Transcriptomic and Metabolomic Analysis Provides Insights into the Fruit Quality and Yield Improvement in Tomato under Soilless Substrate-Based Cultivation
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Production System
2.2. Experimental Design
2.3. Measurement of Biological and Physiological Growth Parameters
2.4. Tomato Fruit Quality Measurement
2.5. Differential Gene and Metabolites Screening
2.6. Quantitative PCR Verification
3. Results
3.1. The Effects of Different Treatments on Tomato Growth, Yield and Quality
3.2. Transcriptional Analysis of Tomato Fruits under Substrate-Based and Soil Cultivation Conditions
3.2.1. Pre-Processing of Transcriptomics Data
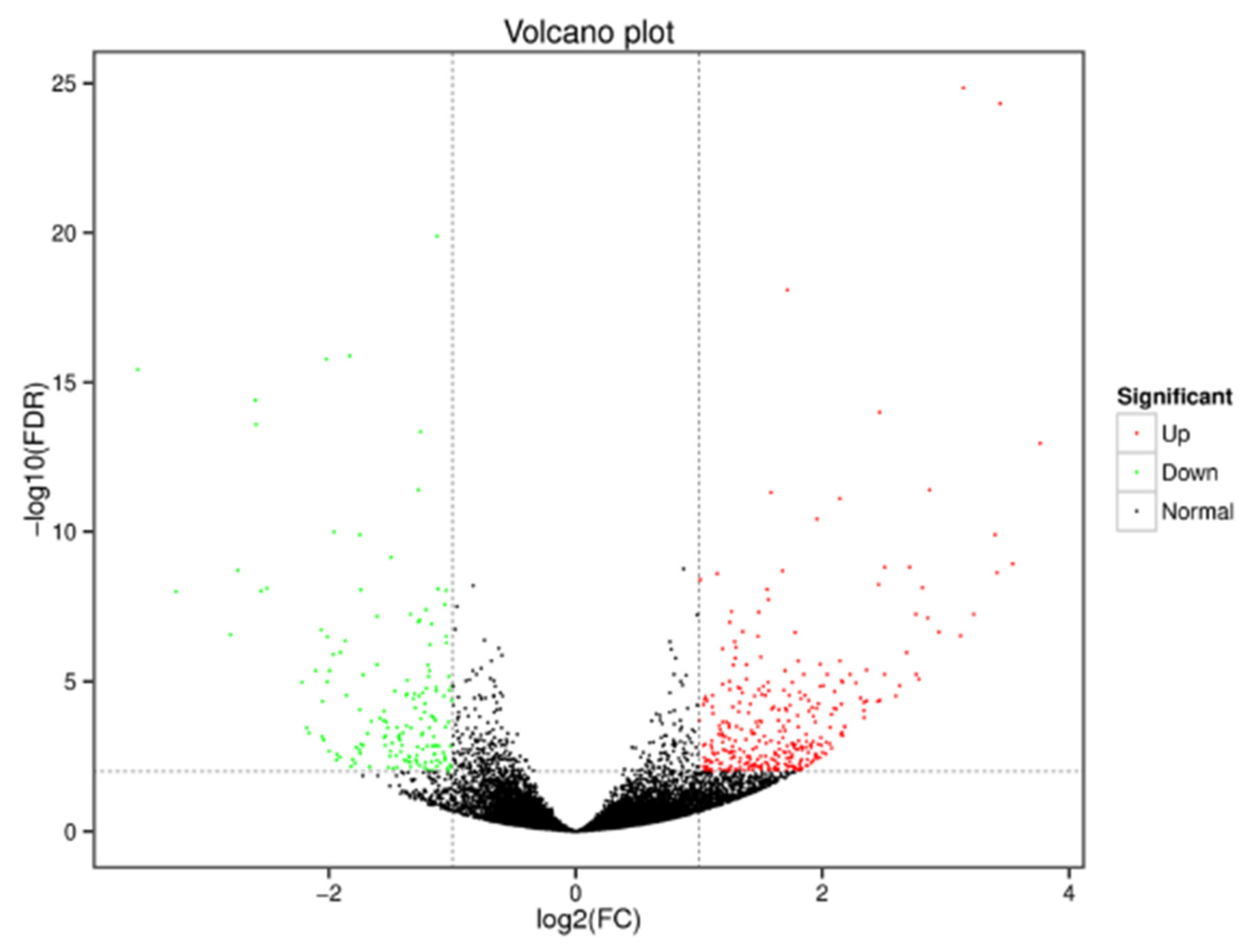
3.2.2. Identification of Differentially Expressed Genes in Tomato Fruits under Substrate-Based and Soil Cultivation Conditions
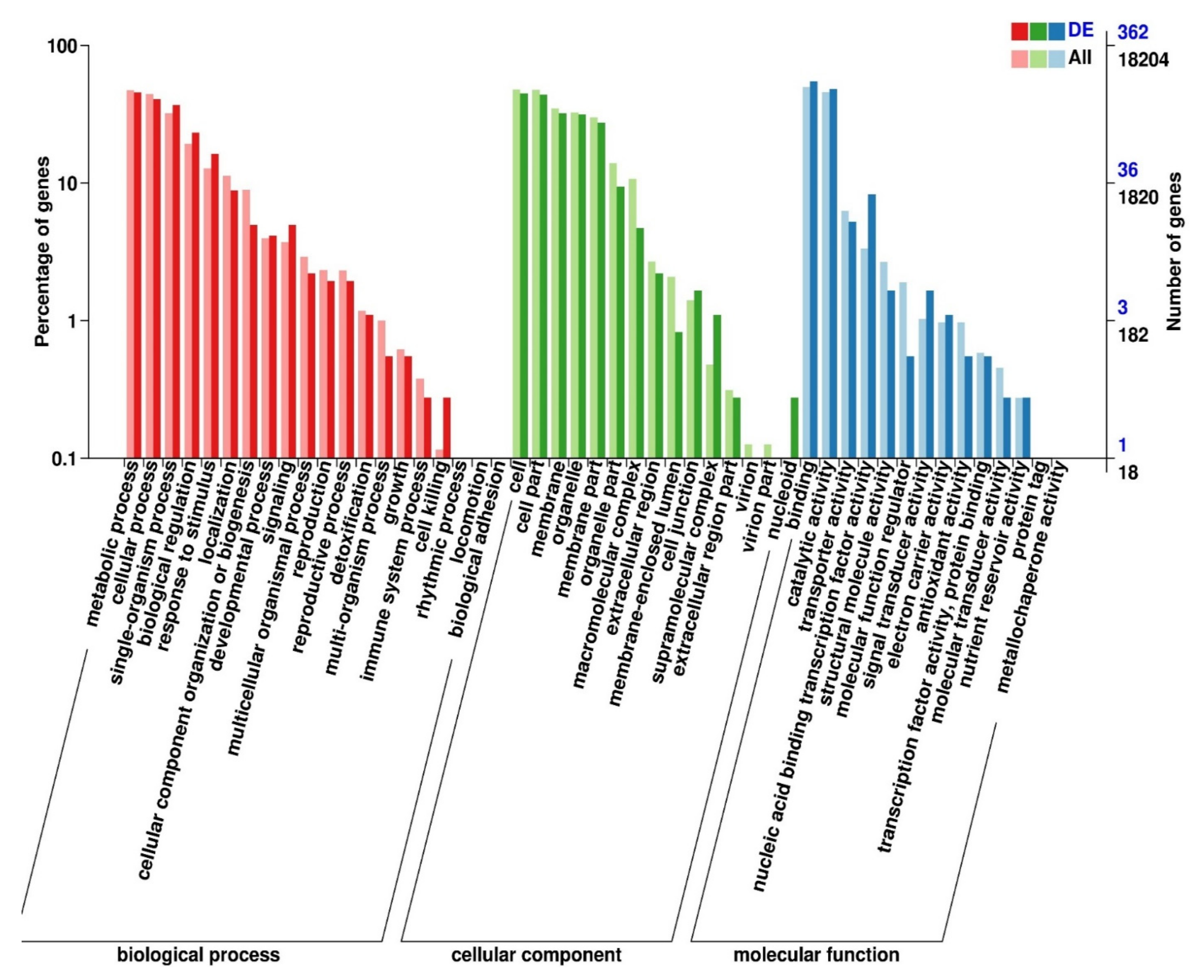
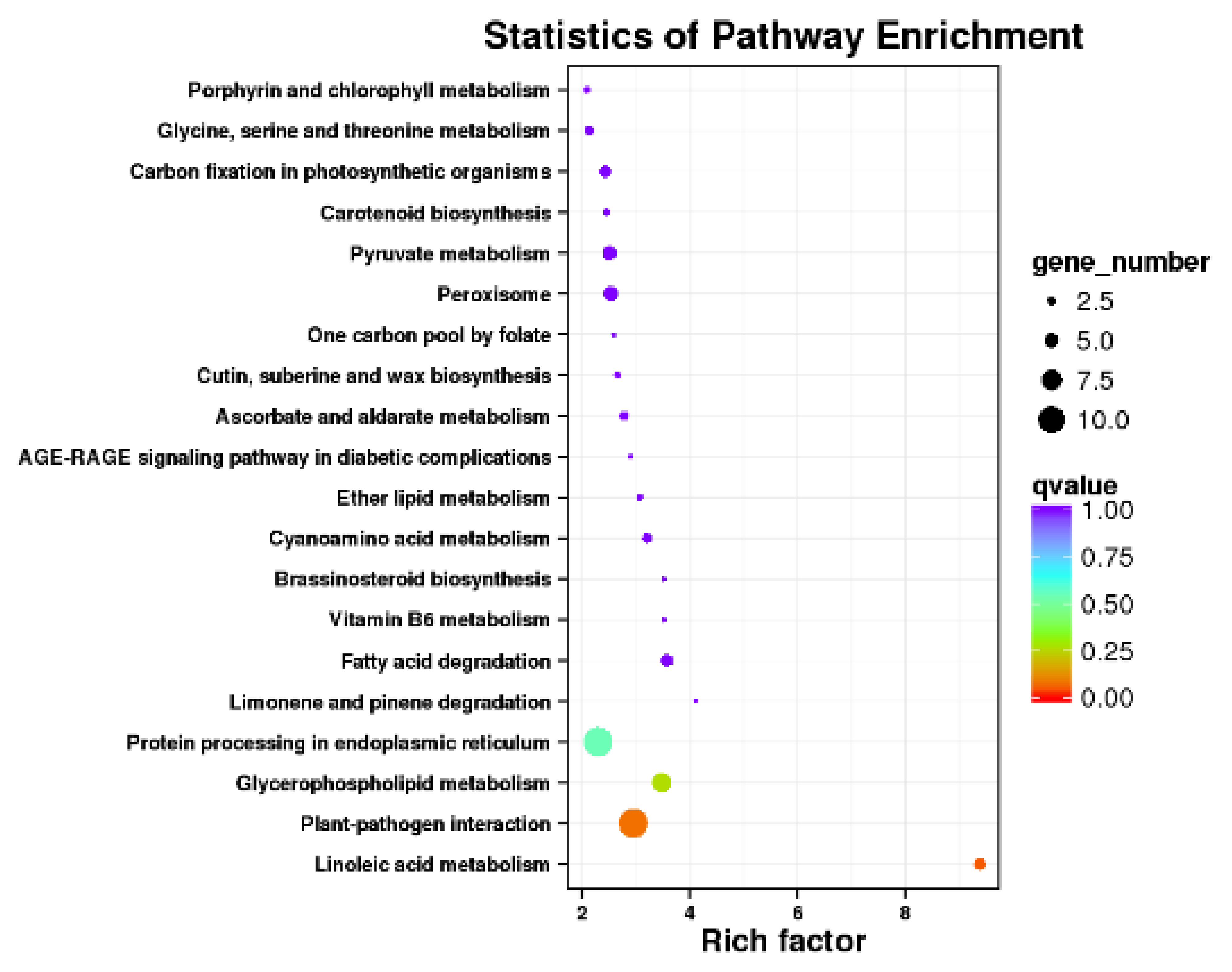
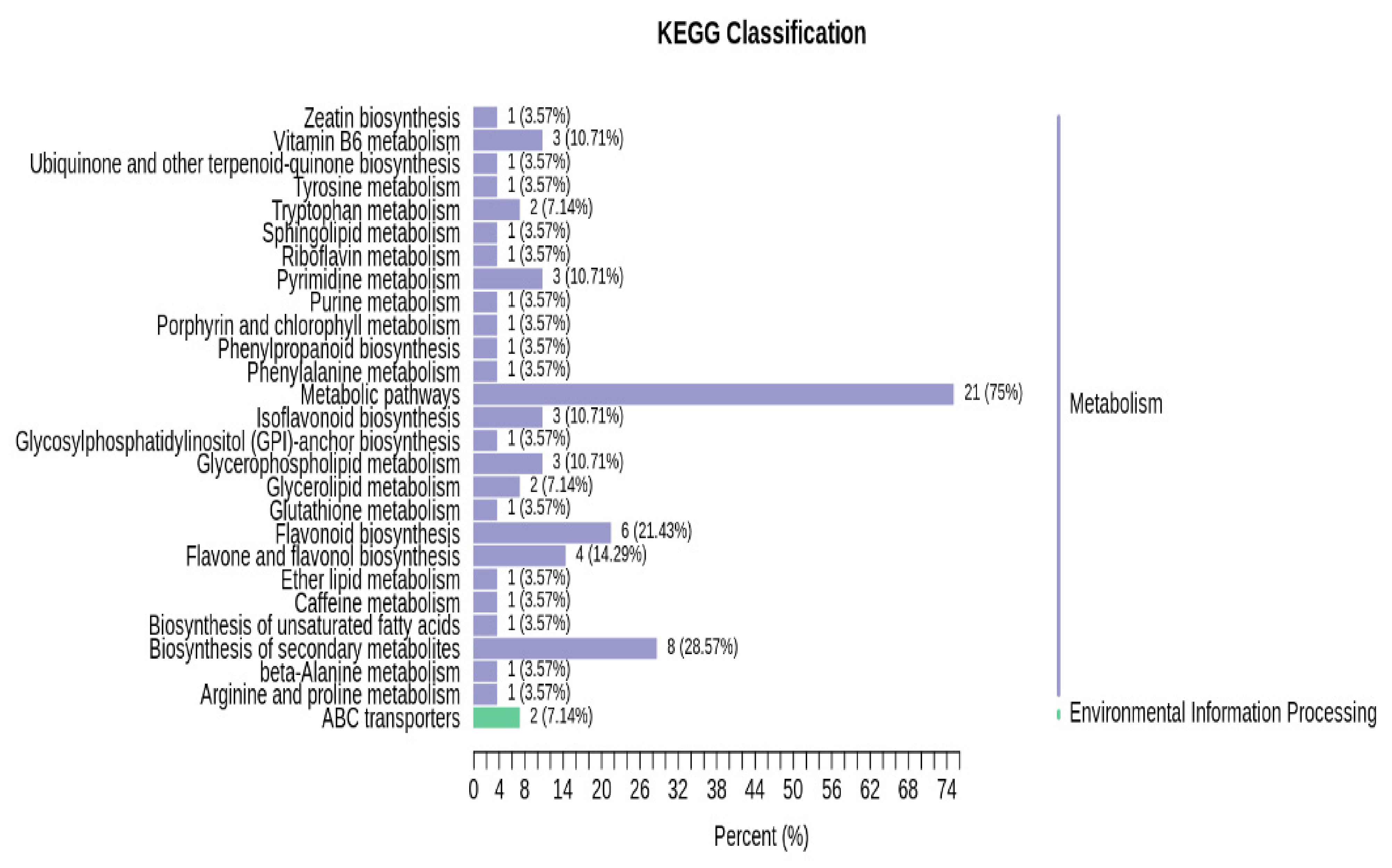
3.2.3. Functional Annotation of Significantly Expressed Genes
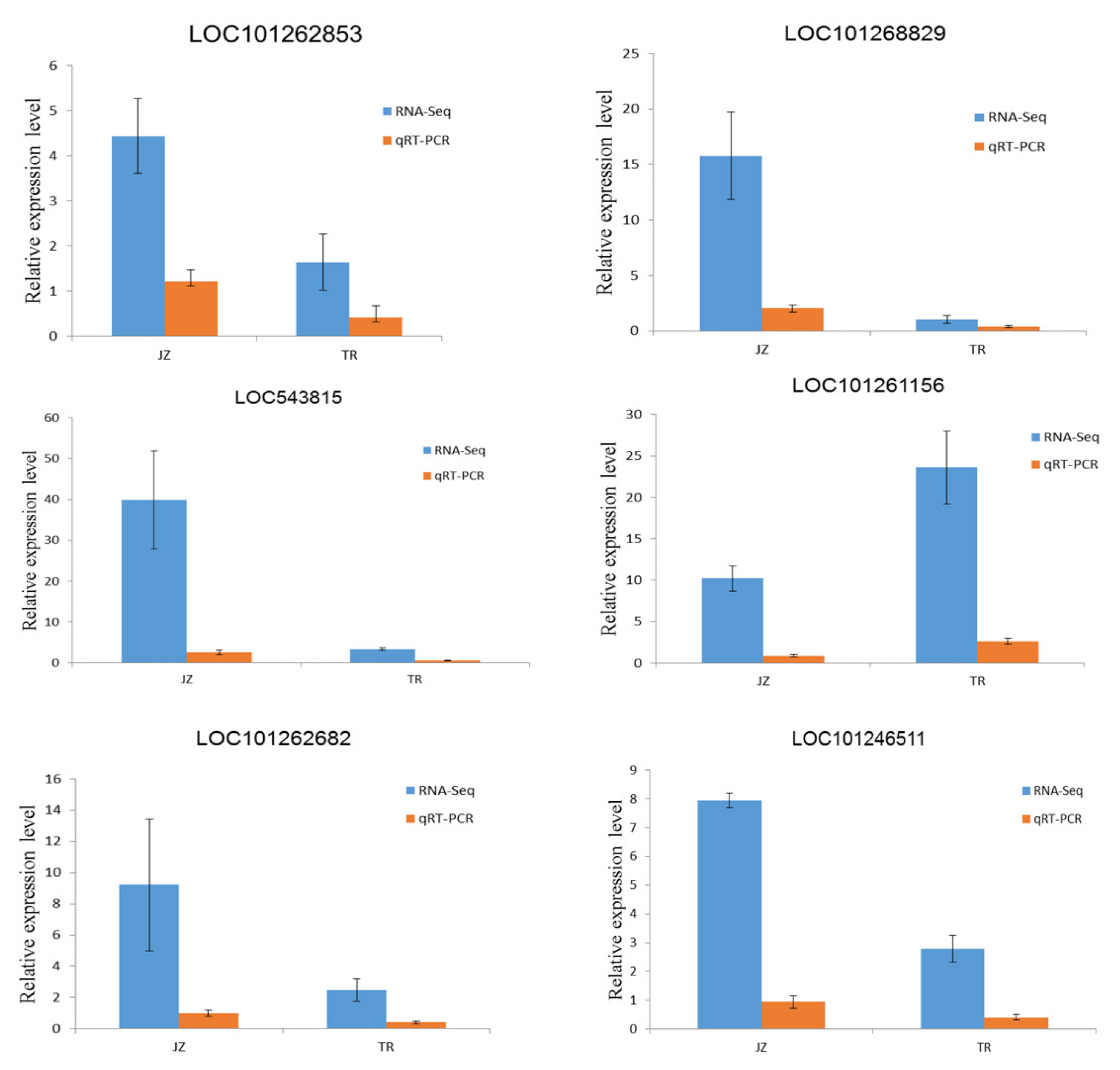
3.2.4. qRT-PCR Assay for Verification of Differentially Expressed Genes
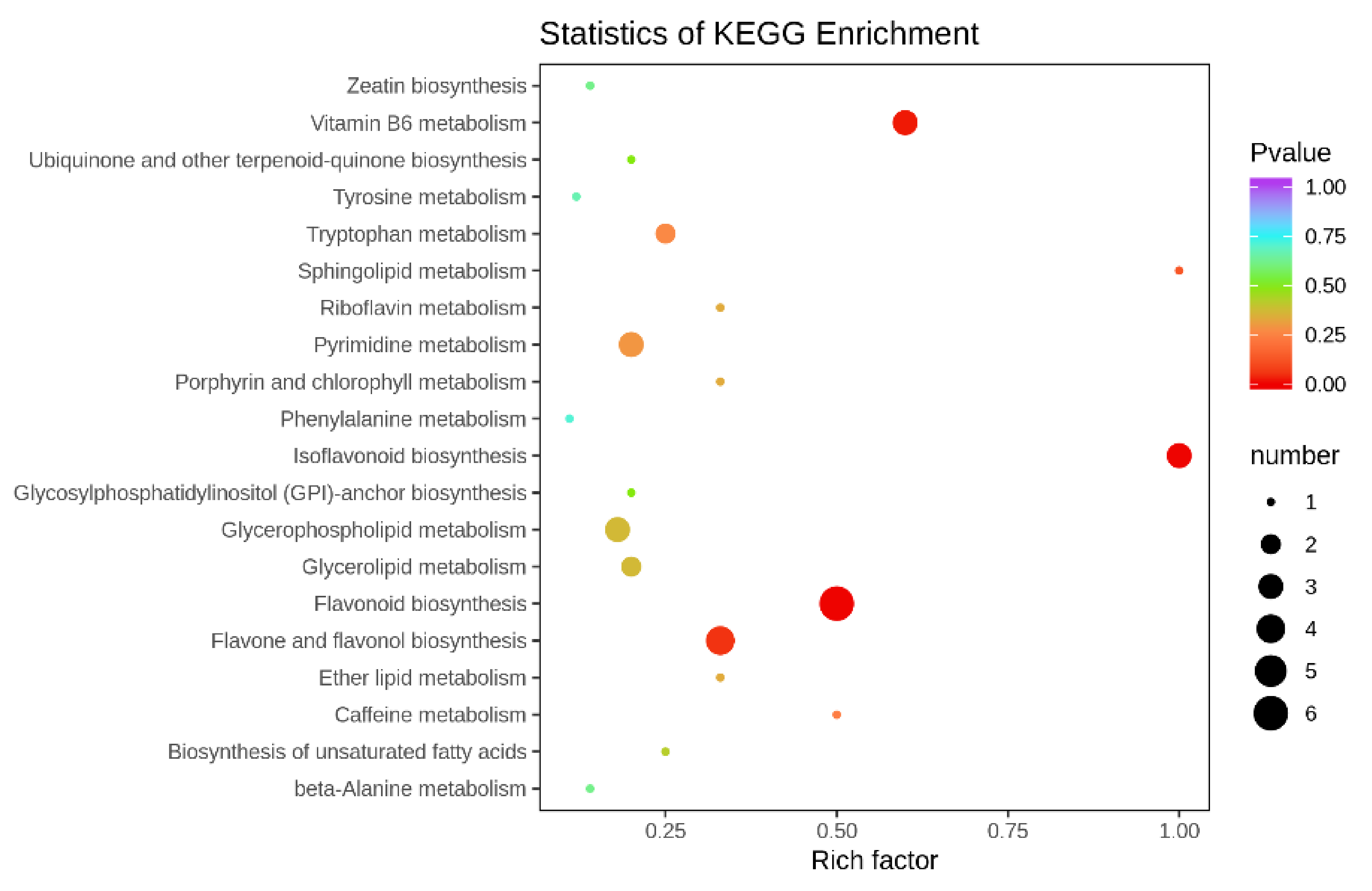
3.3. Screening Results of Different Metabolites in Tomato Fruits under Different Cultivation Conditions
3.3.1. Principal Component Analysis of Differential Metabolites
3.3.2. Identification of Differentially Available Metabolites in Tomato Fruits
3.4. Combined Metabolomics and Transcriptomics Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xu, S.; Li, J.; Ma, X.; Gao, Y.L. Effects of leaf spraying stevia on growth and quality of cherry tomato. North. Hortic. 2015, 18, 48–51. [Google Scholar]
- Perveen, R.; Suleria, H.; Anjum, F.; Butt, M.; Pasha, I.; Ahmad, S. Tomato (Solanum lycopersicum) carotenoids and lycopenes chemistry; metabolism, absorption, nutrition, and allied health claims—A comprehensive review. Crit. Rev. Food Sci. Nutr. 2015, 55, 919–929. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Li, J.; Gao, Y. Effects of root restriction on growth and quality of cherry tomato in solar greenhouse. J. Northwest Agric. Sci. 2014, 23, 131–137. [Google Scholar]
- Zhong, C. Nutrition composition and health care function of tomato. Inner Mongolia Sci. Technol. Econ. 2011, 22, 105–107. [Google Scholar]
- Zhang, C.; Song, S.; Zhao, C.; Wen, T.; Sun, K. Analysis and evaluation of nutritional quality of different varieties of tomato. China Veg. 2011, 18, 68–73. [Google Scholar]
- Yang, F.; An, Z.; Yang, W. Effects of tomato continuous cropping on soil microbial and physicochemical properties in solar greenhouse. Chin. Soil Fertilizer Sci. 2016, 1, 42–46. [Google Scholar]
- Lian, Y.; Lin, X.; Yamada, S.; Inoue, M.; Inosako, K. Soil degradation and prevention in greenhouse production. Springerplus 2013, 2, 1–5. [Google Scholar]
- Shi, L.; Dong, L. Practical Technology of Soilless Cultivation of Vegetable Production; Hebei Science and Technology: Shijiazhuang, China, 2021; p. 178. [Google Scholar]
- Li, P.; Zhu, Y. Advances in the development and application of plant culture substrate based on agricultural and forestry wastes. J. Nanjing For. Univ. 2015, 39, 161–168. [Google Scholar]
- Li, T.; Ma, R.; Cheng, Y.; Wu, H.; Jiao, Y.; Qiao, N. Research progress of vegetable substrate-based cultivation in China. Acta Agric. Sin. 2013, 3, 30–34. [Google Scholar]
- Mao, B.; He, M.; Chen, L.; Chen, Z. Effects of mushroom bran complex substrate-based on nutrient growth and photosynthesis of tomato. J. Nuclear Agric. Sci. 2015, 29, 1821–1827. [Google Scholar]
- Li, M.; Du, J.; Shu, S.; Guo, S. Study on the matrix formula of cherry tomato vinegar grains. J. Shenyang Agric. Univ. 2015, 46, 19–25. (In Chinese) [Google Scholar]
- Cai, D.; Li, J.; Fan, X.; Li, H.; Kong, Z.; Zhang, X. Effects of nutrient solution nitrogen, phosphorus and potassium supplementation on nutrient absorption and yield and quality of tomato. J. Northeast Agric. Univ. 2017, 48, 7–14. [Google Scholar]
- Sunera, A.; Saqib, S.; Uddin, S.; Zaman, W.; Ullah, F.; Ayaz, A.; Asghar, M.; Rehman, S.; Munis, M.; Chaudhary, H. Characterization and phytostimulatory activity of bacteria isolated from tomato (Lycopersicon esculentum Mill.) rhizosphere. Microb. Pathog. 2020, 140, 103966. [Google Scholar] [CrossRef]
- Yang, J.; Wang, C.; Shi, R.; Yu, D.; Gao, M. Effects of substrate-based culture on yield and quality of tomato: A meta-analysis. China Melons Veg. 2011, 34, 47–53. [Google Scholar]
- Wang, P.; Wang, J.; Liu, S.; Yao, T.; Li, W.; Wang, J.; Zhang, J. Effects of Different organic matrix ratio on growth, yield and fruit quality of tomato. Jiangsu Agric. Sci. 2016, 44, 211–213. [Google Scholar]
- Zou, Q. Experimental Guidance of Plant Physiology; Agriculture Press: Beijing, China, 2000; pp. 62–63. [Google Scholar]
- Li, H.; Sun, Q.; Zhao, S. Experimental Principles and Techniques of Plant Physiology and Biochemistry; Higher Education Press: Beijing, China, 2000; pp. 123–248. [Google Scholar]
- Li, W.; Feng, X.; Wang, B. Determination of titratable acid in fruits with automatic potentiometric titrator. Food Sci. 2009, 30, 247–249. [Google Scholar]
- Love, M.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Yang, W.; Hu, X. Research progress of effect of nitrogen fertilizer on Vc and nitrate content in vegetables. J. Hui Agric. Sci. 2006, 34, 5924–5925. [Google Scholar]
- Chen, Z.; Feng, D. Changes of nitrate and nitrite contents and their chemical regulation of leafy vegetables. Chin. Bull. Bot. 1994, 11, 25–26. [Google Scholar]
- Zheng, N.; Ma, J.; Wang, X.; Ye, Z.; Li, T.; Mao, Q.; Wu, X. Effects of combined application of bacterial residue and chemical fertilizers on photosynthetic performance and yield of rice flag leaves. J. Zhejiang Agric. 2013, 25, 603–608. [Google Scholar]
- Guan, Y.; Lin, B.; Ling, B. Effects of light, nitrogen and their interaction on photosynthesis and carbon and nitrogen metabolism of maize seedling leaves. Acta Agron. Sin. 2000, 26, 806–812. [Google Scholar]
- Han, Q.M. Height-related decreases in mesophyll conductance, leaf photosynthesis and compensating adjustments associated with leaf nitrogen concentrations in Pinus densiflora. Tree Physiol. 2011, 31, 976–984. [Google Scholar] [CrossRef] [PubMed]
- Lebaudy, A.; Vavasseur, A.; Hosy, E.; Dreyer, I.; Leonhardt, N.; Thibaud, J.B.; Véry, A.A.; Simonneau, T.; Sentenac, H. Plant adaptation to fluctuating environment and biomass production are strongly dependent on guard cell potassium channels. Proc. Natl. Acad. Sci. USA 2008, 105, 5271–5276. [Google Scholar] [CrossRef]
- Zhang, E.; Zhang, S.; Li, T.; Ge, X. Current status and prospects of vegetable potassium nutrition research. Chin. Agric. Sci. Bull. 2005, 8, 265–268. [Google Scholar] [CrossRef]
- Chen, H.; Tang, Y.; Pan, Y.; Guo, X. Research progress on the metabolic basis and regulation mechanism of fruit flavor. Bull. Bot. Res. 2021, 41, 474–480. [Google Scholar]
- Cao, S.; Zhang, C.; Tang, Y.; Qi, H. Characteristics of plant lipoxygenase protein and its role in fruit ripening and senescence and adversity stress. Acta Phytophysiol. 2014, 50, 1096–1108. [Google Scholar]
- Tangy, F.; Zhang, C.; Cao, S.X.; Wang, X.; Qi, H. The effect of CmLOXs on the production of volatile organic compounds in four aroma types of melon (Cucumis melo). PLoS ONE 2015, 10, e0143567. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Cao, S.; Jin, Y.; Ju, L.; Chen, Q.; Xing, Q.; Qi, H. Melon13-lipoxygenase CmLOX18 may be involved in C6 volatiles biosynthesis in fruit. Sci. Rep. 2017, 7, 2816. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Tieman, D.; Jones, J.B.; Taylor, M.G.; Schmelz, E.; Huffaker, A.; Bies, D.; Chen, K.; Klee, H.J. A 13-lipoxygenase, TomloxC, is essential for synthesis of C5 flavour volatiles in tomato. J. Exp. Bot. 2014, 65, 419–428. [Google Scholar] [CrossRef]
- Meng, K.; Hou, Y.L.; Han, Y.; Ban, Q.; He, Y.; Suo, J.; Rao, J. Exploring the functions of 9-Lipoxygenase (DkLOX3) in ultrastructural changes and hormonal stress response during persimmon fruit storage. Int. J. Mol. Sci. 2017, 18, 589. [Google Scholar] [CrossRef]
- Oenel, A.; Fekete, A.; Krischke, M.; Faul, S.C.; Gresser, G.; Havaux, M.; Mueller, M.J.; Berger, S. Enzymatic and non-enzymatic mechanisms contribute to lipid oxidation during seed aging. Plant Cell Physiol. 2017, 2058, 925–933. [Google Scholar] [CrossRef]
- Zhang, L.; Hu, Z.; Shao, J.; Geng, X. A review on the composition of tomato fruit flavor and its influencing factors. Gansu Agric. Sci. Technol. 2020, 12, 85–91. [Google Scholar]
- Liaquat, F.; Munis, M.; Arif, S.; Haroon, U.; Shi, J.; Saqib, S.; Zaman, W.; Che, S.; Liu, Q. PacBio single-molecule long-read sequencing reveals genes tolerating manganese stress in Schima superba Saplings. Front. Genet. 2021, 12, 635043. [Google Scholar] [CrossRef]
- Zhang, W.; Yang, J.; Wang-Pruski, G.; Hodge, M.; Zhao, Y.; Liang, G. GalUR expression and vitamin C content of different transgenic tomatoes. J. Southwest Univ. 2008, 30, 43–47. [Google Scholar]
- Linster, C.L.; van Schaftingen, E. Vitamin C: Biosynthesis, recycling and degradation in mammals. FEBS J. 2007, 274, 1–22. [Google Scholar] [CrossRef]
- Datko, A.H.; Mudd, S.H. Phosphatidylcholine synthesis. Differing patterns in soybean and carrot. Plant Physiol. 1988, 88, 854–861. [Google Scholar] [CrossRef]
- Nuccio, M.L.; McNeil, S.D.; Ziemak, M.J.; Hanson, A.D.; Jain, R.K.; Selvaraj, G. Choline import into chloroplasts limits glycine betaine synthesis in tobacco; analysis of plants engineered with a chloroplastic or a cytosolic pathway. Metab. Eng. 2000, 2, 300–311. [Google Scholar] [CrossRef]
- Smith, D.D.; Summers, P.S.; Weretilnyk, E.A. Phosphatidylcholine synthesis in spinach: Characterization of Phosphoethanolamine N-methyl-transferase. Plant Physiol. 2000, 108, 286–294. [Google Scholar] [CrossRef]
- Summers, P.S.; Weretilnyk, E.A. Choline synthesis in spinach in relation to salt stress. Plant Physiol. 1993, 103, 1269–1276. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Feng, B.; Yu, J.; Zhang, B.; Song, Z.; Chen, J.; Liao, H.; Zhou, J. Bioinformatics analysis of plant phosphoethanolamine methyltransferase. J. Southwest Agric. 2013, 26, 499–504. [Google Scholar]
- Sun, M.; Peng, F.; Xiao, Y.; Yu, W.; Zhang, Y.; Gao, H. Exogenous phosphatidylcholine treatment alleviates drought stress and maintains the integrity of root cell membranes in peach. Sci. Hortic. 2020, 259, 108821. [Google Scholar] [CrossRef]
- Troesch, B.; Hoeft, B.; McBurney, M.; Eggersdorfer, M.; Weber, P. Dietary surveys indicate vitamin intakes below recommendations are common in representative Western countries. Br. J. Nutri. 2012, 108, 692–698. [Google Scholar] [CrossRef] [PubMed]
- De Moura, F.F.; Palmer, A.C.; Finkelstein, J.L.; Haas, J.D.; Murray-Kolb, L.E.; Wenger, M.J.; Birol, L.; Boy, E.; Peña-Rosas, J.P. Are biofortified staple food crops improving vitamin A and iron status in women and children. New Evidence Efficacy Trials. Adv. Nutr. 2014, 5, 568–570. [Google Scholar] [CrossRef]
- Asensi-Fabado, M.A.; Munné-Bosch, S. Vitamins in plants: Occurrence, biosynthesis and antioxidant function. Trends Plant Sci. 2010, 15, 582–592. [Google Scholar] [CrossRef]




| Cultivation | pH Value | Nitrogen, Nitrate (mg/kg) | Phosphorus (mg/kg) | Potassium (mg/kg) | EC (uS/cm) |
|---|---|---|---|---|---|
| Soilless substrate | 7.1 | 378.25 | 320.2 | 500 | 470 |
| Soil | 7.66 | 213.12 | 378.5 | 250 | 473 |
| Substrate-Based Cultivation | Fertilizer Type | Amount of Fertilizer (kg/667 m2/day) | Amount of Water (1000 Liters/Day) | Day |
|---|---|---|---|---|
| Early fruit set stage (1–2 fruits) | Balanced water-soluble fertilizer 20-20-20N/K = 1:1 | 1~1.2 | 1~1.2 | 20 |
| Fruit set period (3–6 fruits) | High potassium water soluble fertilizer 15-5-30 N/K = 1:2 | 1.2~1.5 | 1.2~1.5 | 30~40 |
| Middle element water-soluble fertilizer | 0.5 | |||
| Harvesting stage | no fertilizer | 1~1.2 | 10 |
| Gene ID | Primer Name | Primer Sequence (5′ to 3′) | Tm (°C) |
|---|---|---|---|
| 1 | 18S-F | CAACCATAAACGATGCCGA | 60 |
| 18S-R | AGCCTTGCGACCATACTCC | ||
| 2 | 101268829-F | CACTGTCTTCATCATCATCTTCTG | |
| 101268829-R | GTAATCGCCTCGGATCTTCT | ||
| 3 | 101261156-F | GATCACCTCCTGGTTGCCTTAG | |
| 101261156-R | AAGAAGTTGGATTTGCCCTGAAGA | ||
| 4 | 101262682-F | ACTGGTAGATCGTCTGCTA | |
| 101262682-R | CACATTCCTCATCGGTCAA | ||
| 5 | 101246511-F | GATGCTCTTTGCCAGTTTGTG | |
| 101246511-R | GCCAACTCCTTCATGTCCAA | ||
| 6 | 543815-F | CACTCCTCCCTGAAGATCCTTAT | |
| 543815-R | TTGCTCCACTCCGACCTT | ||
| 7 | 101262853-F | CCAGATACTGTGCCAATCAGAACT | |
| 101262853-R | ACCAATGCCTATGTCGTAGAATCC |
| Cultivation | Plant Height (cm) | Stem Width (mm) | Fresh Root Weight (kg) | Fresh Stem Weight (kg) | Leaf Weight (kg) | Root Volume (mL) | Yield (kg/667 m2) | Chlorophyll Content (%) |
|---|---|---|---|---|---|---|---|---|
| Substrate | 100.07 a | 8.71 a | 0.073 a | 0.46 a | 1.28 a | 76.67 a | 7177.50 a | 52.25 a |
| Soil | 72.87 b | 7.27 b | 0.029 b | 0.30 b | 0.59 b | 20.00 b | 6517.99 b | 45.33 b |
| Difference | 27.2 | 1.44 | 0.044 | 0.16 | 0.69 | 56.67 | 659.51 | 6.92 |
| Increase% | 37.3 | 19.8 | 151.7 | 53.3 | 116.9 | 283.4 | 10.1 | 15.3 |
| Cultivation | Photosynthetic Rate (μmol/m2/s) | Transpiration Rate (mmol/m2/s) | Stomatal Conductance (mol/m2/s) | Root Vitality µg·g−1·h−1 | Intercellular CO2 Concentration (μmol/mol) |
|---|---|---|---|---|---|
| Substrate | 15.05 a | 7.42 a | 0.59 a | 373.24 a | 300.78 a |
| Soil | 11.67 b | 6.12 b | 0.41 b | 202.34 b | 307.78 a |
| Difference | 3.38 | 1.3 | 0.18 | 170.9 | −7 |
| Increase% | 29.0 | 21.2 | 43.9 | 84.5 | −2.3 |
| Cultivation | Total Soluble Sugar Content (%) | Soluble Solid Content (%) | Vitamin C (mg/100 g) | Sugar-to-Acid Ratio | Nitrate (mg/kg) | Titratable Acid (%) |
|---|---|---|---|---|---|---|
| Substrate cultivation | 40.84 a | 5.23 a | 28.33 a | 9.08 a | 202.99 b | 0.45 b |
| Soil cultivation | 30.09 b | 4.37 b | 23.97 b | 5.91 b | 287.45 a | 0.51 a |
| Difference | 10.75 | 0.86 | 4.36 | 3.17 | −84.46 | −0.06 |
| Increase% | 35.7 | 19.7 | 18.2 | 53.6 | −29.4 | −11.8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, J.; Dong, L.; Kandel, S.L.; Jiao, Y.; Shi, L.; Yang, Y.; Shi, A.; Mou, B. Transcriptomic and Metabolomic Analysis Provides Insights into the Fruit Quality and Yield Improvement in Tomato under Soilless Substrate-Based Cultivation. Agronomy 2022, 12, 923. https://doi.org/10.3390/agronomy12040923
Guo J, Dong L, Kandel SL, Jiao Y, Shi L, Yang Y, Shi A, Mou B. Transcriptomic and Metabolomic Analysis Provides Insights into the Fruit Quality and Yield Improvement in Tomato under Soilless Substrate-Based Cultivation. Agronomy. 2022; 12(4):923. https://doi.org/10.3390/agronomy12040923
Chicago/Turabian StyleGuo, Jinghua, Lingdi Dong, Shyam L. Kandel, Yonggang Jiao, Linqi Shi, Yubo Yang, Ainong Shi, and Beiquan Mou. 2022. "Transcriptomic and Metabolomic Analysis Provides Insights into the Fruit Quality and Yield Improvement in Tomato under Soilless Substrate-Based Cultivation" Agronomy 12, no. 4: 923. https://doi.org/10.3390/agronomy12040923
APA StyleGuo, J., Dong, L., Kandel, S. L., Jiao, Y., Shi, L., Yang, Y., Shi, A., & Mou, B. (2022). Transcriptomic and Metabolomic Analysis Provides Insights into the Fruit Quality and Yield Improvement in Tomato under Soilless Substrate-Based Cultivation. Agronomy, 12(4), 923. https://doi.org/10.3390/agronomy12040923







