Abstract
Olive quick decline syndrome (OQDS), which is caused by Xylella fastidiosa, poses a severe threat to the agriculture of Mediterranean countries and causes severe damage to the olive trees in Italy. Since no effective control measures are currently available, the objective of this study was the screening of antagonistic bacteria that are potentially deployable as biocontrol agents against X. fastidiosa. Therefore, two approaches were used, i.e., the evaluation of the antagonistic activity of (i) endophytic bacteria isolated from two different cultivars of olive trees (Leccino and Ogliarola salentina) and (ii) epiphytic bacteria isolated from the phyllospheres of different host plant species of X. fastidiosa. In vitro dual culture tests showed that 12 out of 200 isolates inhibited X. fastidiosa growth, with appearances of clear zones between 4.0 and 38.6 mm. 16S rRNA gene sequencing revealed different species of the genera Paenibacillus, Bacillus, Pantoea, Microbacterium, Stenotrophomonas, Delftia, and Pseudomonas. Furthermore, an investigation for antimicrobial activity identified 5 out of the 12 antagonistic bacteria, Paenibacillus rigui, Bacillus subtilis, Bacillus pumilus, Microbacterium oxydans, and Stenotrophomonas rhizophila, that were able to produce culture filtrates with inhibitory activities. Our results are promising for further investigation to develop an eco-sustainable strategy to control X. fastidiosa using biocontrol agents or their secreted metabolites.
1. Introduction
In autumn 2013, a disease outbreak affecting olive trees was reported in the region of Apulia (south of Italy). It was named Olive Quick Decline Syndrome (OQDS) disease and is caused by Xylella fastidiosa subspecies pauca ST53 De Donno strain, a xylem-limited phytopathogenic bacterium [,,,]. Xylella fastidiosa has very high pathogenicity on the local olive cultivars ‘Cellina di Nardò’ and ‘Ogliarola salentina’ and different ornamental and perennial hosts []. In the plant, X. fastidiosa shares many characteristics with vascular pathogens in terms of symptom development, beginning with leaf scorch and ending dramatically with total decay []. The incidence of the disease has increased rapidly through the heavily olive-grown countryside of the Salento peninsula. The epidemic has caused the decline of millions of olive trees, despite available containment measures being taken. Unfortunately, there are still no effective measures against the bacterium. Thus, there is an increased interest in biological control through antagonistic microorganisms [,]. Some bacteria may possess direct antagonistic activities against pathogens through hyperparasitism or antibiosis. Meanwhile, indirect antagonists can control diseases by inducing or enhancing plant resistance to pathogen infections or competing for nutrients []. Bacterial antagonists may be rhizospheric, phyllospheric, or endophytic microorganisms, which are classified according to their inhibiting environment. Direct antagonists, which actively produce a broad spectrum of antimicrobial metabolites, are considered the most effective against competitors, allowing advantages for antibiotic-producing microorganisms in resource-limited environments. There are a large number of known antibiotics produced in small amounts by many endophytic microorganisms and released into the environment []. The production of antimicrobial metabolites, mostly with broad-spectrum activities, has been reported for biocontrol bacteria belonging to Agrobacterium, Bacillus, Pantoea, Pseudomonas, and many other genera. Bacillus genera have been observed to produce several antimicrobial substances such as non-ribosomal lipopeptides (LPs), iturins, fengycins, surfactins, and serine proteinase subtilisin [,], while Pseudomonas genera have been more associated with antibiotic metabolites such as pyrrolnitrin and phenazine []. Indeed, the lack of any therapeutic formulation for curing infected olives further emphasizes the need to develop effective and sustainable control strategies. So far, few studies have been conducted to retrieve direct antagonists for a better biocontrol strategy of X. fastidiosa. Particularly, the endophyte Curtobacterium flaccumfaciens was found to limit the in vitro growth of X. fastidiosa and reduce the symptoms generated in Catharanthus roseus []. Moreover, the endophytic bacterium Paraburkholderia phytofirmans strain PsJN was reported to control X. fastidiosa infections in the grapevine []. Recently published studies have stated the absence of native antagonists isolated from Apulian olive trees to inhibit X. fastidiosa ST53 growth in vitro []. Furthermore, other research explored the involvement of microbial endophytes residing in the sapwood of Apulian olive cultivars that might be a promising control strategy for xylem-colonizing pathogens such as X. fastidiosa and in the expression of resistance characteristics against OQDS []. In our study, we targeted olive endophytic and epiphytic bacterial populations inhabiting the leaf surfaces of different host species by in vitro screening assays for their direct antagonistic activities. Moreover, we tested their abilities to produce antimicrobial substances against X. fastidiosa subspecies pauca ST53 in liquid culture.
2. Materials and Methods
2.1. Sampling and Isolation of Endophytic and Epiphytic Bacteria
A sampling of plant material within the Apulian demarcated area of X. fastidiosa was carried out at two sites, an Xf-free area and an Xf-infected zone, as defined in the Commission implementing Decision (EU) 2018/927 and in the Decision of the Regional Plant protection Service. A total of 16 samples were collected from different host species: Olea europaea, Polygala myrtifolia, Rosmarinus officinalis, Nerium oleander, Laurus nobilis, Myrtus communis, Prunus dulcis, and Prunus avium (2 samples from each plant species, 1 sample per site). Moreover, a total of 20 symptomatic and asymptomatic olive trees were randomly selected at the two sites. The trees under study were 10 from the resistant cv. Leccino (5 trees per site) and 10 susceptible olive trees from the cv. Ogliarola salentina (5 trees per site). For the endophytic bacterial isolation, four twigs were sampled from each olive sample tree in relation to the different cardinal points of the canopy. After washing with running tap water, 8–10 cm long twigs were surface-sterilized using 2% sodium hypochlorite for 2 min and 70% ethanol for 2 min and rinsed three times in sterilized water. Thereafter, plant sap was obtained through the patented sap extraction method (CIHEAM/MAIB, Patent number WO2017017555A1). With this method, twigs and sterilized rubber tubes were attached with parafilm to a syringe containing 2 mL of PBS buffer or sterile water. The syringe was gently and slowly pressed to push the plant sap out of the twig, which was collected in an Eppendorf tube. Aliquots of 0.1 mL from a 10-fold serial dilution of the extracted sap were plated onto nutrient agar and King B media [,]. For the epiphytic bacteria isolation, 10 leaves were picked randomly from each plant species, washed separately in flasks containing 50 mL of sterile distilled water and gently shaken for 2 h at 150 rpm [,]. Subsequently, a dilution series was made of each washing solution and, as previously described, 100 µL were spread onto king B and nutrient agar media. After 3 days of growth at 25 °C, the single bacterial colonies were purified by repeated streaking on the same medium.
2.2. Screening of Antagonistic Activity In Vitro
The antagonistic activity of the isolated bacteria against X. fastidiosa was studied on BCYE solid nutrient medium by the modified dual culture method previously described by Zicca et al. []. To evaluate the antagonistic activity, bacteria were co-plated in the following order: Three drops of X. fastidiosa suspension (106 CFU/mL), each containing 20 µL, were placed at the top of the petri dish, 1 cm apart, and allowed to slowly flow down to the opposite side of the plate, resulting in three parallel rows of X. fastidiosa cultures. After 24 h of incubation at 28 °C, a 5 μL aliquot of a suspension of the potential BCA (108 CFU/mL) was placed on top of each row of X. fastidiosa cultures, and ampicillin (20 μg/mL) was used as a positive control. After 7–12 days of incubation at 28 °C, the antagonistic activity was detected as an inhibition zone of X. fastidiosa growth, which was measured as the distance between the edges of X. fastidiosa growth and the growth of the tested strain. The tests were performed in triplicate.
2.3. Molecular Characterization and Identification of Antagonistic Bacteria
DNA fingerprinting was performed to investigate the degree of differentiation in antagonistic bacterial isolates. ERIC-PCR was performed on bacterial DNA extracted by the classical phenol-chloroform method [] and used as a template in a PCR reaction with the primer pair ERIC1R/ERIC2 (5′-ATGTAAGCTCCTGGGGATTCAC-3′/5′-AAGTAAGTGACTGGGGTGAGCG-3′) []. The PCR reaction was performed in a total volume of 25 µL and contained 12.5 µL of 2X Master Mix PCR (ThermoFisher Scientific, Milan, Italy), 0.5 µL of each forward and reverse primer (10 mM), and 2 µL of genomic DNA. Amplifications were performed, starting with an initial denaturation at 95 °C for 7 min, followed by 30 cycles (94 °C for 1 min, 52 °C for 1 min, and 65 °C for 8 min) and a final step at 65 °C for 16 min before cooling at 4 °C. Amplicons were separated by electrophoresis in a 2% TBE agarose gel, and banding patterns were visualized using the Gel Doc EZ system (BIORAD, Milan, Italy). The gel image was exported to Gel-Quest software for smoothing, baselining, peak detection, and fragment size determination. Meanwhile, the software Cluster-Vis was extensively used to construct a phylogenetic tree (1000 bootstrap repeats) based on the amplified bands, which were analyzed and scored (0) for absence and (1) for presence among the lanes (Supplementary File S1). For bacterial identification, the 16S rDNA gene was amplified by polymerase chain reaction (PCR) using approximately 50 ng of genomic DNA in a final volume of 25 μL. Almost the entire gene (approximately 1300 bp) was amplified using primer pairs 63f (5′-CAGGCCTAACACATGCAAGTC-3′) and reverse primer 1387r (5′-GGG CGGWGTGTACAAGGC-3′) []. The PCR mixtures contained 2 µL of 50 ng/µL template DNA, 5 µL of 5X Phusion Green HF buffer (ThermoFisher Scientific, Milan, Italy), 0.5 µL of 50 mM MgCl2, 0.5 µL of 10 mM dNTP, 0.4 µL of 10 µM solutions of each primer, 0.6 µL of DMSO, 0.25 µL of 2.0 U/µL Phusion DNA polymerase (ThermoFisher Scientific), and nuclease-free water to a reaction volume of 25 µL. The PCR cycle parameters were as follows: 98 °C for 30 s, followed by 35 cycles at 98 °C for 10 s, at 55 °C for 30 s, and at 72 °C for 45 s, and a final extension at 72 °C for 7 min. The reaction products were analyzed by electrophoresis in 1.2% TAE agarose gel, and DNA bands were visualized on the Gel Doc EZ system (BIORAD, Milan, Italy). The amplification products were sequenced in both directions using Eurofins Genomics (https://www.eurofinsgenomics.eu/, accessed on 26 January 2021). The accuracy of the obtained sequences was evaluated using FinchTV (version 1.4.0, Denver, CO, USA) software (http://www.geospiza.com/finchTV, accessed on 26 January 2021). The sequences were submitted to the National Center for Biotechnology Information (NCBI) BLAST online search engine. The assigned sequences that had ≥ 98% identity to a valid sequence were deposited in NCBI under specific accession numbers.
2.4. Antimicrobial Activity against X. fastidiosa in Culture Filtrates of Antagonistic Strains
All the antagonistic strains used in this study were evaluated for their antimicrobial compound production in liquid culture against X. fastidiosa ST53 by a well diffusion test [,]. In detail, a 2% bacterial suspension (106 CFU/mL) was inoculated in 20 mL of PD3 broth and incubated under shaking (150 rpm) at 26 °C. After 48h of incubation, the cultures were sampled and cell-free culture filtrates were obtained by centrifugation (10,000 rpm, 4 °C, 10 min) and filtration through 0.22 μm filters. Thereafter, PD3 agar plates were seeded with an X. fastidiosa suspension to obtain three rows, as previously described. Subsequently, at the end of the middle row, a well (8 mm diameter) was made and filled with 200 μL of culture filtrate. Tests were performed in triplicate. Plates were incubated at 28 °C for at least 6 days and the antimicrobial activity was detected as an area of X. fastidiosa growth inhibition and was measured as the distance between the well and the X. fastidiosa growth.
2.5. Statistical Analysis
Statistical analysis was performed using GraphPad software (Version, 8.0.2, San Diego, CA, USA). Data concerning antagonistic activity were compared by applying a one-way ANOVA followed by Tukey’s test to determine significantly different values (p < 0.05).
3. Results
3.1. In Vitro Antagonistic Activity against X. fastidiosa
Seasonally, an approximate average of 3400 bacterial isolates were obtained, belonging to the sampled olive varieties. Among them, 111 isolates were selected as the most frequently isolated, and based on the morphological properties, they were clustered into 16 groups. Additionally, a total of 89 bacterial epiphytic isolates were obtained from the different sampled host plant species. Overall, differences were observed between the bacterial population sizes of various leaves. The highest bacterial population was recorded on Polygala myrtifolia leaves (1.23 × 105 CFU/cm2), and the lowest was observed on Laurus nobilis leaves (2.30 × 103 CFU/cm2). A total of 200 endophytic and epiphytic bacteria were screened for their ability to inhibit X. fastidiosa by in vitro dual culture assay. The results showed that 12 of the tested isolates had antagonistic activities against X. fastidiosa (Table 1).

Table 1.
Epiphytic and endophytic bacteria isolates and their antagonistic activities against X. fastidiosa ST53. Different letters indicate statistically different antagonistic activities of epiphytic and endophytic isolates with p < 0.05 as determined by one-way analysis of variance (ANOVA) followed by Tukey’s test.
For epiphytic bacteria, isolates BA69, BA104, and BA102 showed the highest activities, causing very large inhibition halos (Figure 1), with mean growth zones of 34, 16.6, and 12.3 mm, respectively. BA91, BA2, and BA111 isolates were weak inhibitors of the pathogen, with mean inhibition zones of 4, 6, and 7 mm, respectively. The antagonistic activities of endophytic bacteria were also remarkable. L49 isolate exhibited a complete inhibition on s BCYE agar plate (Figure 1). Furthermore, S55, L36, and L77 isolates were able to strongly curtail X. fastidiosa growth, as indicated by the clearing zones of 21.6, 19, and 15 mm, respectively (Table 1), suggesting that these isolates could efficiently inhibit the growth of X. fastidiosa.

Figure 1.
Dual-culture assay of antagonistic activities of epiphytic and endophytic bacteria isolated from different host plant species against X. fastidiosa ST53.
3.2. Molecular Characterization and Identification of Antagonistic Bacteria
From the sequences and BLAST results, epiphytic antagonistic bacteria were isolated as follows: M. oleivorans (BA91) from Ogliarola salentina, M. phyllosphaerae (BA111) and M. oxydans (BA104) from P. myrtifolia. Two species of the genus Bacillus, subtilis and pumilus, were isolated from Leccino plants. Endophytic bacteria S55 and L49 showed the highest similarity with Peanbacillus rigui (99.25%) and P. naphthalenovorans (99.91%). The genus Pseudomonas was also isolated from the cvs. Ogliarola salentina and Rosmarinus officinalis, with isolates belonging to the species garminis and hibiscicola. Furthermore, other species assigned to the genera Delftia, Pantoea, and Stenotrophomonas were isolated from different ornamental plants, namely, R. officinalis, N. oleander, and P. myrtifolia. The results from the ERIC-PCR analysis highlighted diverse ERIC-PCR profiles, indicating the genetic diversity of the epiphytic and endophytic bacterial antagonistic isolates, even if obtained from the same host plant and belonging to the same genera (Figure 2) (Supplementary File S1).
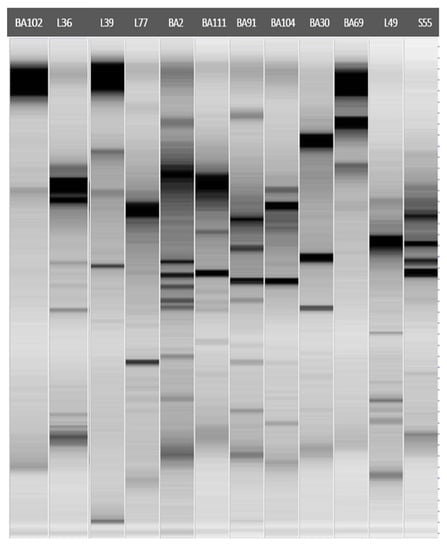
Figure 2.
Gel-like image of ERIC-PCR profiles of endophytic bacteria isolated from olive plants and epiphytic bacteria isolated from different host plants (Table 1).
3.3. Antimicrobial Activity against X. fastidiosa in Culture Filtrates of Antagonistic Strains
All the antagonistic isolates used in this study were also tested for the production of antimicrobial compounds against X. fastidiosa ST53 in liquid culture. Our results indicated that the cell-free supernatants (CFS) of B. subtilis L39, B. pumilus L36, P. rigui S55, S. rhizophila BA102, and M. oxydans BA104 showed relevant antimicrobial activities. These activities were statistically different between the species (Figure 3). The lowest antimicrobial activity was recorded from M. oxydans BA104, whereas the highest inhibition of X. fastidiosa was observed in the presence of CFS of B. subtilis L39, with an inhibition zone of more than 30 mm, followed by S. rhizophila BA102, B. pumilis L36, and P. rigui S55, respectively. Cell-free supernatants from these three isolates formed inhibition halos of between 15.0 and 25.0 mm on X. fastidiosa cultures. Interestingly, the isolates proved to be strongly inhibitory in the direct antagonism assay (Table 1) and produced CFSs with no visible inhibitory activities against X. fastidiosa ST53.
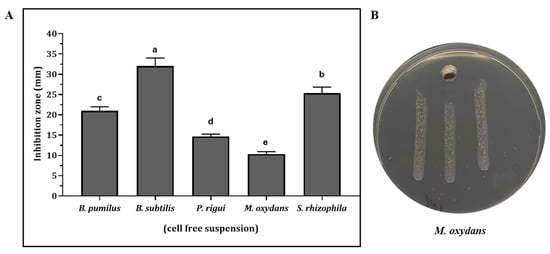
Figure 3.
(A) Antimicrobial activity against X. fastidiosa ST53 of the culture filtrates of B. pumilus L36, B. subtilis L39, P. rigui S55, M. oxydans BA 104, and S. rhizophila BA 102. Different letters indicate statistically different antimicrobial activities with p < 0.05 as determined by one-way analysis of variance (ANOVA) followed by Tukey’s test. (B) Well diffusion test of antimicrobial activity of M. oxydans BA 104 against X. fastidiosa ST53 on PD3 medium.
4. Discussion
Microbial communities inhabit both the external surfaces (epiphytes) and internal spaces (endophytes) of crops, playing a key role in protecting them against plant pathogens [,]. A high number of epiphytic bacteria have already been reported successfully as biocontrol agents (BCAs) against several phytopathogens [,,]. The implementation of sustainable approaches in the plant protection sector has become a major research topic over the last years, and new strategies, such as nanotechnology tools combined with the circular economy starting from agro-industrial waste, could support the exploitation of active compounds produced by BCAs such as those isolated in this study [,,]. Indeed, there is also increasing evidence of the beneficial effects of bacterial endophytes on the host plants, as they are considered to be able to promote plant growth and health through indirect mechanisms, such as plant pathogen inhibition [,]. This last feature makes endophytic bacteria promising biocontrol agent candidates that are potentially able to compete with plant pathogens inhabiting the same niche. Our results clearly indicate the in vitro antagonistic activity of seven epiphytic bacteria belonging to different species of the genera Stenotrophomonas, Pantoea, Microbacterium, Pseudomonas, and Delftia.
It is interesting to note that these species were previously reported from diverse plants and microenvironments. They are also able to live in the endosphere of plants, [,,] and, in the case of X. fastidiosa, it is essential to select a potential biocontrol agent among the epiphytic bacteria able to colonize the xylem. Therefore, further research is necessary to ascertain the efficiency of endophytic colonization by the epiphytic antagonists identified in this study. Microbes arrive on the phyllosphere rather stochastically via the air, soil, rain, or insects, and only selected taxa successfully colonize the phyllosphere []. Frequently occurring genera in phyllosphere communities are Methylobacterium, Sphingomonas, Pseudomonas, and Pantoea [,,]. In these findings, Stenotrophomonas, Microbacterium, and Delftia genera were successfully isolated from the leaf surfaces of Polygala myrtifolia, Olea europaea, and Rosmarinus officinalis. Among the different epiphytic antagonists observed in our results, Pantoea agglomerans BA69 demonstrated the strongest antagonistic activity against X. fastidiosa ST53 in a dual-culture bioassay. The genus Pantoea contains several plant pathogens as well as biocontrol agents that are effective against a range of pathogens, such as Botrytis cinerea, Xanthomonas campestris, and the most extensively studied, Erwinia amylovora []. In our results, a high level of antimicrobial activity was observed in two epiphytic isolates, BA102 and BA104, which were closely related to S. rhizophila (100%) and M. oxydans (99%), respectively. This result is in agreement with previous studies about the potential to control pests and the antimicrobial activity of M. oxydans [,].
Other studies have reported the capacity of S. rhizophila to antagonize different plant pathogens in vitro, such as Verticillium dahliae, Pythium ultimum, Rhizoctonia solani, Colletotrichum gloeosporioides, and Sclerotinia sclerotiorum, among others [,]. In addition, the detection of protease [] and VOCs was previously reported for S. rhizophila. In fact, bacterial VOCs can have direct antagonistic effects against other bacteria. For instance, Pseudomonas fluorescens WR-1 produces volatiles such as benzothiazole and 1-methyl naphthalene with bacteriostatic effects against the tomato pathogen Ralstonia solanacearum []. Indeed, VOC production by S. rhizophila has already been identified as an antagonist mechanism toward phytopathogens, mainly by the production of b-phenylethanol and dodecanal, although there are still different VOCs produced by bacteria that have not yet been identified []. In particular, the antimicrobial properties of b-phenylethanol have been found to alter the permeability of the plasma membrane, disrupt amino acids and the sugar transport system, and inhibit macromolecular synthesis, preventing phytopathogen growth []. Although it remains to be ascertained in further studies, the involvement of one or more VOCs produced by S. rhizophila in the antagonistic activity against X. fastidiosa is plausible. Moreover, the findings of this study provide evidence of the antagonistic capacity of the species P. garminis BA2, M. oleivorans BA91, M. phyllospahrea BA111, and D. acidovorans BA30 against X. fastidiosa ST53. In general, these species have been cited in previous studies for their potential antagonistic abilities against other phytopathogens [,,,,]. To our knowledge, this is the first report about the potential antagonistic activities of epiphytes against the phytopathogen X. fastidiosa ST53.
Importantly, our results point out the highest in vitro antagonistic activity of Bacillus isolates and P. rigui, even with the production of culture filtrates that inhibit pathogen growth, likely by secreting active substances in the liquid medium. Those isolates, together with the above-mentioned M. oxydans and S. rhizophila and differently from the other isolates, show antagonistic and antimicrobial capacities, suggesting that the mode of action of these isolates is different from the other bacteria used in this study. In general, Bacillus strains are well-known for their ability to secrete a variety of antimicrobial compounds, and some of them have already been considered and registered as biocontrol agents against various plant pathogens. As an example, the strain Q713 of B. subtilis produces lipopeptides belonging to the families of the surfactins, iturins, and fengycins []. Moreover, the genes responsible for the production of these substances as well as antibiotics, such as macrolactin, bacilysin, and difficidin, have been identified in its genome []. These findings coincide with recent results, which concluded that several Bacillus strains were able to inhibit the in vitro growth of X. fastidiosa ST53 []. Furthermore, the results of this study provide evidence that P. rigui is able to produce substances with inhibitory effects in liquid culture. Many Paenibacillus species compete with other microorganisms through the production of a wide range of antimicrobial compounds such as peptides that are extremely significant for biocontrol in agriculture []. Overall, different studies have indicated the effects of pH, temperature, and enzymatic degradation on the activities of culture supernatants []. In this regard, several Bacillus species were shown to be insensitive to different enzymes and chemicals and were heat-stable and active in a wide pH range []. Temperature and pH stability were also observed in Peanbacillus species []. In this context, future experiments are recommended to confirm the stability of the antimicrobial activity of the culture supernatant identified in this study.
It is noteworthy that P. naphthalenovorans showed complete antagonistic inhibition in vitro; however, no antimicrobial activity was observed. Moreover, P. agglomerans, P. graminis, M. oleivorans, M. phyllospharaea, D. acidovorans, and P. hibiscola showed only antagonistic activities without inhibitory effects by cell-free suspensions. For these isolates, it is unlikely that the production of antimicrobial compounds is important because growth inhibition was only observed when there was a direct cell-to-cell interaction. A possible explanation for this result is that the antagonistic activities were mediated by mechanisms such as lytic enzymes, space, and nutrient competition [,]. Moreover, the production of secondary metabolites in dual in vitro culture assays also depends on the nutrient concentration and composition of the chosen medium. In particular, the fastidious growth of X. fastidiosa and the faster growth rate of antagonistic bacteria allow them to deplete the nutrient availability and thus limit X. fastidiosa growth [].
5. Conclusions
In this study, bacterial control agents against X. fastidiosa were successfully isolated from different host plants. These agents were different endophytic and epiphytic species belonging to the genera Paenibacillus, Bacillus, Pantoea, Microbacterium, Stenotrophomonas, Delftia, and Pseudomonas. More specifically, M. oxydans, S. rhizophila, B. subtilis, B. pumilus, and P. rigui were also able to secrete inhibitory substances in cell-free suspensions. Therefore, their mode of action requires further study. In addition, research on the secreted antibacterial compounds is also recommended in natural field conditions.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/agronomy12061266/s1.
Author Contributions
Conceptualization, formal analysis, methodology, writing—original draft preparation, investigation, and data curation, M.M., A.H. and F.V.; supervision and writing—review and editing, F.V. and G.M.B.; writing—review and editing, A.M.D. and S.W.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Cure XF (H2020-MSCA-RISE Grant Agreement no. 734353) project in the framework of Capacity Building and Raising Awareness in Europe and Third Countries to Cope with Xylella fastidiosa.
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and/or its Supplementary Materials.
Acknowledgments
The authors thank the Cure XF project for funding the research and all the experts who have added significant knowledge and contributions that allowed us to reach these findings.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of the data; in the writing of the manuscript; or in the decision to publish the results.
References
- Saponari, M.; Boscia, D.; Nigro, F.; Martelli, G.P. Identification of DNA Sequences Related to Xylella fastidiosa in Oleander, Almond and Olive Trees Exhibiting Leaf Scorch Symptoms in Apulia (Southern Italy). J. Plant Pathol. 2013, 95, 659–668. [Google Scholar]
- Loconsole, G.; Potere, O.; Boscia, D.; Altamura, G.; Djelouah, K.; Elbeaino, T.; Frasheri, D.; Lorusso, D.; Palmisano, F.; Pollastro, P.; et al. Detection of Xylella fastidiosa in Olive Trees by Molecular and Serological Methods. J. Plant Pathol. 2014, 96, 8. [Google Scholar]
- Giampetruzzi, A.; Saponari, M.; Loconsole, G.; Boscia, D.; Savino, V.N.; Almeida, R.P.P.; Zicca, S.; Landa, B.B.; Chacón-Diaz, C.; Saldarelli, P. Genome-Wide Analysis Provides Evidence on the Genetic Relatedness of the Emergent Xylella fastidiosa Genotype in Italy to Isolates from Central America. Phytopathology 2017, 107, 816–827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elbeaino, T.; Valentini, F.; Abou Kubaa, R.; Moubarak, P.; Yaseen, T.; Digiaro, M. Multilocus Sequence Typing of Xylella fastidiosa Isolated from Olive Affected by” Olive Quick Decline Syndrome” in Italy. Phytopathol. Mediterr. 2014, 53, 533–542. [Google Scholar]
- Saponari, M.; Boscia, D.; Altamura, G.; Loconsole, G.; Zicca, S.; D’Attoma, G.; Morelli, M.; Palmisano, F.; Saponari, A.; Tavano, D.; et al. Isolation and Pathogenicity of Xylella fastidiosa Associated to the Olive Quick Decline Syndrome in Southern Italy. Sci. Rep. 2017, 7, 17723. [Google Scholar] [CrossRef]
- Martelli, G.P. The Current Status of the Quick Decline Syndrome of Olive in Southern Italy. Phytoparasitica 2016, 44, 1–10. [Google Scholar] [CrossRef]
- Maggiore, G.; Semeraro, T.; Aretano, R.; De Bellis, L.; Luvisi, A. GIS Analysis of Land-Use Change in Threatened Landscapes by Xylella fastidiosa. Sustainability 2019, 11, 253. [Google Scholar] [CrossRef] [Green Version]
- EFSA Panel on Plant Health (EFSA PLH Panel); Bragard, C.; Dehnen-Schmutz, K.; Di Serio, F.; Gonthier, P.; Jacques, M.; Jaques Miret, J.A.; Justesen, A.F.; MacLeod, A.; Magnusson, C.S.; et al. Effectiveness of in Planta Control Measures for Xylella fastidiosa. EFSA J. 2019, 17, 5666. [Google Scholar] [CrossRef]
- Pal, K.K.; McSpadden Gardener, B. Biological Control of Plant Pathogens. Plant Health Instr. 2006, 2, 1117–1141. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Klimova, E.; Rodríguez-Peña, K.; Sánchez, S. Endophytes as Sources of Antibiotics. Biochem. Pharmacol. 2017, 134, 1–17. [Google Scholar] [CrossRef]
- Caulier, S.; Nannan, C.; Gillis, A.; Licciardi, F.; Bragard, C.; Mahillon, J. Overview of the Antimicrobial Compounds Produced by Members of the Bacillus Subtilis Group. Front. Microbiol. 2019, 10, 302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biondi, E.; Gallipoli, L.; Mazzaglia, A.; Fuentealba, S.P.; Kuzmanović, N.; Bertaccini, A.; Balestra, G. Bacillus-Based Products for Management of Kiwifruit Bacterial Canker. Phytopathol. Mediterr. 2021, 60, 215–228. [Google Scholar] [CrossRef]
- Huang, R.; Feng, Z.; Chi, X.; Sun, X.; Lu, Y.; Zhang, B.; Lu, R.; Luo, W.; Wang, Y.; Miao, J.; et al. Pyrrolnitrin Is More Essential than Phenazines for Pseudomonas Chlororaphis G05 in Its Suppression of Fusarium graminearum. Microbiol. Res. 2018, 215, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Lacava, P.T.; Li, W.; Arau, W.L.; Hartung, J.S. The Endophyte Curtobacterium flaccumfaciens Reduces Symptoms Caused by Xylella fastidiosa in Catharanthus Roseus. Microbiology 2007, 45, 7. [Google Scholar]
- Baccari, C.; Antonova, E.; Lindow, S. Biological Control of Pierce’s Disease of Grape by an Endophytic Bacterium. Phytopathology 2019, 109, 248–256. [Google Scholar] [CrossRef] [Green Version]
- Zicca, S.; De Bellis, P.; Masiello, M.; Saponari, M.; Saldarelli, P.; Boscia, D.; Sisto, A. Antagonistic Activity of Olive Endophytic Bacteria and of Bacillus Spp. Strains against Xylella fastidiosa. Microbiol. Res. 2020, 236, 126467. [Google Scholar] [CrossRef]
- Hanani, A.; Valentini, F.; Sanzani, S.M.; Santoro, F.; Minutillo, S.A.; Gallo, M.; Cavallo, G.; Mourou, M.; El Moujabber, M.; D’Onghia, A.M.; et al. Community Analysis of Culturable Sapwood Endophytes from Apulian Olive Varieties with Different Susceptibility to Xylella fastidiosa. Agronomy 2021, 12, 9. [Google Scholar] [CrossRef]
- Brown, A.; Smith, H. Benson’s Microbiological Applications, Laboratory Manual in General Microbiology, Short Version; McGraw-Hill Education: New York, NY, USA, 2014. [Google Scholar]
- Etminani, F.; Harighi, B. Isolation and Identification of Endophytic Bacteria with Plant Growth Promoting Activity and Biocontrol Potential from Wild Pistachio Trees. Plant Pathol. J. 2018, 34, 208–217. [Google Scholar] [CrossRef]
- Balestra, G.M.; Agostini, R.; Bellincontro, A.; Mencarelli, F.; Varvaro, L. Bacterial Populations Related to Gerbera (Gerbera Jamesonii L.) Stem Break. Phytopathol. Mediterr. 2005, 44, 9. [Google Scholar]
- Schreiber, L.; Krimm, U.; Knoll, D.; Sayed, M.; Auling, G.; Kroppenstedt, R.M. Plant–Microbe Interactions: Identification of Epiphytic Bacteria and Their Ability to Alter Leaf Surface Permeability. New Phytol. 2005, 166, 589–594. [Google Scholar] [CrossRef]
- Minas, K.; McEwan, N.R.; Newbold, C.J.; Scott, K.P. Optimization of a High-Throughput CTAB-Based Protocol for the Extraction of QPCR-Grade DNA from Rumen Fluid, Plant and Bacterial Pure Cultures. FEMS Microbiol. Lett. 2011, 325, 162–169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Versalovic, J.; Schneider, M.; De Bruijn, F.J.; Lupski, J.R. Genomic Fingerprinting of Bacteria Using Repetitive Sequence-Based Polymerase Chain Reaction. Methods Mol. Cell. Biol. 1994, 5, 25–40. [Google Scholar]
- Marchesi, J.R.; Sato, T.; Weightman, A.J.; Martin, T.A.; Fry, J.C.; Hiom, S.J.; Dymock, D.; Wade, W.G. Design and Evaluation of Useful Bacterium-Specific PCR Primers That Amplify Genes Coding for Bacterial 16S RRNA. Appl. Environ. Microbiol. 1998, 64, 2333. [Google Scholar] [CrossRef] [Green Version]
- Dagher, F.; Olishevska, S.; Philion, V.; Zheng, J.; Déziel, E. Development of a Novel Biological Control Agent Targeting the Phytopathogen Erwinia amylovora. Heliyon 2020, 6, e05222. [Google Scholar] [CrossRef]
- Borriss, R. Use of Plant-Associated Bacillus Strains as Biofertilizers and Biocontrol Agents in Agriculture. In Bacteria in Agrobiology: Plant Growth Responses; Springer: Berlin, Germany, 2011; pp. 41–76. [Google Scholar]
- Gardener, B.B.M.; Fravel, D.R. Biological Control of Plant Pathogens: Research, Commercialization, and Application in the USA. Plant Health Prog. 2002, 3, 17. [Google Scholar] [CrossRef] [Green Version]
- Legein, M.; Smets, W.; Vandenheuvel, D.; Eilers, T.; Muyshondt, B.; Prinsen, E.; Samson, R.; Lebeer, S. Modes of Action of Microbial Biocontrol in the Phyllosphere. Front. Microbiol. 2020, 11, 1619. [Google Scholar] [CrossRef]
- Thomashow, L.S.; Weller, D.M. Current Concepts in the Use of Introduced Bacteria for Biological Disease Control: Mechanisms and Antifungal Metabolites. In Plant-Microbe Interactions; Springer: New York, NY, USA, 1996; pp. 187–235. [Google Scholar]
- Daranas, N.; Roselló, G.; Cabrefiga, J.; Donati, I.; Francés, J.; Badosa, E.; Spinelli, F.; Montesinos, E.; Bonaterra, A. Biological Control of Bacterial Plant Diseases with Lactobacillus Plantarum Strains Selected for Their Broad-Spectrum Activity. Ann. Appl. Biol. 2019, 174, 92–105. [Google Scholar] [CrossRef] [Green Version]
- Schiavi, D.; Balbi, R.; Giovagnoli, S.; Camaioni, E.; Botticella, E.; Sestili, F.; Balestra, G.M. A Green Nanostructured Pesticide to Control Tomato Bacterial Speck Disease. Nanomaterials 2021, 11, 1852. [Google Scholar] [CrossRef]
- Schiavi, D.; Ronchetti, R.; Di Lorenzo, V.; Salustri, M.; Petrucci, C.; Vivani, R.; Giovagnoli, S.; Camaioni, E.; Balestra, G.M. Circular Hazelnut Protection by Lignocellulosic Waste Valorization for Nanopesticides Development. Appl. Sci. 2022, 12, 2604. [Google Scholar] [CrossRef]
- Schiavi, D.; Francesconi, S.; Taddei, A.R.; Fortunati, E.; Balestra, G.M. Exploring Cellulose Nanocrystals Obtained from Olive Tree Wastes as Sustainable Crop Protection Tool against Bacterial Diseases. Sci. Rep. 2022, 12, 6149. [Google Scholar] [CrossRef]
- Afzal, I.; Shinwari, Z.K.; Sikandar, S.; Shahzad, S. Plant Beneficial Endophytic Bacteria: Mechanisms, Diversity, Host Range and Genetic Determinants. Microbiol. Res. 2019, 221, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Santoyo, G.; Moreno-Hagelsieb, G.; del Carmen Orozco-Mosqueda, M.; Glick, B.R. Plant Growth-Promoting Bacterial Endophytes. Microbiol. Res. 2016, 183, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Lumactud, R.; Shen, S.Y.; Lau, M.; Fulthorpe, R. Bacterial Endophytes Isolated from Plants in Natural Oil Seep Soils with Chronic Hydrocarbon Contamination. Front. Microbiol. 2016, 7, 755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmidt, C.S.; Mrnka, L.; Lovecká, P.; Frantík, T.; Fenclová, M.; Demnerová, K.; Vosátka, M. Bacterial and Fungal Endophyte Communities in Healthy and Diseased Oilseed Rape and Their Potential for Biocontrol of Sclerotinia and Phoma Disease. Sci. Rep. 2021, 11, 3810. [Google Scholar] [CrossRef]
- Woźniak, M.; Gałązka, A.; Tyśkiewicz, R.; Jaroszuk-Ściseł, J. Endophytic Bacteria Potentially Promote Plant Growth by Synthesizing Different Metabolites and Their Phenotypic/Physiological Profiles in the Biolog GEN III MicroPlateTM Test. Int. J. Mol. Sci. 2019, 20, 5283. [Google Scholar] [CrossRef] [Green Version]
- Maignien, L.; DeForce, E.A.; Chafee, M.E.; Eren, A.M.; Simmons, S.L. Ecological Succession and Stochastic Variation in the Assembly of Arabidopsis Thaliana Phyllosphere Communities. MBio 2014, 5, e00682-13. [Google Scholar] [CrossRef] [Green Version]
- Delmotte, N.; Knief, C.; Chaffron, S.; Innerebner, G.; Roschitzki, B.; Schlapbach, R.; von Mering, C.; Vorholt, J.A. Community Proteogenomics Reveals Insights into the Physiology of Phyllosphere Bacteria. Proc. Natl. Acad. Sci. USA 2009, 106, 16428–16433. [Google Scholar] [CrossRef] [Green Version]
- Lindow, S.E.; Brandl, M.T. Microbiology of the Phyllosphere. Appl Env. Microbiol 2003, 69, 9. [Google Scholar] [CrossRef] [Green Version]
- Vorholt, J.A. Microbial Life in the Phyllosphere. Nat. Rev. Microbiol. 2012, 10, 828–840. [Google Scholar] [CrossRef]
- Walterson, A.M.; Stavrinides, J. Pantoea: Insights into a Highly Versatile and Diverse Genus within the Enterobacteriaceae. FEMS Microbiol. Rev. 2015, 39, 968–984. [Google Scholar] [CrossRef] [Green Version]
- Irshad, A.; Ahmad, I.; Kim, S.B. Isolation, Characterization and Antimicrobial Activity of Halophilic Bacteria in Foreshore Soils. Afr. J. Microbiol. Res. 2013, 7, 164–173. [Google Scholar]
- Ozsahin, E.; Sezen, K.; Demir, I.; Demirbag, Z. Bacterial Isolates from Palomena prasina (Hemiptera: Pentatomidae) Include Potential Microbial Control Agents. Biocontrol Sci. Technol. 2014, 24, 1039–1051. [Google Scholar] [CrossRef]
- Kai, M.; Effmert, U.; Berg, G.; Piechulla, B. Volatiles of Bacterial Antagonists Inhibit Mycelial Growth of the Plant Pathogen Rhizoctonia solani. Arch. Microbiol. 2007, 187, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Perez, J.J.; Hernandez-Montiel, L.G.; Vero, S.; Noa-Carrazana, J.C.; Quiñones-Aguilar, E.E.; Rincón-Enríquez, G. Postharvest Biocontrol of Colletotrichum gloeosporioides on Mango Using the Marine Bacterium Stenotrophomonas rhizophila and Its Possible Mechanisms of Action. J. Food Sci. Technol. 2019, 56, 4992–4999. [Google Scholar] [CrossRef] [PubMed]
- Vida, C.; Cazorla, F.M.; de Vicente, A. Characterization of Biocontrol Bacterial Strains Isolated from a Suppressiveness-Induced Soil after Amendment with Composted Almond Shells. Res. Microbiol. 2017, 168, 583–593. [Google Scholar] [CrossRef] [PubMed]
- Raza, W.; Ling, N.; Liu, D.; Wei, Z.; Huang, Q.; Shen, Q. Volatile Organic Compounds Produced by Pseudomonas Fluorescens WR-1 Restrict the Growth and Virulence Traits of Ralstonia solanacearum. Microbiol. Res. 2016, 192, 103–113. [Google Scholar] [CrossRef]
- Enespa; Chandra, P. Microbial Volatiles as Chemical Weapons Against Pathogenic Fungi. In Volatiles and Food Security; Choudhary, D.K., Sharma, A.K., Agarwal, P., Varma, A., Tuteja, N., Eds.; Springer: Singapore, 2017; pp. 227–254. ISBN 978-981-10-5552-2. [Google Scholar]
- Etschmann, M.; Bluemke, W.; Sell, D.; Schrader, J. Biotechnological Production of 2-Phenylethanol. Appl. Microbiol. Biotechnol. 2002, 59, 1–8. [Google Scholar]
- Cho, K.M.; Hong, S.Y.; Lee, S.M.; Kim, Y.H.; Kahng, G.G.; Lim, Y.P.; Kim, H.; Yun, H.D. Endophytic Bacterial Communities in Ginseng and Their Antifungal Activity Against Pathogens. Microb. Ecol. 2007, 54, 341–351. [Google Scholar] [CrossRef]
- Collazo, C.; Abadias, M.; Aguiló-Aguayo, I.; Alegre, I.; Chenoll, E.; Viñas, I. Studies on the Biocontrol Mechanisms of Pseudomonas graminis Strain CPA-7 against Food-Borne Pathogens in Vitro and on Fresh-Cut Melon. LWT Food Sci. Technol. 2017, 85, 301–308. [Google Scholar] [CrossRef] [Green Version]
- Han, J.; Sun, L.; Dong, X.; Cai, Z.; Sun, X.; Yang, H.; Wang, Y.; Song, W. Characterization of a Novel Plant Growth-Promoting Bacteria Strain Delftia tsuruhatensis HR4 Both as a Diazotroph and a Potential Biocontrol Agent against Various Plant Pathogens. Syst. Appl. Microbiol. 2005, 28, 66–76. [Google Scholar] [CrossRef]
- Sahu, K.P.; Kumar, A.; Patel, A.; Kumar, M.; Gopalakrishnan, S.; Prakash, G.; Rathour, R.; Gogoi, R. Rice Blast Lesions: An Unexplored Phyllosphere Microhabitat for Novel Antagonistic Bacterial Species against Magnaporthe oryzae. Microb. Ecol. 2021, 81, 731–745. [Google Scholar] [CrossRef]
- Szentes, S.; Radu, G.-L.; Laslo, É.; Lányi, S.; Mara, G. Selection and Evaluation of Potential Biocontrol Rhizobacteria from a Raised Bog Environment. Crop. Prot. 2013, 52, 116–124. [Google Scholar] [CrossRef]
- Fiedler, S.; Heerklotz, H. Vesicle Leakage Reflects the Target Selectivity of Antimicrobial Lipopeptides from Bacillus subtilis. Biophys. J. 2015, 109, 2079–2089. [Google Scholar] [CrossRef] [Green Version]
- Pandin, C.; Le Coq, D.; Deschamps, J.; Védie, R.; Rousseau, T.; Aymerich, S.; Briandet, R. Complete Genome Sequence of Bacillus velezensis QST713: A Biocontrol Agent That Protects Agaricus Bisporus Crops against the Green Mould Disease. J. Biotechnol. 2018, 278, 10–19. [Google Scholar] [CrossRef]
- Grady, E.N.; MacDonald, J.; Liu, L.; Richman, A.; Yuan, Z.-C. Current Knowledge and Perspectives of Paenibacillus: A Review. Microb. Cell Factories 2016, 15, 203. [Google Scholar] [CrossRef] [Green Version]
- Rojas-Rojas, F.U.; Salazar-Gómez, A.; Vargas-Díaz, M.E.; Vásquez-Murrieta, M.S.; Hirsch, A.M.; De Mot, R.; Ghequire, M.G.K.; Ibarra, J.A.; Estrada-de los Santos, P. Broad-Spectrum Antimicrobial Activity by Burkholderia cenocepacia TAtl-371, a Strain Isolated from the Tomato Rhizosphere. Microbiology 2018, 164, 1072–1086. [Google Scholar] [CrossRef]
- Korenblum, E.; von Der Weid, I.; Santos, A.L.S.; Rosado, A.S.; Sebastián, G.V.; Coutinho, C.; Magalhaes, F.C.M.; De Paiva, M.M.; Seldin, L. Production of Antimicrobial Substances by Bacillus Subtilis LFE-1, B. Firmus H2O-1 and B. Licheniformis T6-5 Isolated from an Oil Reservoir in Brazil. J. Appl. Microbiol. 2005, 98, 667–675. [Google Scholar] [CrossRef]
- Von der Weid, I.; Alviano, D.S.; Santos, A.L.S.; Soares, R.M.A.; Alviano, C.S.; Seldin, L. Antimicrobial Activity of Paenibacillus peoriae Strain NRRL BD-62 against a Broad Spectrum of Phytopathogenic Bacteria and Fungi. J. Appl. Microbiol. 2003, 95, 1143–1151. [Google Scholar] [CrossRef]
- Compant, S.; Duffy, B.; Nowak, J.; Clément, C.; Barka, E.A. Use of Plant Growth-Promoting Bacteria for Biocontrol of Plant Diseases: Principles, Mechanisms of Action, and Future Prospects. Appl. Environ. Microbiol. 2005, 71, 4951–4959. [Google Scholar] [CrossRef] [Green Version]
- Poppe, L.; Vanhoutte, S.; Höfte, M. Modes of Action of Pantoea agglomerans CPA-2, an Antagonist of Postharvest Pathogens on Fruits. Eur. J. Plant Pathol. 2003, 109, 963–973. [Google Scholar] [CrossRef]
- Köhl, J.; Kolnaar, R.; Ravensberg, W.J. Mode of Action of Microbial Biological Control Agents Against Plant Diseases: Relevance Beyond Efficacy. Front. Plant Sci. 2019, 10, 845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).