Ectopic Expression of Kenaf (Hibiscus cannabinus L.) HcWRKY50 Improves Plants’ Tolerance to Drought Stress and Regulates ABA Signaling in Arabidopsis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. RNA Isolation and qRT-PCR
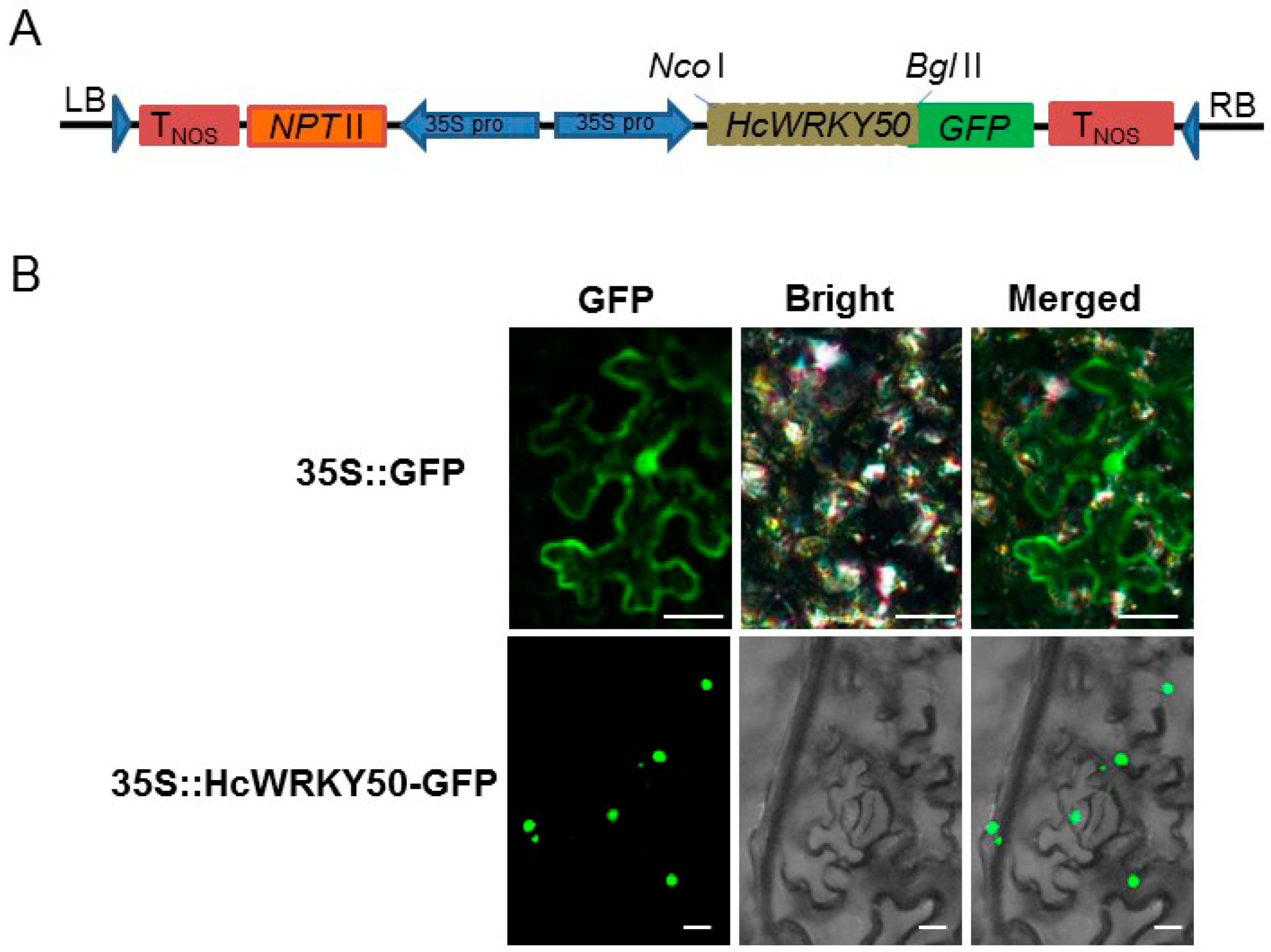
2.3. HcWRKY50 Sequence Cloning and Subcellular Localization
2.4. Plant Transformation and Stress Treatments
2.5. MDA Content, Electrolyte Leakage, and Water Loss Rate
2.6. Stomatal Conductance and Density
2.7. Statistical Analysis
3. Results
3.1. Identification and Sequence Characteristic of HcWRKY50 Gene
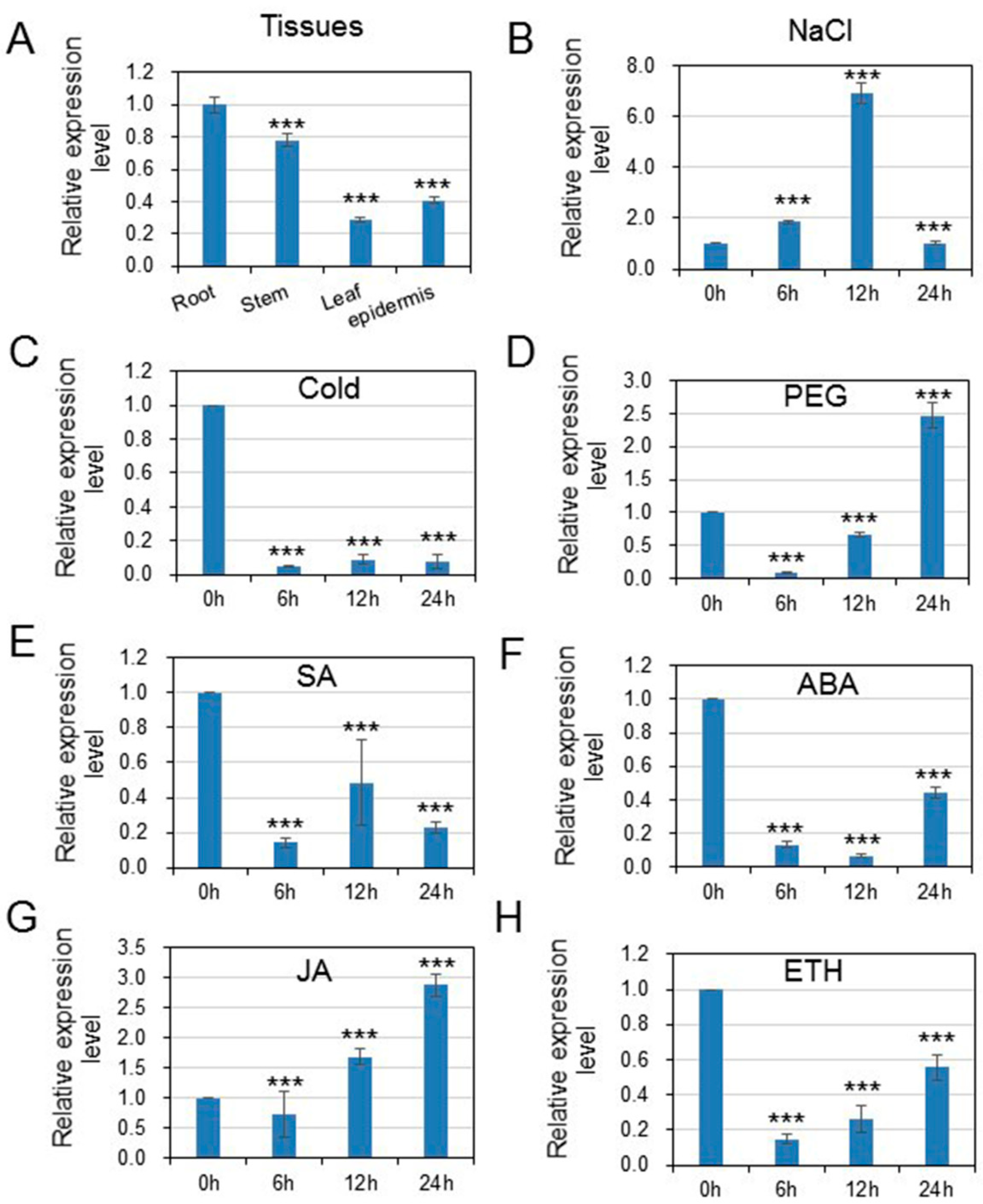
3.2. HcWRKY50 Was Differentially Expressed in Tissues and Induced by Different Stresses
3.3. HcWRKY50 Protein Localizes in Cell Nuclei
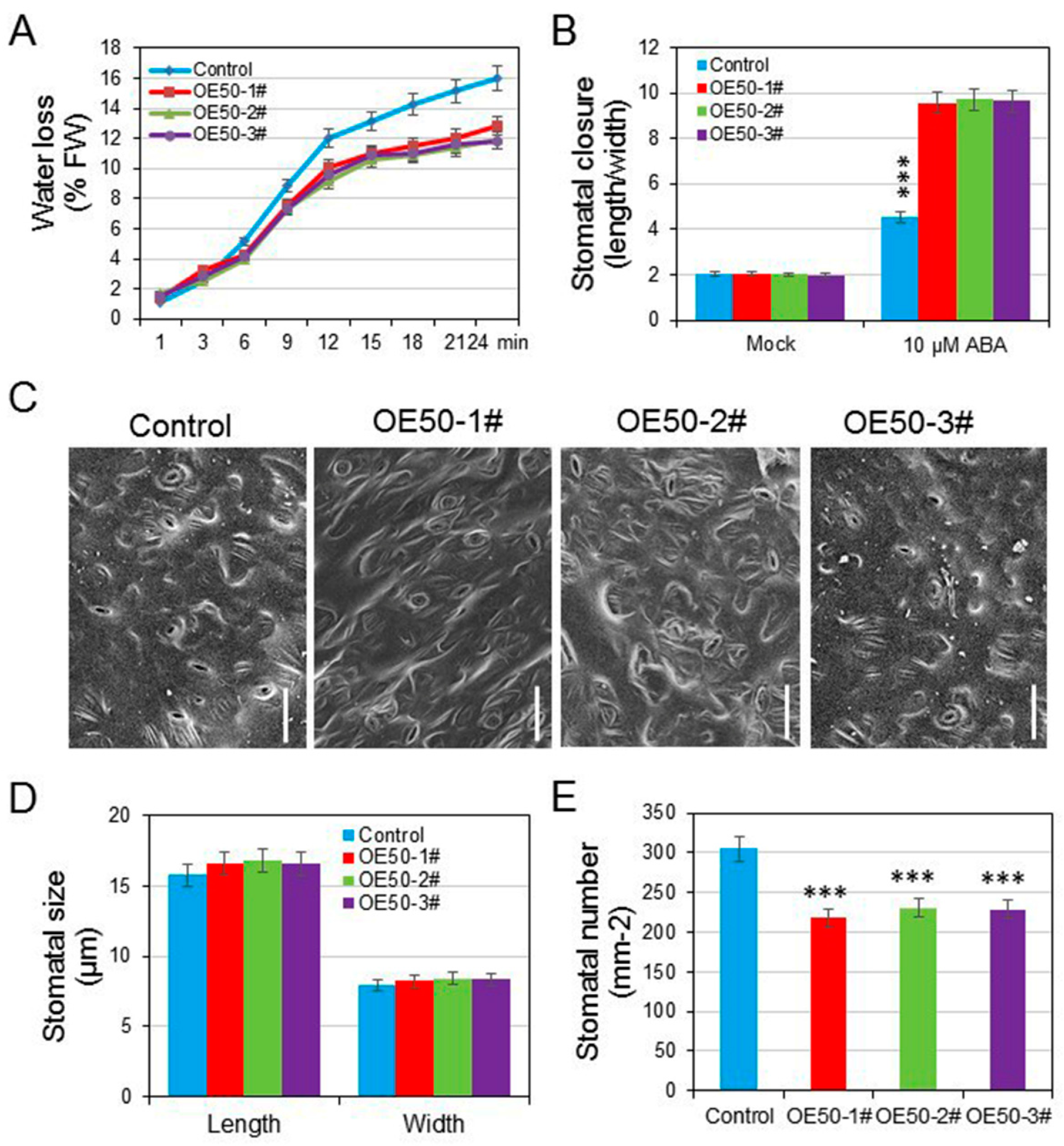
3.4. HcWRKY50 Overexpression Plants Had Decreased Sensitivity to ABA
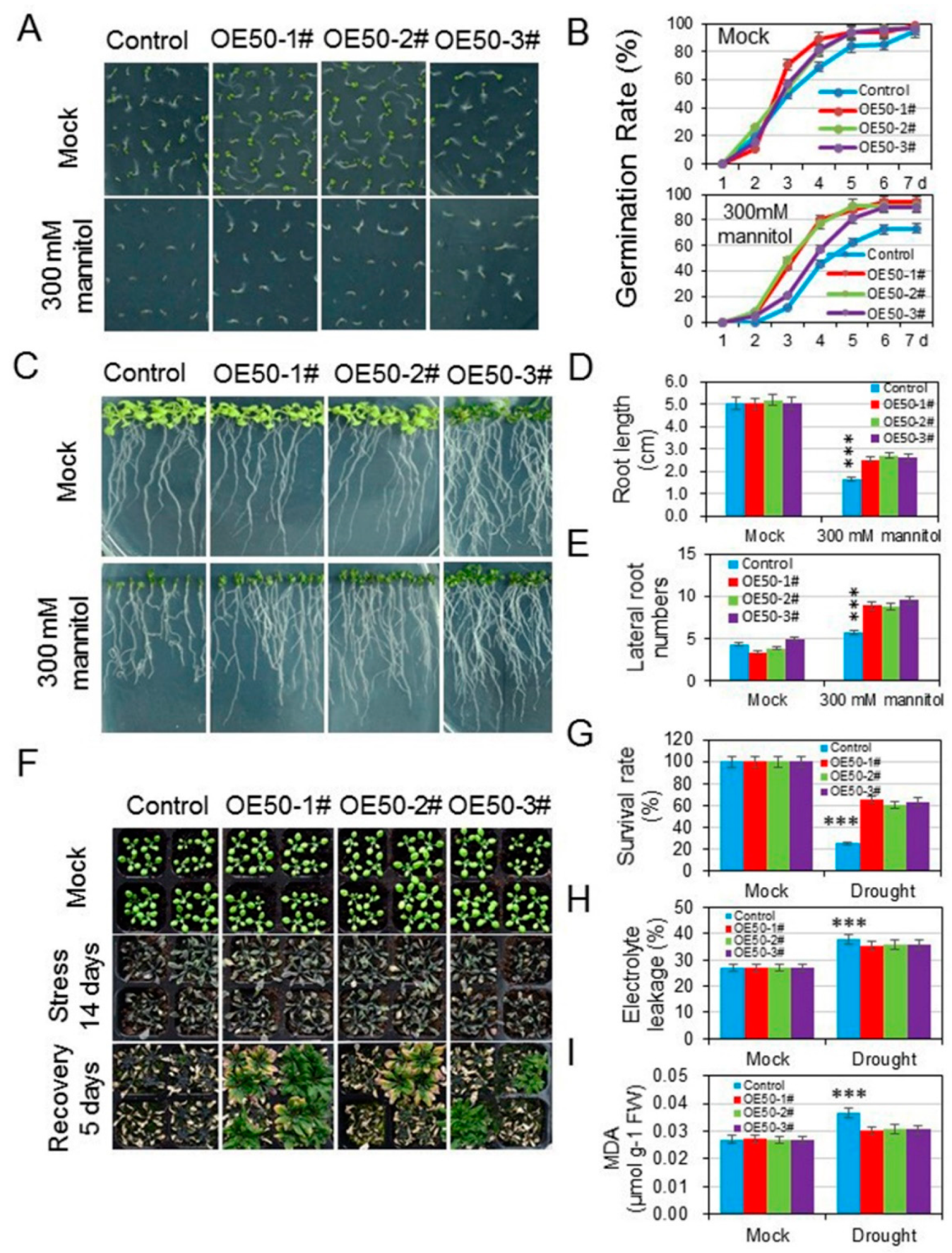
3.5. HcWRKY50 Overexpression Plants Increased the Tolerance to Drought Stress
3.6. HcWRKY50 Regulated ABA-Induced Stomatal Closure and Decreased the Stomatal Density
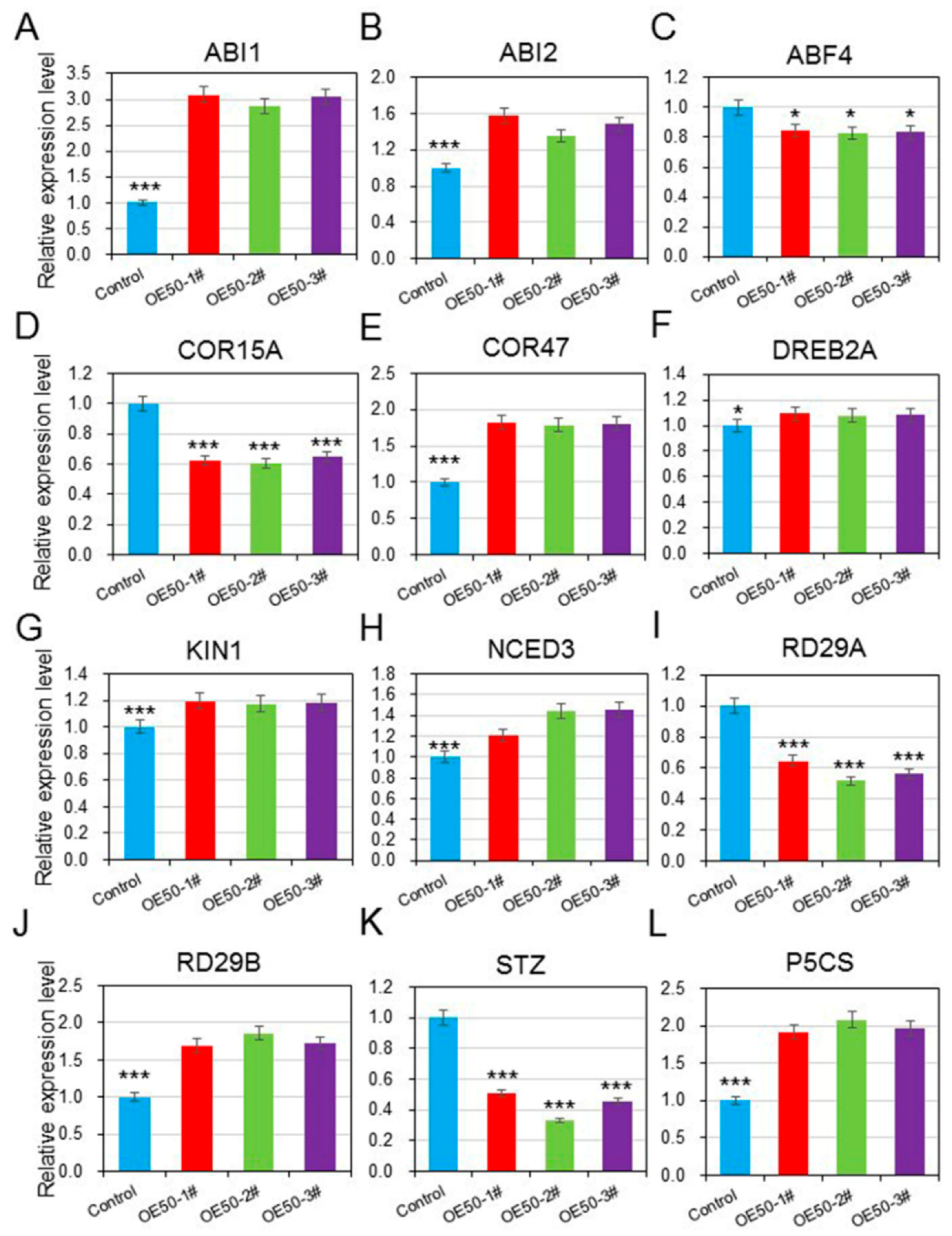
3.7. HcWRKY50 Positively Regulated the Expression of ABA- and Stress-Responsive Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, H.; Lai, L.; Li, L.; Liu, L.; Jakada, B.H.; Huang, Y.; He, Q.; Chai, M.; Niu, X.; Qin, Y. AcoMYB4, an Ananas comosus L. MYB Transcription Factor, Functions in Osmotic Stress through Negative Regulation of ABA Signaling. Int. J. Mol. Sci. 2020, 21, 5727. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dang, F.F.; Wang, Y.N.; Yu, L.; Eulgem, T.; Lai, Y.; Liu, Z.Q.; Wang, X.; Qiu, A.L.; Zhang, T.X.; Lin, J.; et al. CaWRKY40, a WRKY protein of pepper, plays an important role in the regulation of tolerance to heat stress and resistance to Ralstonia solanacearum infection. Plant Cell Environ. 2013, 36, 757–774. [Google Scholar] [CrossRef] [PubMed]
- Bakshi, M.; Oelmuller, R. WRKY transcription factors: Jack of many trades in plants. Plant Signal Behav. 2014, 9, e27700. [Google Scholar] [CrossRef] [Green Version]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef]
- Wani, S.H.; Anand, S.; Singh, B.; Bohra, A.; Joshi, R. WRKY transcription factors and plant defense responses: Latest discoveries and future prospects. Plant Cell Rep. 2021, 40, 1071–1085. [Google Scholar] [CrossRef]
- Jiang, Y.; Liang, G.; Yu, D. Activated expression of WRKY57 confers drought tolerance in Arabidopsis. Mol. Plant. 2012, 5, 1375–1388. [Google Scholar] [CrossRef] [Green Version]
- Ren, X.; Chen, Z.; Liu, Y.; Zhang, H.; Zhang, M.; Liu, Q.; Hong, X.; Zhu, J.K.; Gong, Z. ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J. 2010, 63, 417–429. [Google Scholar] [CrossRef] [Green Version]
- Xie, Z.; Zhang, Z.L.; Zou, X.; Huang, J.; Ruas, P.; Thompson, D.; Shen, Q.J. Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol. 2005, 137, 176–189. [Google Scholar] [CrossRef] [Green Version]
- Niu, C.F.; Wei, W.; Zhou, Q.Y.; Tian, A.G.; Hao, Y.J.; Zhang, W.K.; Ma, B.; Lin, Q.; Zhang, Z.B.; Zhang, J.S.; et al. Wheat WRKY genes TaWRKY2 and TaWRKY19 regulate abiotic stress tolerance in transgenic Arabidopsis plants. Plant Cell Environ. 2012, 35, 1156–1170. [Google Scholar] [CrossRef]
- Zou, X.; Seemann, J.R.; Neuman, D.; Shen, Q.J. A WRKY gene from creosote bush encodes an activator of the abscisic acid signaling pathway. J. Biol. Chem. 2004, 279, 55770–55779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, X.; Bai, X.; Sun, X.; Zhu, D.; Liu, B.; Ji, W.; Cai, H.; Cao, L.; Wu, J.; Hu, M.; et al. Expression of wild soybean WRKY20 in Arabidopsis enhances drought tolerance and regulates ABA signalling. J. Exp. Bot. 2013, 64, 2155–2169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic acid: Emergence of a core signaling network. Annu Rev. Plant. Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rushton, D.L.; Tripathi, P.; Rabara, R.C.; Lin, J.; Ringler, P.; Boken, A.K.; Langum, T.J.; Smidt, L.; Boomsma, D.D.; Emme, N.J.; et al. WRKY transcription factors: Key components in abscisic acid signalling. Plant Biotechnol. J. 2012, 10, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Sim, Y.Y.; Nyam, K.L. Hibiscus cannabinus L. (kenaf) studies: Nutritional composition, phytochemistry, pharmacology, and potential applications. Food Chem. 2021, 344, 128582. [Google Scholar] [CrossRef]
- Chen, P.; Chen, T.; Li, Z.; Jia, R.; Luo, D.; Tang, M.; Lu, H.; Hu, Y.; Yue, J.; Huang, Z. Transcriptome analysis revealed key genes and pathways related to cadmium-stress tolerance in Kenaf (Hibiscus cannabinus L.). Ind. Crop Prod. 2020, 158, 112970. [Google Scholar] [CrossRef]
- Niu, X.; Qi, J.; Chen, M.; Zhang, G.; Tao, A.; Fang, P.; Xu, J.; Onyedinma, S.A.; Su, J. Reference genes selection for transcript normalization in kenaf (Hibiscus cannabinus L.) under salinity and drought stress. PeerJ 2015, 3, e1347. [Google Scholar] [CrossRef] [Green Version]
- Danalatos, N.G.; Archontoulis, S.V. Growth and biomass productivity of kenaf (Hibiscus cannabinus L.) under different agricultural inputs and management practices in central Greece. Ind. Crop Prod. 2010, 32, 231–240. [Google Scholar] [CrossRef]
- Ramesh, M. Kenaf (Hibiscus cannabinus L.) fibre based bio-materials: A review on processing and properties. Prog. Mater. Sci. 2016, 78–79, 1–92. [Google Scholar] [CrossRef]
- Niu, X.; Chen, M.; Tao, A.; Qi, J. Salinity and Drought Treatment Assays in Kenaf (Hibiscus cannabinus L.). Bio-Protocol 2016, 6, 1918. [Google Scholar] [CrossRef]
- Zhang, L.; Xu, Y.; Zhang, X.; Ma, X.; Zhang, L.; Liao, Z.; Zhang, Q.; Wan, X.; Cheng, Y.; Zhang, J.; et al. The genome of kenaf (Hibiscus cannabinus L.) provides insights into bast fibre and leaf shape biogenesis. Plant Biotechnol. J. 2020, 18, 1796–1809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bechtold, N.; Pelletier, G. In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 1998, 82, 259–266. [Google Scholar] [PubMed]
- Krasensky, J.; Jonak, C. Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J. Exp. Bot. 2012, 63, 1593–1608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, H.; Chen, X.; Hong, Y.Y.; Wang, Y.; Xu, P.; Ke, S.D.; Liu, H.Y.; Zhu, J.K.; Oliver, D.J.; Xiang, C.B. Activated expression of an Arabidopsis HD-START protein confers drought tolerance with improved root system and reduced stomatal density. Plant Cell 2008, 20, 1134–1151. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.Q.; Yan, L.; Wu, Z.; Mei, C.; Lu, K.; Yu, Y.T.; Liang, S.; Zhang, X.F.; Wang, X.F.; Zhang, D.P. Cooperation of three WRKY-domain transcription factors WRKY18, WRKY40, and WRKY60 in repressing two ABA-responsive genes ABI4 and ABI5 in Arabidopsis. J. Exp. Bot. 2012, 63, 6371–6392. [Google Scholar] [CrossRef] [Green Version]
- Shang, Y.; Yan, L.; Liu, Z.Q.; Cao, Z.; Mei, C.; Xin, Q.; Wu, F.Q.; Wang, X.F.; Du, S.Y.; Jiang, T.; et al. The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell 2010, 22, 1909–1935. [Google Scholar] [CrossRef] [Green Version]
- Shen, Y.Y.; Wang, X.F.; Wu, F.Q.; Du, S.Y.; Cao, Z.; Shang, Y.; Wang, X.L.; Peng, C.C.; Yu, X.C.; Zhu, S.Y.; et al. The Mg-chelatase H subunit is an abscisic acid receptor. Nature 2006, 443, 823–826. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Hu, L.; Zhang, S.; Zhang, M.; Jiang, W.; Wu, T.; Du, X. Rice OsWRKY50 Mediates ABA-Dependent Seed Germination and Seedling Growth, and ABA-Independent Salt Stress Tolerance. Int. J. Mol. Sci. 2021, 22, 8625. [Google Scholar] [CrossRef]
- Sakuma, Y.; Maruyama, K.; Osakabe, Y.; Qin, F.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 2006, 18, 1292–1309. [Google Scholar] [CrossRef] [Green Version]
- Jia, H.Y.; Zhang, S.J.; Ruan, M.Y.; Wang, Y.L.; Wang, C.Y. Analysis and application of RD29 genes in abiotic stress response. Acta Physiol. Plant 2012, 34, 1239–1250. [Google Scholar] [CrossRef]
- Gao, Y.F.; Liu, J.K.; Yang, F.M.; Zhang, G.Y.; Wang, D.; Zhang, L.; Ou, Y.B.; Yao, Y.A. The WRKY transcription factor WRKY8 promotes resistance to pathogen infection and mediates drought and salt stress tolerance in Solanum lycopersicum. Physiol. Plantarum 2020, 168, 98–117. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R.; Kim, Y.; Song, L.; Coutu, J.; Coutu, A.; Ciftci-Yilmaz, S.; Lee, H.; Stevenson, B.; Zhu, J.K. Gain- and loss-of-function mutations in Zat10 enhance the tolerance of plants to abiotic stress. FEBS Lett. 2006, 580, 6537–6542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Q.Y.; Tian, A.G.; Zou, H.F.; Xie, Z.M.; Lei, G.; Huang, J.; Wang, C.M.; Wang, H.W.; Zhang, J.S.; Chen, S.Y. Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol. J. 2008, 6, 486–503. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niu, X.; Chen, M.; She, Z.; Aslam, M.; Qi, J.; Qin, Y. Ectopic Expression of Kenaf (Hibiscus cannabinus L.) HcWRKY50 Improves Plants’ Tolerance to Drought Stress and Regulates ABA Signaling in Arabidopsis. Agronomy 2022, 12, 1176. https://doi.org/10.3390/agronomy12051176
Niu X, Chen M, She Z, Aslam M, Qi J, Qin Y. Ectopic Expression of Kenaf (Hibiscus cannabinus L.) HcWRKY50 Improves Plants’ Tolerance to Drought Stress and Regulates ABA Signaling in Arabidopsis. Agronomy. 2022; 12(5):1176. https://doi.org/10.3390/agronomy12051176
Chicago/Turabian StyleNiu, Xiaoping, Meixia Chen, Zeyuan She, Mohammad Aslam, Jianmin Qi, and Yuan Qin. 2022. "Ectopic Expression of Kenaf (Hibiscus cannabinus L.) HcWRKY50 Improves Plants’ Tolerance to Drought Stress and Regulates ABA Signaling in Arabidopsis" Agronomy 12, no. 5: 1176. https://doi.org/10.3390/agronomy12051176
APA StyleNiu, X., Chen, M., She, Z., Aslam, M., Qi, J., & Qin, Y. (2022). Ectopic Expression of Kenaf (Hibiscus cannabinus L.) HcWRKY50 Improves Plants’ Tolerance to Drought Stress and Regulates ABA Signaling in Arabidopsis. Agronomy, 12(5), 1176. https://doi.org/10.3390/agronomy12051176






