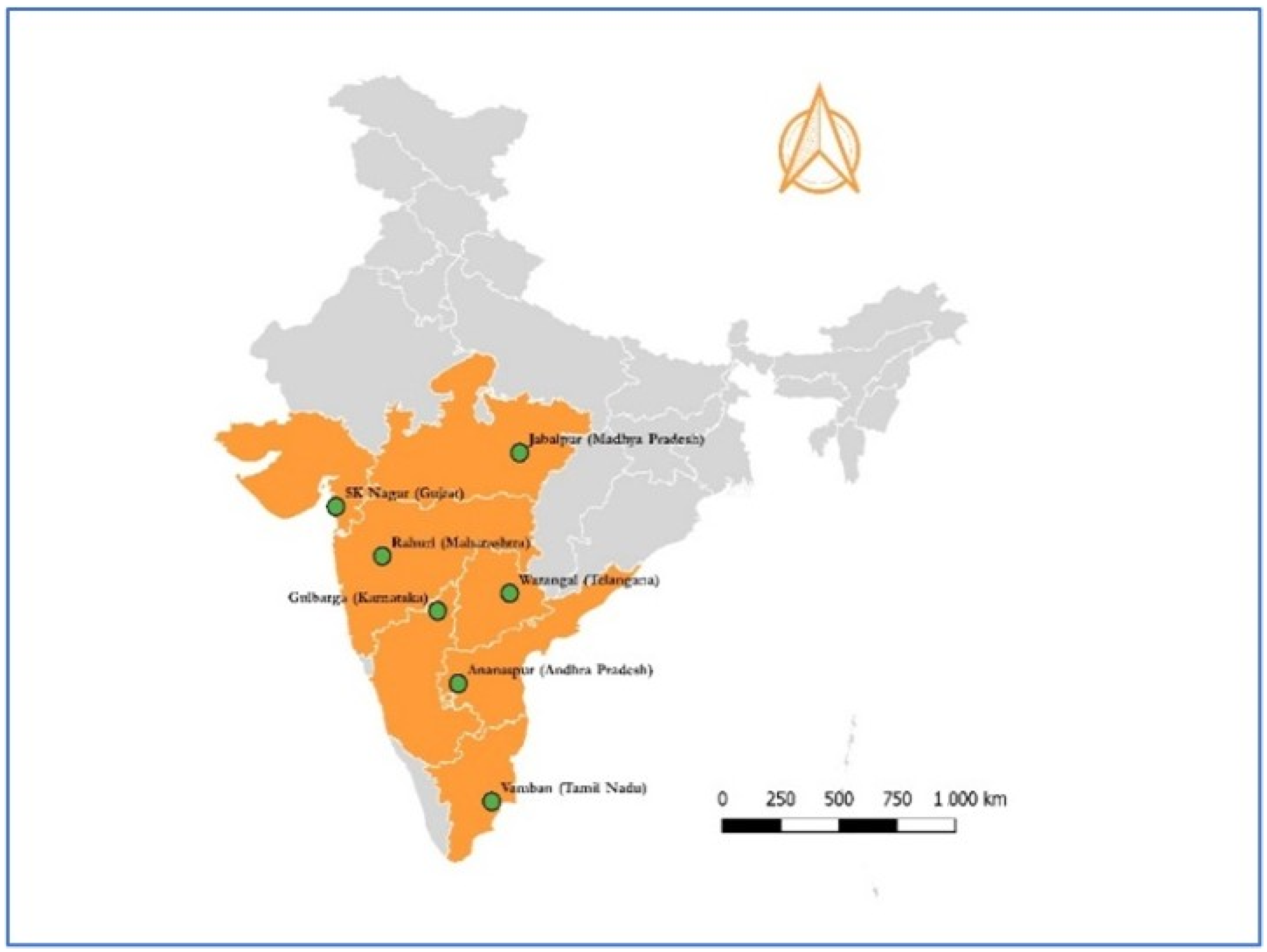
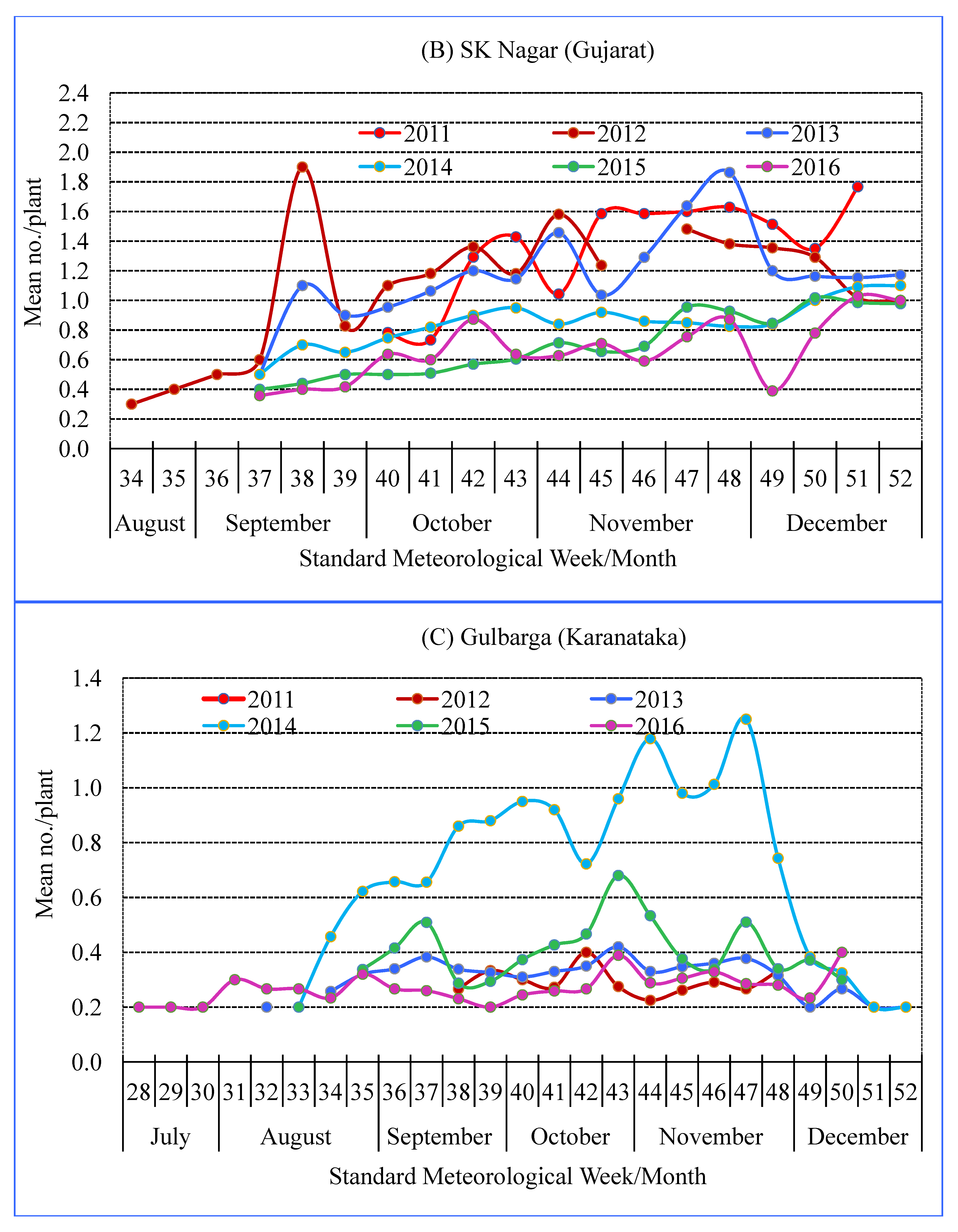
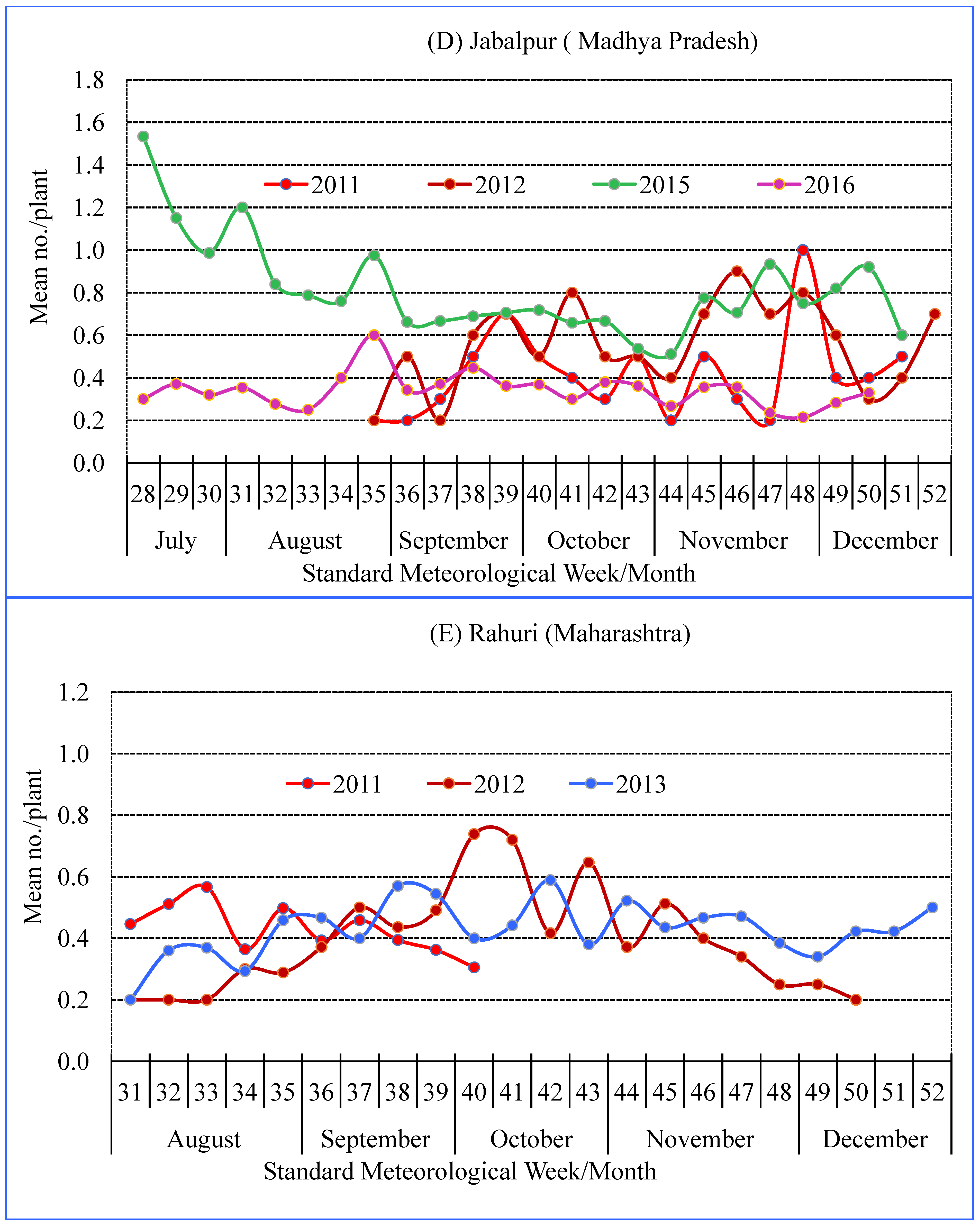
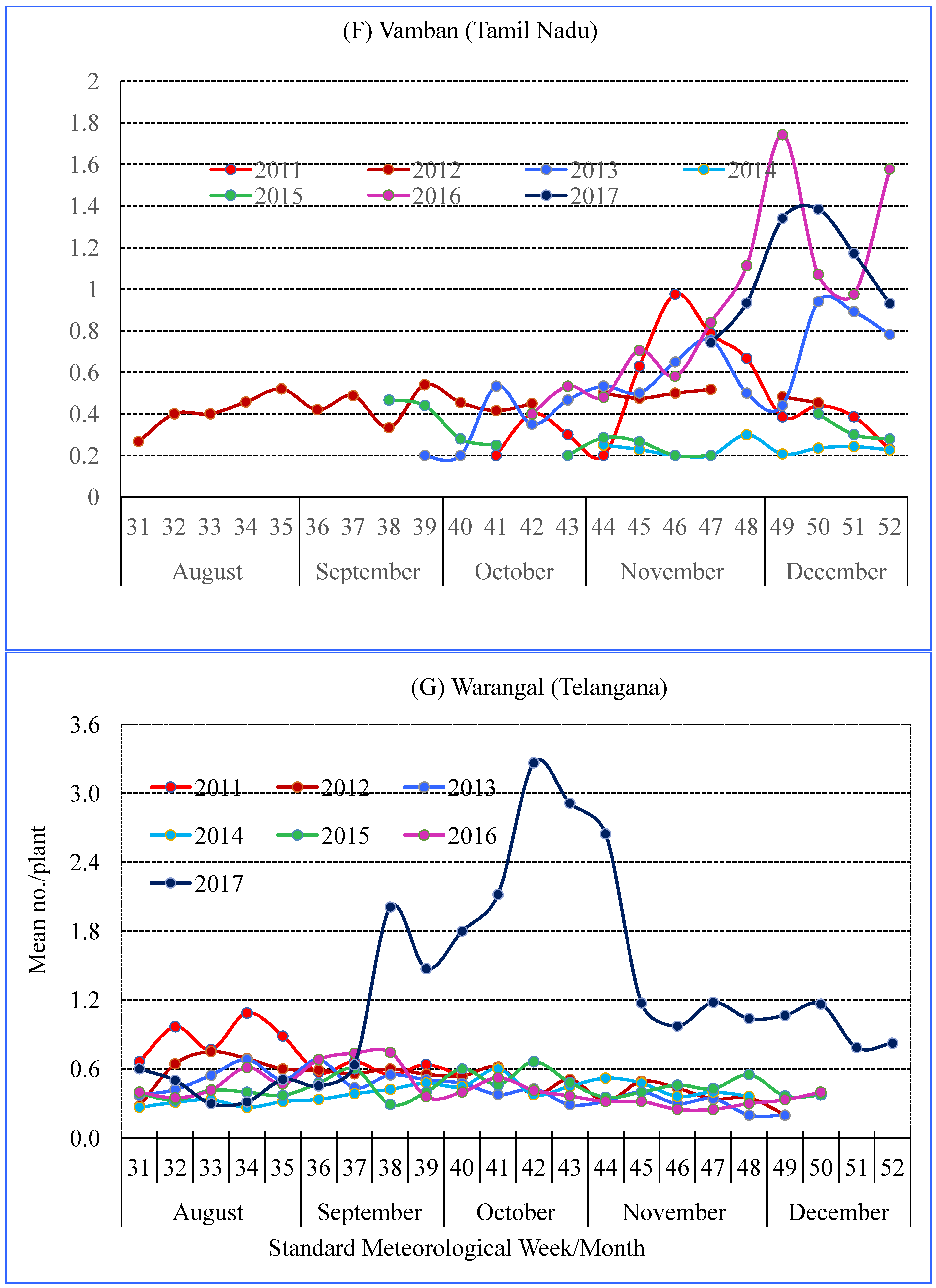

| Location | Agro-Ecological Region | Agro-Climate Zone | GPS Co-Ordinates | Study Period | Crop Season (SMW) |
|---|---|---|---|---|---|
| Anantapur | Deccan plateau and central highland, hot arid ecoregion | Southern Plateau and Hills Region | 14°43′ N, 77°40′ E | 2013–2016 | 30–52 |
| SK Nagar | Western plain, Kachhh and part of Kathiawar peninsula, hot arid ecoregion | Gujarat Plains and Hills Region | 21°10′ N, 72°51′ E | 2011–2016 | 37–52 |
| Gulbarga | Deccan plateau Aravallis, hot semi-arid ecoregion | Southern Plateau and Hills Region | 17°21′ N, 76°48′ E | 2012–2016 | 28–52 |
| Jabalpur | Central highland (Malwa, Bundelkhand, and eastern Satpura), hot semi-humid ecoregion | Central Plateau and Hills Region | 23°10′ N, 79°59′ E | 2011,12,15 &16 | 26–51 |
| Rahuri | Deccan plateau Aravallis, hot semi-arid ecoregion | Western Plateau and Hills Region | 19°22′ N, 74°39′ E | 2011–2013 | 31–52 |
| Vamban | Eastern ghat, TN upland and decan plateau, hot semi-arid ecoregion | East Coast Plains and Hills Region | 10°21′ N, 78°54′ E | 2011–2017 | 30–52 |
| Warangal | Decan plateau and eastern ghat, hot semi-arid ecoregion | Southern Plateau and Hills Region | 18°00′ N, 79°36′ E | 2011–2017 | 33–52 |
| Statistical Measures | Spiders (Response Variable) | ||||||
|---|---|---|---|---|---|---|---|
| Anantapur (AP) | SK Nagar (GJ) | Gulbarga (KA) | Jabalpur (MP) | Rahuri (MH) | Vamban (TN) | Warangal (TS) | |
| Mean | 0.76 | 0.89 | 0.50 | 0.74 | 0.50 | 0.63 | 0.77 |
| Median | 0.60 | 0.80 | 0.40 | 0.60 | 0.40 | 0.40 | 0.60 |
| Maximum | 5.40 | 5.20 | 5.40 | 5.60 | 6.00 | 7.00 | 10.20 |
| Minimum | 0.10 | 0.20 | 0.20 | 0.20 | 0.10 | 0.10 | 0.20 |
| SD # | 0.54 | 0.65 | 0.45 | 0.54 | 0.44 | 0.56 | 0.68 |
| CV # (%) | 71.64 | 73.69 | 89.95 | 73.10 | 87.83 | 88.42 | 88.90 |
| Skewness | 2.54 | 1.43 | 4.00 | 2.34 | 4.63 | 3.64 | 5.73 |
| Kurtosis | 12.17 | 2.84 | 4.00 | 10.70 | 38.48 | 25.35 | 59.45 |
| Location | Kolmogorov–Smirnov | Anderson–Darling | ||
|---|---|---|---|---|
| Statistic | p-Value | Statistic | p-Value | |
| Anantapur | 0.16 | <0.010 | 33.05 | <0.005 |
| SK Nagar | 0.16 | <0.010 | 31.03 | <0.005 |
| Gulbarga | 0.28 | <0.010 | 82.83 | <0.005 |
| Jabalpur | 0.17 | <0.001 | 36.66 | <0.005 |
| Rahuri | 0.24 | <0.010 | 68.80 | <0.005 |
| Vamban | 0.25 | <0.010 | 63.76 | <0.005 |
| Warangal | 0.20 | <0.010 | 52.52 | <0.005 |
| Location | MaxT (°C) | MinT (°C) | RHM (%) | RHE (%) | RF (mm) | SS (h/day) | Wind (km/h) | RD (No. of Days) |
|---|---|---|---|---|---|---|---|---|
| Anantapur | 35.66–28.46 | 25.5–14.11 | 99–71.86 | 68.29–21.71 | 168.3–0 | 19.43–0.29 | 19.57–2 | 6–0 |
| SK Nagar | 38.84–25.21 | 27.14–4.94 | 95.25–8.9 | 89.43–18 | 383.6–0 | 10.14–10.43 | 14.1–0.38 | 5–0 |
| Gulbarga | 33.19–26.26 | 26.93–9.46 | 94.04–53.07 | 80.17–24.27 | 195–0 | Not available | 52.29–0 | 5–0 |
| Jabalpur | 35.1–23.36 | 24.54–4.18 | 9571–77.71 | 88.86–22 | 221.6–0 | 9.71–0 | 8.43–1.43 | 7–0 |
| Rahuri | 33.66–28.23 | 22.63–7.40 | 87.57–46.29 | 70.57–24.86 | 118.6–0 | 9.86–2.14 | 8.14–0.14 | 5–0 |
| Vamban | 38.36–27.00 | 25.86–16.20 | 96.25–72.43 | 92–59.86 | 256–0 | 8.29–0 | 6–0.71 | 6–0 |
| Warangal | 32.86–27.88 | 24.93–12.75 | 91.86–82 | 73.14–38.75 | 117.4–0 | 7.86–1 | - | 4–0 |
| Weather Parameters | Anantapur | SK Nagar | Gulbarga | Jabalpur | Rahuri | Vamban | Warangal |
|---|---|---|---|---|---|---|---|
| MaxT-1 | −0.11 * | −0.11 *** | 0.12 *** | 0.15 *** | 0.01 | −0.14 *** | 0.29 *** |
| MinT-1 | −0.18 *** | −0.28 *** | 0.20 *** | 0.19 *** | 0.09 * | 0.10 ** | 0.15 *** |
| RHM-1 | −0.09 * | −0.05 * | −0.04 | −0.19 ** | 0.10 ** | −0.05 | 0.07 ** |
| RHE-1 | −0.001 | −0.35 *** | −0.10 ** | 0.12 ** | 0.08 * | 0.13 *** | −0.01 |
| RF-1 | −0.01 | −0.03 | −0.04 | 0.07 * | 0.16 *** | −0.09 ** | −0.001 |
| SS-1 | −0.33 *** | 0.22 *** | − | −0.09 * | −0.03 | 0.12 ** | −0.13 *** |
| Wind-1 | −0.06 | 0.02 | 0.28 *** | 0.19 *** | −0.09 * | −0.08 * | − |
| RD-1 | −0.03 | −0.08 ** | 0.06 * | 0.08 * | 0.17 *** | 0.001 | 0.005 |
| Location | Model Equation |
|---|---|
| Anantapur | 1.09 2212 0.014 MaxT-1 − 0.009 SS-1 |
| SK Nagar | 1.64 − 0.014 MaxT-1 − 0.005 RHE-1 + 0.005 RF-1 + 0.03 RD-1 + 0.004 SS-1 + 0.05 Wind-1 |
| Gulbarga | 0.21 + 0.01 MaxT-1 + 0.003 MinT-1 + 0.01 RF-1 + 0.005 Wind-1 |
| Jabalpur | 1.65 + 0.01 MinT-1 − 0.007 RHM-1 − 0.003 RHE-1 − 0.0003 RF-1 − 0.01 SS-1 + 0.02 Wind-1 |
| Rahuri | 0.91 + 0.002 MinT-1 + 0.01 RD-1 − 0.01 Wind-1 |
| Vamban | 1.20 − 0.02 MaxT-1 + 0.02 MinT-1 − 0.01 RD-1 + 0.01 SS-1 |
| Warangal | −0.38 + 0.04 MaxT-1 +0.008 RHM-1 − 0.001 RHE-1 − 0.06 RD-1 − 0.05SS-1 |
| Location | W1 | W2 | W3 | W4 | W5 | V | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| # L | # HN | # L | # HN | # L | # HN | # L | # HN | # L | # HN | # L | # HN | |
| Anantapur | 1 | 1 | 1 | 1 | 1 | 1 | 4 | 2 | 4 | 2 | 6 | 3 |
| SK Nagar | 1 | 1 | 1 | 1 | 1 | 1 | 3 | 4 | 3 | 4 | 1 | 1 |
| Gulbarga | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Jabalpur | 1 | 1 | 2 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Rahuri | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Vamban | 1 | 1 | 1 | 1 | 2 | 1 | 4 | 1 | 4 | 1 | 1 | 1 |
| Warangal | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Location | No. of Observations Used for | RMSE | MAPE (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Estimation | Validation | LR | ANN | W–LR | W–ANN | LR | ANN | W–LR | W–ANN | |
| Anantapur | 363 | 40 | 0.079 | 0.801 | 0.079 | 0.064 | 9.3 | 9.2 | 9.1 | 8.0 |
| SK Nagar | 1427 | 159 | 0.117 | 0.113 | 0.106 | 0.104 | 8.6 | 8.5 | 8.5 | 7.3 |
| Gulbarga | 981 | 109 | 0.112 | 0.110 | 0.108 | 0.065 | 11.2 | 11.0 | 10.5 | 6.4 |
| Jabalpur | 659 | 73 | 0.147 | 0.143 | 0.141 | 0.134 | 8.5 | 8.2 | 8.1 | 6.3 |
| Rahuri | 545 | 61 | 0.138 | 0.135 | 0.133 | 0.105 | 11.3 | 11.1 | 11.0 | 7.4 |
| Vamban | 690 | 77 | 0.391 | 0.386 | 0.381 | 0.185 | 20.8 | 20.3 | 19.8 | 10.1 |
| Warangal | 1600 | 178 | 0.666 | 0.664 | 0.663 | 0.625 | 31.1 | 31.0 | 30.9 | 27.5 |
| Combinations | Alternative Hypothesis | D-M Statistic | p-Value |
|---|---|---|---|
| Anantapur | |||
| ANN and LR | Predictive accuracy of LR is less than that of ANN | 0.70 | 0.76 |
| W–LR and LR | Predictive accuracy of LR is less than that of W–LR | 6.64 | >0.99 |
| W–LR and ANN | Predictive accuracy of ANN is less than that of W–LR | 7.02 | >0.99 |
| W–ANN and LR | Predictive accuracy of LR is less than that of W–ANN | 0.33 | 0.63 |
| W–ANN and ANN | Predictive accuracy of ANN is less than that of W–ANN | −0.64 | 0.26 |
| W–ANN and W–LR | Predictive accuracy of W–LR is less than that of W–ANN | −6.62 | <0.0001 |
| SK Nagar | |||
| ANN and LR | Predictive accuracy of LR is less than that of ANN | −1.70 | 0.05 |
| W–LR and LR | Predictive accuracy of LR is less than that of W–LR | −1.72 | 0.04 |
| W–LR and ANN | Predictive accuracy of ANN is less than that of W–LR | 1.63 | 0.95 |
| W–ANN and LR | Predictive accuracy of LR is less than that of W–ANN | −2.02 | 0.02 |
| W–ANN and ANN | Predictive accuracy of ANN is less than that of W–ANN | −1.72 | 0.04 |
| W–ANN and W–LR | Predictive accuracy of W–LR is less than that of W–ANN | −1.89 | 0.02 |
| Gulbarga | |||
| ANN and LR | Predictive accuracy of LR is less than that of ANN | 4.60 | >0.99 |
| W–LR and LR | Predictive accuracy of LR is less than that of W–LR | 3.44 | 0.99 |
| W–LR and ANN | Predictive accuracy of ANN is less than that of W–LR | 5.17 | >0.99 |
| W–ANN and LR | Predictive accuracy of LR is less than that of W–ANN | −5.89 | <0.0001 |
| W–ANN and ANN | Predictive accuracy of ANN is less than that of W–ANN | −4.92 | <0.0001 |
| W–ANN and W–LR | Predictive accuracy of W–LR is less than that of W–ANN | −5.97 | <0.0001 |
| Jabalpur | |||
| ANN and LR | Predictive accuracy of LR is less than that of ANN | −1.94 | 0.03 |
| W–LR and LR | Predictive accuracy of LR is less than that of W–LR | −2.03 | 0.02 |
| W–LR and ANN | Predictive accuracy of ANN is less than that of W–LR | 1.72 | 0.96 |
| W–ANN and LR | Predictive accuracy of LR is less than that of W–ANN | −1.59 | 0.05 |
| W–ANN and ANN | Predictive accuracy of ANN is less than that of W–ANN | 1.55 | 0.94 |
| W–ANN and W–LR | Predictive accuracy of W–LR is less than that of W–ANN | −0.96 | 0.16 |
| Rahuri | |||
| ANN and LR | Predictive accuracy of LR is less than that of ANN | −0.30 | 0.38 |
| W–LR and LR | Predictive accuracy of LR is less than that of W–LR | 0.004 | 0.50 |
| W–LR and ANN | Predictive accuracy of ANN is less than that of W–LR | 0.28 | 0.61 |
| W–ANN and LR | Predictive accuracy of LR is less than that of W–ANN | −4.93 | <0.0001 |
| W–ANN and ANN | Predictive accuracy of ANN is less than that of W–ANN | −1.78 | 0.04 |
| W–ANN and W–LR | Predictive accuracy of W–LR is less than that of W–ANN | −6.99 | <0.0001 |
| Vamban | |||
| ANN and LR | Predictive accuracy of LR is less than that of ANN | −9.59 | <0.0001 |
| W–LR and LR | Predictive accuracy of LR is less than that of W–LR | −7.38 | <0.0001 |
| W–LR and ANN | Predictive accuracy of ANN is less than that of W–LR | 9.36 | >0.99 |
| W–ANN and LR | Predictive accuracy of LR is less than that of W–ANN | −6.48 | <0.0001 |
| W–ANN and ANN | Predictive accuracy of ANN is less than that of W–ANN | −4.91 | <0.0001 |
| W–ANN and W–LR | Predictive accuracy of W–LR is less than that of W–ANN | −6.35 | <0.0001 |
| Warangal | |||
| ANN and LR | Predictive accuracy of LR is less than that of ANN | 10.93 | >0.99 |
| W–LR and LR | Predictive accuracy of LR is less than that of W–LR | −5.07 | <0.0001 |
| W–LR and ANN | Predictive accuracy of ANN is less than that of W–LR | −11.92 | <0.0001 |
| W–ANN and LR | Predictive accuracy of LR is less than that of W–ANN | −13.07 | <0.0001 |
| W–ANN and ANN | Predictive accuracy of ANN is less than that of W–ANN | −17.56 | <0.0001 |
| W–ANN and W–LR | Predictive accuracy of W–LR is less than that of W–ANN | −12.97 | <0.0001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paul, R.K.; Vennila, S.; Yeasin, M.; Yadav, S.K.; Nisar, S.; Paul, A.K.; Gupta, A.; Malathi, S.; Jyosthna, M.K.; Kavitha, Z.; et al. Wavelet Decomposition and Machine Learning Technique for Predicting Occurrence of Spiders in Pigeon Pea. Agronomy 2022, 12, 1429. https://doi.org/10.3390/agronomy12061429
Paul RK, Vennila S, Yeasin M, Yadav SK, Nisar S, Paul AK, Gupta A, Malathi S, Jyosthna MK, Kavitha Z, et al. Wavelet Decomposition and Machine Learning Technique for Predicting Occurrence of Spiders in Pigeon Pea. Agronomy. 2022; 12(6):1429. https://doi.org/10.3390/agronomy12061429
Chicago/Turabian StylePaul, Ranjit Kumar, Sengottaiyan Vennila, Md Yeasin, Satish Kumar Yadav, Shabistana Nisar, Amrit Kumar Paul, Ajit Gupta, Seetalam Malathi, Mudigulam Karanam Jyosthna, Zadda Kavitha, and et al. 2022. "Wavelet Decomposition and Machine Learning Technique for Predicting Occurrence of Spiders in Pigeon Pea" Agronomy 12, no. 6: 1429. https://doi.org/10.3390/agronomy12061429
APA StylePaul, R. K., Vennila, S., Yeasin, M., Yadav, S. K., Nisar, S., Paul, A. K., Gupta, A., Malathi, S., Jyosthna, M. K., Kavitha, Z., Mathukumalli, S. R., & Prabhakar, M. (2022). Wavelet Decomposition and Machine Learning Technique for Predicting Occurrence of Spiders in Pigeon Pea. Agronomy, 12(6), 1429. https://doi.org/10.3390/agronomy12061429






