Abstract
As the global population continues to grow, food demand will be reaching levels which current agricultural practices cannot meet. This projected demand combined with the negative impacts of climate change on crop production calls for more careful breeding efforts to develop better adapted plants more tolerant to climate fluctuations. Fortunately, the development of molecular biology techniques like genome, transcriptome and epigenome sequencing now offer new approaches to help classical breeding meet these challenges. This review focuses on the potential of epigenetic approaches, particularly the creation of epigenetic markers (epi-markers) for guiding the selection process in breeding programs. Many studies have indeed successfully linked stable epigenetic modifications to different plant traits of interest but research on the applicability of using epi-markers in breeding programs is still scarce. This review emphasises the current progress that has been made with regards to the usefulness of epi-markers in selective plant breeding programs and the gaps in knowledge that still need to be addressed. It highlights the importance of pursuing research efforts to confirm the value of epi-markers for crop development in the years to come in order to meet the agricultural challenges of the 21st century.
1. Introduction
Plant breeding has allowed for tremendous advancements in crop quality and yield improvement for more than a century. However, rapidly growing global population and intensifying climate change magnify the need for high quality, high-yield crops with low requirements for inputs such as fertilizers and pesticides [1]. As the global population will approach 10 billion people over the next 30 years, it is increasingly evident that global food demands are reaching levels that current agricultural practices cannot meet [1]. Areas for improvement will include, but will not be limited to pest and disease management, reduced inputs (ex.: nitrogen and phosphorus) and adaptation to new growing areas and changing environments. In order to do this, new approaches need to be integrated into classical breeding programs. These include molecular approaches such as genomics, transcriptomics, and epigenetics. This review will introduce these concepts and focus on how the emerging field of epigenetics can support the development of more sustainable crops.
2. Breeding Methods
2.1. Classical and Genomic Breeding
Natural selection explains that individuals with better adapted genes for a given environment are more likely to produce progeny that will inherit those genes. Artificial selection does the same andhuman beings can select individuals equipped with desirable traits and underlying genes they deem to be more useful. Therefore, selective propagation of plants with desirable traits has long predated our modern understanding of genetics [2]. Important considerations into the evolutionary background, gene flow processes, mating system, and population structure must be factored into classical and genomic breeding programs [3,4]. Because the transfer of genetic material is mostly done using the natural crossing process, crops produced via classical breeding are not considered transgenic, which is a benefit in terms of public acceptance in some markets [5].
Historically, classical breeding has been carried out by crossing plants with desirable traits to a produce a progeny population (F1) that displays a range of the desirable phenotypes. The selected individuals can then be selfed and selected further until the phenotype stabilizes with desirable alleles having reached homozygosity. It can also be necessary to cross the new stable lines back to their elite parent, a process known as backcrossing. Backcrossing helps reduce linkage drag, which is a natural phenomenon during crossover events by which the genetic material flanking the gene(s) of interest is simultaneously introduced to progeny [6]. These flanking regions may contain agronomically undesirable genetic material that will not segregate independently from the gene of interest because of their physical proximity on the chromosome, and therefore will slowly separate/decay through multiple generations of selective backcrosses [5,7,8]. Linkage drag can therefore increase significantly the time required for producing new cultivated varieties (cultivars) and even preclude the introgression of certain desirable traits.
As the foundations of genetic inheritance became clearer, backcrossing was refined as a technique to be more strategic. Desirable plant traits started to be quantified more precisely and associated to increasingly more precise regions on the chromosomes, giving rise to the concept of quantitative trait loci (QTL) [9]. Together with the increasing availability of genomic markers, it was now possible to accelerate the selection of a desired trait [5]. Classical markers refers to any type of clue that can reveal the presence of a trait of interest, they can be morphological, cytological, biochemical and/or DNA/molecular markers: restriction fragment length polymorphism (RFLP), single-nucleotide polymorphism (SNP), simple sequence repeat (SSR), amplified fragment length polymorphism (AFLP), insertions, deletions, etc. that are associated with a particular trait [10,11]. Marker-assisted selection (MAS) is a benchmark practice in genomic or molecular plant breeding and plays a vital role in introgression of exotic genes from wild parents to cultivars through backcrossing (Figure 1) [12]. Ideally molecular markers are codominant with the trait of interest and highly reproducible. Molecular markers are becoming an important tool in breeding practices because they provide valuable information on genotypic and phenotypic diversity and classification allowing for more efficient and precise breeding [13,14].
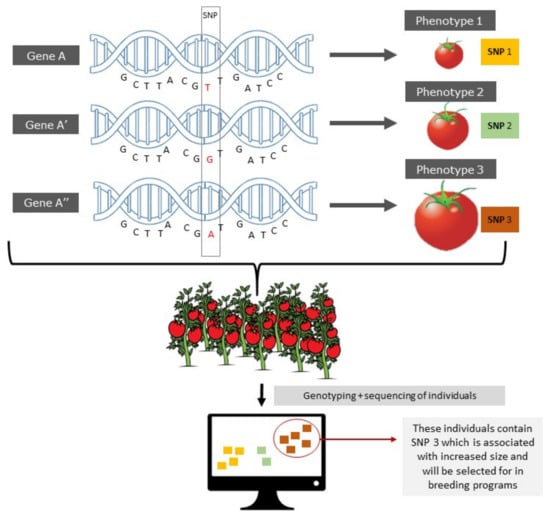
Figure 1.
Marker-assisted selection of a quantitative trait in tomato using a SNP marker.
2.2. Transcriptomic Tools
Over the past two decades transcriptomic techniques have brought a wealth of information to support strategic plant breeding. For a given gene to influence plant development, it needs to be transcribed into messenger RNA (mRNA) molecules at the right time and at the right place. By studying these molecules, one can gain insight into the molecular mechanisms underlying a trait of interest. When a gene of interest is identified, its corresponding mRNA becomes a de facto marker of the activity of that gene that can guide breeding efforts. This was made possible by the advancement of transcriptomic studies from single known genes to massively high-throughput investigations including known and unknown genes [15]. Small scale investigations of differential expression of a few genes became possible through reverse transcription (RT) polymerase chain reaction (PCR) and subsequently RT quantitative PCR (RT-qPCR) and digital PCR (RT-dPCR). However, these methods are relatively low throughput (relative to today’s standards) and detection of transcripts from unknown genes can be difficult and expensive.
Sequencing and cloning short fragments of transcripts or tags led to expressed sequence tag (EST) sequencing [16]. EST sequencing expanded possibilities for high throughput transcriptomics, even if at an expensive cost considering its relatively low accuracy of transcript detection and a poor sensitivity. Improvements were made to overcome these challenges through development of serial analysis of gene expression (SAGE) [17], cap analysis gene expression (CAGE) [18], LongSAGE [19], SuperSAGE [20,21], DeepSAGE [22], and massively parallel signature sequencing (MPSS) [23]. Microarrays [24] represent the next significant advancement in the transcriptomic revolution by which studies could include the entire transcriptome from a plant with a sequenced genome, rather than just covering a few genes. The first plant microarray was conducted using the model plant Arabidopsis thaliana [25], and this technology allowed for expansion into many other agriculturally important plants including strawberry [26], soybean [27], and rice [28] among others. Transcriptomic analysis of plants accelerated breeding strategies by linking complex spatiotemporal changes in gene expression to specific phenotypes, inevitably pointing in the direction of the genes involved in a particular trait. As transcriptomic investigations became more complex, the limitations of microarrays quickly became apparent. This technique is limited by the availability of species specific probes, as it can be expensive to create chips for new transcriptomes and therefore only commercially available probes can be considered for immediate application.
The introduction of next generation sequencing (NGS) laid new groundwork for transcriptomics. RNA-sequencing (RNA-seq) has been the most influential method for transcriptome analysis over the last decade [29,30]. RNA-seq has provided an expansive and less biased view of the transcriptome by simultaneously covering known and unknown transcripts and by greatly increasing the capacity for quantification of lowly and highly expressed genes [31]. RNA-seq expanded the transcriptional atlases of non-model plants without fully sequenced genomes, improving the overall breeding potential for all economically valuable plants. Direct comparisons of expression between control and experimental samples can be made in order to uncover the genes underlying/influencing the particular experimental cue (e.g., growth conditions, environmental stresses, pathogens, etc.). As a consequence, just like genomic markers, mRNA abundance differences can now be associated to traits and become expression QTLs (eQTLs) [32]. Surveying mRNA becomes particularly informative for expression of genes influencing traits that are not readily observable. Moreover, with this information digitally available, sharing of information and potential for meta-RNA-seq analysis becomes within arm’s reach.
3. Epigenetics and Its Potential Applications in Plant Breeding
3.1. Introduction to Epigenetics
Most cells in a given organism share the same genes yet they can be drastically different from one another. This is because not all genes are activated in all cells at any given time. The epigenetic context of a gene, or the accessibility of a gene to transcription, can reveal if the gene is likely to be active or not [33]. Indeed, the nuclear genome is found as a dense mixture of long strings of DNA wrapped around specialized proteins called histones to form superstructures known as nucleosomes [34]. These nucleosomes can be further packed together to make even denser 3D structures at times visible under the microscope which inspired the name “chromatin”. The modifications and proteins associated with the DNA are called epigenetic modifications, or marks, and they can influence gene expression within the cell. Epigenetic modifications can occur spontaneously in the genome and can be lost just as randomly presenting the concept of stability. It follows that the influences of epigenetic modifications on gene expression can only be passed from mother cell to daughter cell and from one generation to the next if the underlying modifications are actively maintained. As a strategy for adaptation, both genome sequence-dependent (genetic) and independent (epigenetic) variability are used in combination to create variability at the phenotypic level [35]. This, in turn, maximizes the chances of survival of at least some progeny under different conditions and provides a wider pool of phenotypic variation to be used for plant breeding [35,36].
3.2. Molecular Mechanisms
Because DNA is densely packed in the nucleus, for any gene that needs to be actively transcribed into mRNA, local DNA has to be unpacked and made accessible to the transcriptional machinery [33,37]. Epigenetic modifications therefore regulate accessibility to the chromatin and gene activity. This can take the form of direct modification to the DNA, the addition of histone variants, histone post-translational modification, chromatin super-structure, etc. [38]. Because of their role, these modifications can provide useful information about gene activity (or lack thereof) at the transcriptional level. Contrary to the actual DNA sequence, epigenetic marks can be removed over time but some of them are maintained through cell divisions and even passed down to the next generation. These epigenetic marks serve in a variety of processes including DNA replication and repair, stem cell maintenance, tissue and organ development and differentiation, as well as initiating responses to environmental stimuli [39]. Indeed, the flexibility of epigenetic modifications makes them an ideal mechanism to rapidly adapt the transcriptional program of a cell to changes in environmental conditions. As such, epigenetic context might be particularly useful when trying to map environment related traits. Finally, repressive epigenetic modifications are used to silence repeated sequences like transposable elements that can jeopardize genomic integrity [40].
3.3. DNA Methylation
Epigenetic marks come in many flavors but this review will focus specifically on the most widely studied epigenetic mark, DNA methylation. DNA methylation is one of the most widely studied epigenetic modifications owing to its abundance and stability as well as the ease at which it can be detected and assessed using various methods [38,41]. In short, a methyl chemical group is covalently bound to the position 5 carbon atom of the aromatic ring of the cytosine base (that therefore becomes 5-methylcytosine). This reaction is catalyzed by specialized enzymes called DNA methyltransferases and can only be reversed by another set of specialized enzymes, making it a very stable modification once applied. However, methylation must be propagated when new strands of DNA are being synthesized during replication or methylation will be lost due to dilution (a form of demethylation) [42]. In plants, DNA methylation is found to occur in three different sequence contexts, CG, CHG, and CHH (H= A, T or C) all of which have their own underlying mechanisms and biological impact [39,43,44]. Consequently, the different contexts are useful to decode the epigenetic information encountered in different genomic locations. Indeed, while mCG is found both on expressed genes and repressed sequences, mCHG and mCHH are mostly found on the latter.
3.4. Quantifying DNA Methylation
Because it is covalently associated with DNA, DNA methylation is more readily quantified than other epigenetic marks and there are different convenient techniques to do this. In order to choose an appropriate technique to quantify DNA methylation, one must consider: (1) the type of biological sample being analyzed, (2) the desired outcome (identification of unknown epigenetic marks vs. assessment of methylated regions within genes of interest), and (3) the availability and cost of the technology [45,46]. Restriction enzyme-based approaches involve the use of methylation-sensitive restriction enzymes whose activity are influenced by the presence of the methyl group on DNA [45]. Restriction endonucleases used in this method can include MspI, HpaII, NotI, SmaI and McrBC. For example, HpaII recognizes the CCGG restriction sites in genomic DNA when it is unmethylated however, the addition of methyl group (CmCGG or mCCGG) to the restriction site prevents HpaII activity. Thus, the different treatment of methylated and non-methylated sequences can be used to assess the methylation status of a locus, as long as the restriction sequence is present. Cleavage of the DNA molecule prevents PCR amplification thus creating a signal that can easily be detected for the presence or absence of DNA methylation. Alternatively, genomic DNA can be treated with sodium bisulfite resulting in the conversion of unmethylated cytosines to uracils [45]. During PCR amplification of the region of interest, uracils are read by DNA polymerase as thymines which can be detected by melting curve analysis, Sanger sequencing or Amplicon sequencing.
When there are many sequences of interest or if they are unknown, a genome-wide approach needs to be considered. Affinity enrichments-based approaches use antibodies designed to bind to methylcytosines and to pull them down so that methylated DNA is enriched [45]. Once the methylated DNA has been isolated, bisulfite sequencing is typically used to sequence the methylated regions. This reduces the cost of sequencing by focusing on regions where high methylation is present. Whole-genome bisulfite sequencing (WGBS) is still the predominant approach used to precisely quantify DNA methylation genome-wide. This method is frequently used because it is easy, fast, and provides highly accurate results that can be used to identify all differentially methylated regions (DMRs) existing between two samples [47]. Although bisulfite sequencing has been used as the gold standard approach for methylome analysis, there are limitations to this technique. Sodium bisulfite is a reactive chemical that damages the DNA molecules which can lead to inaccurate interpretations or destruction of limited samples. Enzymatic methyl-seq is a new approach designed to minimize DNA damage by using enzymes (including TET2 and APOBEC2) rather than chemicals to convert the methylated bases [48,49]. Using this method, DNA integrity can be better maintained resulting in more accurate readings and mapping efficiencies. This newly-established approach is a promising alternative to bisulfite sequencing.
Once information on methylated sites is obtained, downstream analysis can proceed to identify DMRs. This involves comparing methylated sites ratios between samples using a specific distance criteria and statistical tests [50]. Once DMRs have been identified, they can be linked to quantitative traits segregating in a population much like QTLs. Identification of DMRs from sequencing data requires consideration of various factors to optimize accurate interpretation and reduce bias. This includes (1) considering spatial correlation between methylation levels of neighboring methylated sites to more accurately estimate corresponding hypomethylated sites, (2) considering sequencing depth which takes into account sampling variability during sequencing, and (3) considering biological variation among samples which will minimize the number of false positives in the results [50].
3.5. Variation in DNA Methylation Patterns
With all these techniques, it was found that DNA methylation patterns are dynamic and exhibit considerable variation in the levels of methylation and the nature of the sequences being methylated. Indeed, much like the DNA sequence in the genome, the epigenome can also accumulate heritable changes in the lifetime of an organism. However, these epigenetic changes occur at a much faster rate than mutations in the DNA sequence [51]. This has led to the hypothesis that heritable epi-mutations can be selected naturally or artificially like their DNA mutation counterparts. Interestingly, methylation patterns are also shaped by the environment and thus can change in response to abiotic and biotic stress exposure [52,53,54,55]. This brings the possibility to store information about the conditions an organism encountered in its development. Consequently, pattern differences can be divided into two main groups; (1) developmental, whereby individuals exhibit specific patterns associated with different stages of growth and development and (2) acquired, whereby individuals exhibit specific or random patterns related to specific conditions encountered in their environment.
3.5.1. Developmental Epigenetic Modifications
DNA methylation is a feature of constitutive heterochromatin in many eukaryotes where it plays an important structural role. In some organisms, like in flowering plants, it is also involved in developmental processes including reproduction, seed development, germination, tissue differentiation and growth. [56,57,58]. Understanding methylation patterns at different developmental stages is important in linking traits to particular methylation markers. For traits relating to a particular tissue, like seed size, it may be worth looking for specific epigenetic changes in that tissue and even selecting a precise time window when it is most abundant for higher resolution. Indeed, it was recently published that DNA methylation levels differ greatly at different stages of seed development in a variety of species [57,59,60,61,62]. Specifically, DNA methylation levels seem to increase throughout the stages of seed development. Fruit ripening is another developmental trait that comes with important epigenetic modifications and has been studied in various plant species including tomato, apple, and orange [63,64,65].
DNA methylation also exhibits extensive variation among different tissues and cell types particularly in vegetative organs such as leaves, shoots, and roots. Within Arabidopsis, an analysis of the methylation status within the root apical meristem revealed differences in methylation status among various cell types [66]. For example, there is widespread hypermethylation within the columella cells when compared to the epidermis, cortex, endodermic, and stele cells [66]. In soybean, differences in methylation patterns between single-celled root hairs and multicellular stripped roots were assessed [67]. Analyses revealed significant differences in DNA methylation patterns, particularly in the CHH methylation context, between the two types of cells [67]. DNA methylation patterns also greatly differed between roots and shoot tissues in Arabidopsis and also other Brassicaceae [68,69]. In inbred lines of maize, patterns of DNA methylation greatly differed among tassel, bracteal leaf, and ear leaf tissues [70]. Moreover, during fruit development in tomato, the fruit and the leaf tissues displayed different methylation levels [63]. In addition to epigenetic variation among vegetative organs, there also seems to be epigenetic variation among different tissues within the seed. Particularly in seeds of Arabidopsis and rice, the endosperm displayed lower levels of DNA methylation relative to the embryo [71,72]. These examples demonstrate that DNA methylation can play an important role in tissue/cell differentiation. By comparing levels of DNA methylation among various developmental stages and tissue/cell types, we can appreciate the magnitude of epigenetic variation in a developmental and tissue-specific manner. This information is particularly useful when trying to produce high resolution epigenetic studies especially when targeting a specific trait.
3.5.2. Acquired Epigenetic Modifications
Epigenetic marks can also be acquired or lost in response to environmental cues, including abiotic and biotic stresses. These epigenetic modifications can be heritable and allow for plants to rapidly adapt their development to changing environments and play important roles in pathogen defense and abiotic tolerance (salinity, drought, high and low temperatures, etc.) [73]. Environmentally stimulated changes in DNA methylation have been reported in many crop varieties including rice [74], soybean [75], maize [76], and wheat [77]. In soybean, DMRs among root hair cells and stripped root cells were assessed following a heat treatment [67]. At room temperature, root hair cells showed hypermethylation at specific DMRs when compared to stripped root cells which showed hypomethylation. In response to a heat stress, hypomethylation occurred at DMRs in both cell types revealing that root hair cells may be more sensitive to fluctuations in temperature. Furthermore in wheat, individuals with a salt-tolerant genotype have higher methylation levels in the root tissues which has been associated with restricting Na+ entry from the soil into the root tissues [78]. Analyzing these changes can provide candidate markers for detecting plant stress responses within a population.
While these epigenetic changes can function in momentarily tuning gene expression to adapt to a new condition, they can also act as ”stress memory” [79]. This stress memory allows for individuals to respond more quickly to recurring stimuli by priming gene expression patterns for more rapid adjustments [80]. Studies have shown that Arabidopsis exhibits heat-stress memory whereby heat-inducible genes remain activated through hypermethylation for at least two days following removal of the stress [81]. This primes the individual for a quicker response to recurring heat stress. Moreover, epigenetic cross-adaptation can also occur whereby exposure to one stress can lead to resistance to other stresses [82]. This was tested in cold-tolerant Brassica rapa, to see whether individuals also showed heat tolerance. DNA methylation patterns among four candidate genes were compared between control and cold-acclimated (CA) plants [82]. It was found that the promoter regions of these four candidate genes exhibited demethylation resulting in increased gene expression in CA plants [82]. This demethylation was linked to elevated levels of organic acids and enhanced photosynthesis which authors concluded were contributing factors to enhanced heat tolerance and higher growth rates in CA plants [82]. Such study suggests that DNA methylation plays a role in cross-adaptation within plants and can help generate plants resilient to extreme temperatures. They also show that certain epigenetic modifications are only revealed in certain specific conditions.
3.6. Population Epigenetic Diversity
The accumulation of random and environment stimulated epigenetic changes can lead isolated populations to diverge more and more with time if the epigenetic changes are well-maintained and not lost. Following the assessment of 1107 methylomes from 1028 Arabidopsis accessions, considerable epigenetic variation was found [83]. It was proposed that most of the epigenetic variation across accessions were related to differences in geographic location and climate. Survey of multiple populations has also been conducted in maize [84], soybean [85], and rice [86]. These studies provide further evidence that epigenetic changes can accumulate and be selected for within a population [51]. Importantly, these DNA methylation patterns are stable and can segregate as it was demonstrated in RILs of soybean [87]. In this study, leaf tissue was isolated among four lines and leaf methylomes were sequenced. 1416 DMRs at various sequence contexts were identified between parental and RIL lines. DMRs which overlapped important genes were found to correlate with decreasing levels of gene expression. Moreover, methylomes among nine rice strains were examined to determine the existence of epigenetic polymorphisms/epigenetic markers [88]. This study found that between any two strains, 2–3% of the total methylome corresponded to epigenetic polymorphisms/markers. If these epigenetic markers are linked to phenotypic variation, they could be considered in rice breeding programs. These findings suggest that the existence of stable and heritable epigenetic variation between different populations is a prerequisite to contemplate a breeding strategy based on epigenetic markers.
3.6.1. Epialleles and epiQTLs
Two alleles occupying the same locus and sharing the same DNA sequence can exist in different epigenetic context, they are consequently epialleles. As such, this epigenetic context becomes another layer of information that can help explain phenotypic variation [51]. There are three groups of epialleles defined by their relative dependency on an underlying genetic variant; (1) obligate epialleles which completely depend on the presence of a genetic variant (2) pure epialleles which can be maintained without a genetic variant and (3) facilitated epialleles which can be influenced by a genetic variant but not as strongly as obligate epialleles [89]. Epialleles affecting various traits have been reported in model plant organisms such as Arabidopsis thaliana [73] as well as crop plants including oil palm [90], rice [73], and maize [91], and tomato [92]. Epialleles can also either be naturally occurring or artificially induced and can contribute to phenotypic variation. For example, the natural epimutant clark-kent (clk) in Arabidopsis thaliana displays observable phenotypic differences including enhanced stamens and carpels when the SUPERMAN locus is in a hypermethylated state [73]. For epialleles to act as reliable epi-markers they must be relatively stable and heritable. If epialleles can act as reliable epi-markers, then they can be exploited in selective breeding programs. Such an approach is depicted in Figure 2.
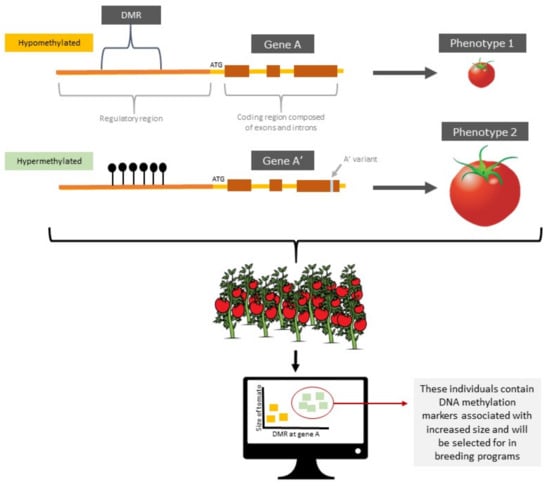
Figure 2.
Marker-assisted selection of tomatoes with increased size using epi-markers detecting a differentially methylation region. In the example, the marker would be a patch of methylation that exists in the regulatory sequence of the gene. The hypermethylated form is associated with a gene variant that influences the desired phenotype.
Stable epialleles can therefore be linked to quantitative trait loci (epiQTL) and become potential epi-markers. To identify epiQTLs, epigenetic recombinant inbred lines (epiRILs) are used. EpiRILs are created using genetically identical parental plants which differ in levels of DNA methylation [93]. These differences in DNA methylation patterns are associated with altered phenotypes [93]. The parents are then crossed and progeny in subsequent generations are scanned to determine the distribution of epialleles. These epialleles are then linked to variation in a phenotypic trait and become epiQTLs. EpiQTLs involved in flowering time [94], root length [94], and pathogen resistance [95] in Arabidopsis thaliana have been identified. EpiQTLs for flowering time and root length were found to explain 60–90% of the heritability of these traits [94]. Another study in Brassica napus identified epiQTLs associated with seven agronomic traits. In the same way that QTLs can become de facto genomic markers, epiQTLs can become epi-markers and inform on the presence of a specific trait.
3.6.2. Epigenome-Wide Association Mapping (EWAS)
In plants, genome-wide association studies (GWAS) continues to be a powerful tool used to understand the underlying genetic architecture governing complex traits and plays an important role in accelerating plant breeding programs [96]. Similarly, epigenome-wide association studies (EWAS) may also prove to be a useful tool to better understand the origin of phenotypic variation. Once complete epigenome profiles have been gathered, researchers can go on to perform EWAS to link a particular phenotypic trait with epigenetic variation. EWAS involves three major steps; (1) epigenome sequencing, (2) phenotyping for desired traits, and (3) bioinformatics/statistical testing to associate both variables on a genomic scale [73]. While EWAS studies have been widely conducted in humans to identify epigenetic variants associated with various diseases, its use in plant populations is lacking [96]. The first EWAS study in plants was performed in oil palm and successfully identified epigenetic variants within the MANTLED gene at the Karma splice site [90]. The Good Karma epiallele predicts a wild type fruit set whereas the Bad Karma epiallele predicts fruit abnormalities associated with loss of yield. As more whole-epigenome datasets become available due to advancements in sequencing technology, EWAS will become easier to perform. In turn, this will promote EWAS research in crop plants and add to the limited information currently available.
3.7. Epigenetic Selection
The prediction value of epigenetic markers has been amply demonstrated for human health and disease prediction but is still lacking in plants [97]. Encouragingly, the publication of more and more plant genomes will offer new opportunities for such approaches. Already, the accumulating epigenetic data in model and crop plants shows a great potential for the use of epigenetic markers in breeding better-adapted crops. As an original proof-of-concept, fundational studies in Arabidopsis thaliana have reported the use of epigenetically-depleted individuals crossed with normal isogenic mates to test the effects and segregation of epigenetic alleles on plant phenotypes [94,98]. It was concluded that a sizable share of segregating traits were explained by the presence of DMRs (in the absence of genetic variation). As an example, it was found that up to 65% of the variation in plant height was accounted for by epigenetic variation [99]. In the same population, it was also found that DMRs associated with root length and flowering time acted as reliable epiQTL markers and accounted for roughly 60 to 90% of the heritability of these two traits [94]. These studies suggest not only that epi-markers could be used to guide selection of desired traits but that it is possible to generate new traits through the artificial creation of epialleles.
Analysis of the methylomes of various crop plants has also revealed an association between DMRs and agronomic traits. In theory, these DMRs could be used as epi-markers to select for individuals displaying a partiular favourable agronomic trait. Recently, a study identified DMRs associated with various metabolic traits in a maize segregating population [100]. These stable DMRs can therefore be used to select and breed individuals with favourable metabolic traits. Importantly, some of this phenotypic variation could not be linked to SNP-based genomic markers highlighting what could be gained in using epi-markers [100]. This study also provides some insight on the causal relationship between DMRs and gene expression. The authors found that compared to SNPs, DMRs are actually more likely to have a causal role either having a negative or positive effect on nearby gene expression [100]. In another groundbreaking example, a reproductive defect of clonally-propagated oil palm was mapped to a single causative DMR within a gene [90]. This study provides an example whereby epigenetic marks can be used for plant health prediction [101]. Other studies have also shown that isogenic plants can be made to develop new traits in different plants and that their underlying cause is in fact epigenetic [102,103,104,105]. These studies demonstrate that increase in yield [102], adaptation to low humidity [106], and changes in flowering time [104] were all made possible by epigenetic modifications. In tomato, DMRs associated with vitamin E content [92], fruit ripening, shelf-life and fruit quality [63] have been identified and may prove to be useful epi-markers for future breeding efforts. Epi-markers have also been used in other crop species including for crop improvement. It therefore appears that there is ample epigenetic variation linked to traits in different plant species for it to be considered as an approach to plant breeding.
3.8. Limitations of Epigenetic Markers
Although there seems to be great potential in using epigenetic markers in trait selection for plant breeding, more work is needed to prove that it can be efficiently applied to breeding programs [107,108]. For one, many epigenetic studies have been conducted in model organisms such as the plant Arabidopsis thaliana, warranting further research in crop plants. Moreover, the most obvious difficulty inherent to epigenetic marks is that they are less stable and heritable than variations in the DNA sequence. This is particularly important in crop breeding which is a multi-year endeavor that needs to produce plants with uniform, stable qualities [108]. To overcome this hurdle, the epigenetic markers selected will have to be tested across different populations and generations to ensure their stable inheritance. The same logic applies for epigenetic modifications in response to stresses, they may only be detectable in specific conditions making them unreliable markers for breeding purposes. In such case however, the epigenetic markers could be used as a large-scale diagnostic test to evaluate the penetrance of a stress response within a population. More studies involving crop plants and more refined modelling of epigenetic inheritance are therefore required to prove that such an approach can benefit the development of new and improved cultivars.
4. Conclusions
As the global population continues to grow, there comes a greater urgency to rapidly develop crops better adapted to their environment [109]. To have a chance to meet the growing demand, new approaches need to be considered in support of existing breeding practices. Epigenetic modifications, such as DNA methylation may prove to be a powerful selection tool in crop breeding. One promising approach would be the creation of epi-markers to complement genomic markers in tagging quantitative traits. Indeed, many plant traits have already been successfully linked to specific epigenetic marks even in cases where no genomic markers are present [100,110]. Further research on the application of this approach for trait selection in crop plants is needed before it can be more widely adopted. This includes further research on epiallele stability using segregating populations of crop plants. Once stable epialleles have been identified and linked to the occurance of a particular trait, they can then be used as epi-markers in breeding practices. Once identified, these epi-markers can then be used as targets for epigenomic editing to induce specific epi-mutations and produce a desired phenotype. That being said, epigenetics opens the door to additional tools that can be used in plant breeding for the selection and editing of epialleles and traits.
Author Contributions
Writing—Original Draft Preparation, H.T., J.H., B.S. and J.-S.P.; Writing—Review & Editing, B.S. and J.-S.P.; Supervision, B.S. and J.-S.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Agriculture and Agri-Food Canada.
Acknowledgments
The authors would like to thank Elroy Cober, from Agriculture and Agri-Food Canada, for helpful discussions and critical review of this manuscript.
Conflicts of Interest
The authors declares no conflict of interest.
References
- Hickey, L.T.; Hafeez, A.N.; Robinson, H.; Jackson, S.A.; Leal-Bertioli, S.C.M.; Tester, M.; Gao, X.; Godwin, I.; Hayes, B.; Wulff, B.B.H. Breeding crops to feed 10 billion. Nat. Biotechnol. 2019, 37, 744–754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piperno, D.R.; Ranere, A.J.; Holst, I.; Iriarte, J.; Dickau, R. Starch grain and phytolith evidence for early ninth millennium B.P. maize from the Central Balsas River Valley, Mexico. Proc. Natl. Acad. Sci. USA 2009, 106, 5019–5024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamrick, J.L. Isozymes and the Analysis of Genetic Structure in Plant Populations. In Isozymes in Plant Biology; Springer: Dordrecht, The Netherlands, 1989; pp. 87–105. [Google Scholar]
- Nadeem, M.A.; Nawaz, M.A.; Shahid, M.Q.; Doğan, Y.; Comertpay, G.; Yıldız, M.; Hatipoğlu, R.; Ahmad, F.; Alsaleh, A.; Labhane, N.; et al. DNA molecular markers in plant breeding: Current status and recent advancements in genomic selection and genome editing. Biotechnol. Biotechnol. Equip. 2018, 32, 261–285. [Google Scholar] [CrossRef] [Green Version]
- Gepts, P. A comparison between crop domestication, classical plant breeding, and genetic engineering. Crop Sci. 2002, 42, 1780–1790. [Google Scholar] [CrossRef] [Green Version]
- Brinkman, M.A.; Frey, K.J. Yield-component Analysis of Oat Isolines that Produce Different Grain Yields. Crop Sci. 1977, 17, 165–168. [Google Scholar] [CrossRef]
- Brown, A.H.D.; Lawrence, G.J.; Jenkin, M.; Douglass, J.; Gregory, E. Linkage drag in backcross breeding in barley. J. Hered. 1989, 80, 234–239. [Google Scholar] [CrossRef]
- Zeven, A.C.; Knott, D.R.; Johnson, R. Investigation of linkage drag in near isogenic lines of wheat by testing for seedling reaction to races of stem rust, leaf rust and yellow rust. Euphytica 1983, 32, 319–327. [Google Scholar] [CrossRef]
- Collard, B.C.Y.; Jahufer, M.Z.Z.; Brouwer, J.B.; Pang, E.C.K. An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: The basic concepts. Euphytica 2005, 142, 169–196. [Google Scholar] [CrossRef]
- Collard, B.C.Y.; Mackill, D.J. Marker-assisted selection: An approach for precision plant breeding in the twenty-first century. Philos. Trans. R. Soc. B Biol. Sci. 2008, 363, 557–572. [Google Scholar] [CrossRef] [Green Version]
- Jiang, G.-L. Molecular Markers and Marker-Assisted Breeding in Plants. In Plant Breeding from Laboratories to Fields; InTech: Rijeka, Croatia, 2013. [Google Scholar]
- Simmonds, N.W. Introgression and incorporation. Strategies for the use of crop genetic resources. Biol. Rev. 1993, 68, 539–562. [Google Scholar] [CrossRef]
- Naeem, M.; Ghouri, F.; Shahid, M.Q.; Iqbal, M.; Baloch, F.; Chen, L.; Ul-Allah, S.; Babar, M.; Rana, M. Genetic diversity in mutated and non-mutated rice varieties. Genet. Mol. Res. GMR 2015, 14, 17109–17123. [Google Scholar] [CrossRef]
- Tiwari, J.K.; Singh, B.P.; Gopal, J.; Patil, V.U. Molecular characterization of the Indian Andigena potato core collection using microsatellite markers. Afr. J. Biotechnol. 2013, 12, 1025–1033. [Google Scholar]
- Agarwal, P.; Parida, S.K.; Mahto, A.; Das, S.; Mathew, I.E.; Malik, N.; Tyagi, A.K. Agarwal 2014—Expanding frontiers in plant transcriptomics in aid of functional genomics and molecular breeding. Biotechnol. J. 2014, 9, 1480–1492. [Google Scholar] [CrossRef]
- Adams, M.D.; Kerlavage, A.R.; Fleischmann, R.D.; Fuldner, R.A. Initial assessment of human gene diversity and expression patterns based upon 83 million nucleotides of cDNA sequence. Nature 1995, 377, 3–174. [Google Scholar]
- Velculescu, V.E.; Vogelsein, B.; Kinzler, K.W.; Zhang, L. Serial analysis of gene expression. Science 1995, 270, 484–487. [Google Scholar] [CrossRef] [Green Version]
- Shiraki, T.; Kondo, S.; Katayama, S.; Waki, K.; Kasukawa, T.; Kawaji, H.; Kodzius, R.; Watahiki, A.; Nakamura, M.; Arakawa, T.; et al. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc. Natl. Acad. Sci. USA 2003, 100, 15776–15781. [Google Scholar] [CrossRef] [Green Version]
- Wei, C.L.; Ng, P.; Chiu, K.P.; Wong, C.H.; Ang, C.C.; Lipovich, L.; Liu, E.T.; Ruan, Y. 5′ Long serial analysis of gene expression (LongSAGE) and 3′ LongSAGE for transcriptome characterization and genome annotation. Proc. Natl. Acad. Sci. USA 2004, 101, 11701–11706. [Google Scholar] [CrossRef] [Green Version]
- Matsumura, H.; Urasaki, N.; Yoshida, K.; Krüger, D.H.; Kahl, G.; Terauchi, R. SuperSAGE: Powerful Serial Analysis of Gene Expression. In RNA Abundance Analysis; Humana Press: Totowa, NJ, USA, 2012; Volume 883, pp. 1–17. [Google Scholar]
- Matsumura, H.; Yoshida, K.; Luo, S.; Kimura, E.; Fujibe, T.; Albertyn, Z.; Barrero, R.A.; Krüger, D.H.; Kahl, G.; Schrot, G.P.; et al. High-throughput superSAGE for digital gene expression analysis of multiple samples using next generation sequencing. PLoS ONE 2010, 5, e12010. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, K.L.; Høgh, A.L.; Emmersen, J. DeepSAGE—Digital transcriptomics with high sensitivity, simple experimental protocol and multiplexing of samples. Nucleic Acids Res. 2006, 34, e133. [Google Scholar] [CrossRef] [Green Version]
- Brenner, S.; Johnson, M.; Bridgham, J.; Golda, G.; Lloyd, D.H.; Johnson, D.; Luo, S.; McCurdy, S.; Foy, S.; Ewan, M.; et al. Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat. Biotechnol. 2000, 18, 630–634. [Google Scholar] [CrossRef]
- Lashkari, D.A.; Derisi, J.L.; McCusker, J.H.; Namath, A.F.; Gentile, C.; Hwang, S.Y.; Brown, P.O.; Davis, R.W. Yeast microarrays for genome wide parallel genetic and gene expression analysis. Proc. Natl. Acad. Sci. USA 1997, 94, 13057–13062. [Google Scholar] [CrossRef] [Green Version]
- Zhu, T.; Wang, X. Large-Scale Profiling of the Arabidopsis Transcriptome. Plant Physiol. 2000, 124, 1472–1476. [Google Scholar] [CrossRef] [Green Version]
- Aharoni, A.; Keizer, L.C.P.; Bouwmeester, H.J.; Sun, Z.; Alvarez-Huerta, M.; Verhoeven, H.A.; Blaas, J.; van Houwelingen, A.M.; De Vos, R.C.; van der Voet, H.; et al. Identification of the SAAT gene involved in strawberry flavor biogenesis by use of DNA microarrays. Plant Cell 2000, 12, 647–661. [Google Scholar] [CrossRef] [Green Version]
- Shoemaker, R.; Keim, P.; Vodkin, L.; Retzel, E.; Clifton, S.W.; Waterston, R.; Smoller, D.; Coryell, V.; Khanna, A.; Erpelding, J.; et al. A compilation of soybean ESTs: Generation and analysis. Genome 2002, 45, 329–338. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.; Chen, C.; Liu, X.; Jiao, Y.; Su, N.; Li, L.; Wang, X.; Cao, M.; Sun, N.; Zhang, X.; et al. A microarray analysis of the rice transcriptome and its comparison to Arabidopsis. Genome Res. 2005, 15, 1274–1283. [Google Scholar] [CrossRef] [Green Version]
- Lister, R.; O’Malley, R.C.; Tonti-Filippini, J.; Gregory, B.D.; Berry, C.C.; Millar, A.H.; Ecker, J.R. Highly Integrated Single-Base Resolution Maps of the Epigenome in Arabidopsis. Cell 2008, 133, 523–536. [Google Scholar] [CrossRef] [Green Version]
- Nagalakshmi, U.; Wang, Z.; Waern, K.; Shou, C.; Raha, D.; Gerstein, M.; Snyder, M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 2008, 320, 1344–1349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kukurba, K.R.; Montgomery, S.B. RNA Sequencing and Analysis. Cold Spring Harb. Protoc. 2015, 11, 951–969. [Google Scholar] [CrossRef] [Green Version]
- Cubillos, F.A.; Coustham, V.; Loudet, O. Lessons from eQTL mapping studies: Non-coding regions and their role behind natural phenotypic variation in plants. Curr. Opin. Plant Biol. 2012, 15, 192–198. [Google Scholar] [CrossRef]
- Klemm, S.L.; Shipony, Z.; Greenleaf, W.J. Chromatin accessibility and the regulatory epigenome. Nat. Rev. Genet. 2019, 20, 207–220. [Google Scholar] [CrossRef]
- Samo, N.; Ebert, A.; Kopka, J.; Mozgová, I. Plant chromatin, metabolism and development—An intricate crosstalk. Curr. Opin. Plant Biol. 2021, 61, 102002. [Google Scholar] [CrossRef] [PubMed]
- Tirnaz, S.; Batley, J. Epigenetics: Potentials and Challenges in Crop Breeding. Mol. Plant 2019, 12, 1309–1311. [Google Scholar] [CrossRef] [PubMed]
- Gallusci, P.; Dai, Z.; Génard, M.; Gauffretau, A.; Leblanc-Fournier, N.; Richard-Molard, C.; Vile, D.; Brunel-Muguet, S. Epigenetics for Plant Improvement: Current Knowledge and Modeling Avenues. Trends Plant Sci. 2017, 22, 610–623. [Google Scholar] [CrossRef] [PubMed]
- Kono, H.; Ishida, H. Nucleosome unwrapping and unstacking. Curr. Opin. Struct. Biol. 2020, 64, 119–125. [Google Scholar] [CrossRef]
- Agarwal, G.; Kudapa, H.; Ramalingam, A.; Choudhary, D.; Sinha, P.; Garg, V.; Singh, V.K.; Patil, G.B.; Pandey, M.K.; Nguyen, H.T.; et al. Epigenetics and epigenomics: Underlying mechanisms, relevance, and implications in crop improvement. Funct. Integr. Genom. 2020, 20, 739–761. [Google Scholar] [CrossRef]
- Bartels, A.; Han, Q.; Nair, P.; Stacey, L.; Gaynier, H.; Mosley, M.; Huang, Q.Q.; Pearson, J.K.; Hsieh, T.F.; Charles An, Y.Q.; et al. Dynamic DNA methylation in plant growth and development. Int. J. Mol. Sci. 2018, 19, 2144. [Google Scholar] [CrossRef] [Green Version]
- Okamoto, H.; Hirochika, H. Silencing of transposable elements in plants. Trends Plant Sci. 2001, 6, 527–534. [Google Scholar] [CrossRef]
- Ito, T.; Nishio, H.; Tarutani, Y.; Emura, N.; Honjo, M.N.; Toyoda, A.; Fujiyama, A.; Kakutani, T.; Kudoh, H. Seasonal stability and dynamics of dna methylation in plants in a natural environment. Genes 2019, 10, 544. [Google Scholar] [CrossRef] [Green Version]
- Bochtler, M.; Kolano, A.; Xu, G.-L. DNA demethylation pathways: Additional players and regulators. Bioessays 2017, 39, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Anderson, S.N.; Zynda, G.J.; Song, J.; Han, Z.; Vaughn, M.W.; Li, Q.; Springer, N.M. Subtle perturbations of the maize methylome reveal genes and transposons silenced by chromomethylase or RNA-directed DNA methylation pathways. G3 Genes Genomes Genet. 2018, 8, 1921–1932. [Google Scholar] [CrossRef] [Green Version]
- Cheng, C.; Tarutani, Y.; Miyao, A.; Ito, T.; Yamazaki, M.; Sakai, H.; Fukai, E.; Hirochika, H. Loss of function mutations in the rice chromomethylase OsCMT3a cause a burst of transposition. Plant J. 2015, 83, 1069–1081. [Google Scholar] [CrossRef] [Green Version]
- Pajares, M.J.; Palanca-Ballester, C.; Urtasun, R.; Alemany-Cosme, E.; Lahoz, A.; Sandoval, J. Methods for analysis of specific DNA methylation status. Methods 2021, 187, 3–12. [Google Scholar] [CrossRef]
- Kurdyukov, S.; Bullock, M. DNA methylation analysis: Choosing the right method. Biology 2016, 5, 3. [Google Scholar] [CrossRef]
- Gravina, S.; Ganapathi, S.; Vijg, J. Single-cell, locus-specific bisulfite sequencing (SLBS) for direct detection of epimutations in DNA methylation patterns. Nucleic Acids Res. 2015, 43, e93. [Google Scholar] [CrossRef] [Green Version]
- Vaisvila, R.; Ponnaluri, V.K.C.; Sun, Z.; Langhorst, B.W.; Saleh, L.; Guan, S.; Dai, N.; Campbell, M.A.; Sexton, B.S.; Marks, K.; et al. Enzymatic methyl sequencing detects DNA methylation at single-base resolution from picograms of DNA. Genome Res. 2021, 31, 1280–1289. [Google Scholar] [CrossRef]
- Suhua, F.; Zhenhui, Z.; Ming, W.; Steven, E.J. Efficient and accurate determination of genome-wide DNA methylation patterns in Arabidopsis with enzymatic methyl sequencing. Epigenetics Chromatin 2021, 13, 1–17. [Google Scholar]
- Shafi, A.; Mitrea, C.; Nguyen, T.; Draghici, S. A survey of the approaches for identifying differential methylation using bisulfite sequencing data. Brief. Bioinform. 2017, 19, 737–753. [Google Scholar] [CrossRef] [Green Version]
- Noshay, J.M.; Springer, N.M. Stories that can’t be told by SNPs; DNA methylation variation in plant populations. Curr. Opin. Plant Biol. 2021, 61, 101989. [Google Scholar] [CrossRef]
- Rajkumar, M.S.; Shankar, R.; Garg, R.; Jain, M. Bisulphite sequencing reveals dynamic DNA methylation under desiccation and salinity stresses in rice cultivars. Genomics 2020, 112, 3537–3548. [Google Scholar] [CrossRef]
- Ashapkin, V.V.; Kutueva, L.I.; Aleksandrushkina, N.I.; Vanyushin, B.F. Epigenetic mechanisms of plant adaptation to biotic and abiotic stresses. Int. J. Mol. Sci. 2020, 21, 7457. [Google Scholar] [CrossRef]
- Kong, L.; Liu, Y.; Wang, X.; Chang, C. Insight into the role of epigenetic processes in abiotic and biotic stress response in wheat and barley. Int. J. Mol. Sci. 2020, 21, 1480. [Google Scholar] [CrossRef] [Green Version]
- Guo, H.; Wu, T.; Li, S.; He, Q.; Yang, Z.; Zhang, W.; Gan, Y.; Sun, P.; Xiang, G.; Zhang, H.; et al. The methylation patterns and transcriptional responses to chilling stress at the seedling stage in rice. Int. J. Mol. Sci. 2019, 20, 5089. [Google Scholar] [CrossRef] [Green Version]
- Grover, J.W.; Kendall, T.; Baten, A.; Burgess, D.; Freeling, M.; King, G.J.; Mosher, R.A. Maternal components of RNA-directed DNA methylation are required for seed development in Brassica rapa. Plant J. 2018, 94, 575–582. [Google Scholar] [CrossRef] [Green Version]
- Kawakatsu, T.; Nery, J.R.; Castanon, R.; Ecker, J.R. Dynamic DNA methylation reconfiguration during seed development and germination. Genome Biol. 2017, 18, 171. [Google Scholar] [CrossRef] [Green Version]
- Ikeuchi, M.; Iwase, A.; Sugimoto, K. Control of plant cell differentiation by histone modification and DNA methylation. Curr. Opin. Plant Biol. 2015, 28, 60–67. [Google Scholar] [CrossRef]
- Lin, J.Y.; Le, B.H.; Chen, M.; Henry, K.F.; Hur, J.; Hsieh, T.F.; Chen, P.Y.; Pelletier, J.M.; Pellegrini, M.; Fischer, R.L.; et al. Similarity between soybean and Arabidopsis seed methylomes and loss of non-CG methylation does not affect seed development. Proc. Natl. Acad. Sci. USA 2017, 114, E9730–E9739. [Google Scholar] [CrossRef] [Green Version]
- Xing, M.Q.; Zhang, Y.J.; Zhou, S.R.; Hu, W.Y.; Wu, X.T.; Ye, Y.J.; Wu, X.X.; Xiao, Y.P.; Li, X.; Xue, H.W. Global analysis reveals the crucial roles of DNA methylation during rice seed development. Plant Physiol. 2015, 168, 1417–1432. [Google Scholar] [CrossRef] [Green Version]
- Bouyer, D.; Kramdi, A.; Kassam, M.; Heese, M.; Schnittger, A.; Roudier, F.; Colot, V. DNA methylation dynamics during early plant life. Genome Biol. 2017, 18, 179. [Google Scholar] [CrossRef]
- Rajkumar, M.S.; Gupta, K.; Khemka, N.K.; Garg, R.; Jain, M. DNA methylation reprogramming during seed development and its functional relevance in seed size/weight determination in chickpea. Commun. Biol. 2020, 3, 340. [Google Scholar] [CrossRef]
- Zhong, S.; Fei, Z.; Chen, Y.R.; Zheng, Y.; Huang, M.; Vrebalov, J.; McQuinn, R.; Gapper, N.; Liu, B.; Xiang, J.; et al. Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat. Biotechnol. 2013, 31, 154–159. [Google Scholar] [CrossRef]
- Daccord, N.; Celton, J.M.; Linsmith, G.; Becker, C.; Choisne, N.; Schijlen, E.; van de Geest, H.; Bianco, L.; Micheletti, D.; Velasco, R.; et al. High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat. Genet. 2017, 49, 1099–1106. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Liu, R.; Niu, Q.; Tang, K.; Zhang, B.; Zhang, H.; Chen, K.; Zhu, J.K.; Lang, Z. Global increase in DNA methylation during orange fruit development and ripening. Proc. Natl. Acad. Sci. USA 2019, 116, 1430–1436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawakatsu, T.; Stuart, T.; Valdes, M.; Breakfield, N.; Schmitz, R.J.; Nery, J.R.; Urich, M.A.; Han, X.; Lister, R.; Benfey, P.N.; et al. Unique cell-type-specific patterns of DNA methylation in the root meristem. Nat. Plants 2016, 2, 16058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hossain, M.S.; Kawakatsu, T.; Kim, K.D.; Zhang, N.; Nguyen, C.T.; Khan, S.M.; Batek, J.M.; Joshi, T.; Schmutz, J.; Grimwood, J.; et al. Divergent cytosine DNA methylation patterns in single-cell, soybean root hairs. New Phytol. 2017, 214, 808–819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Widman, N.; Feng, S.; Jacobsen, S.E.; Pellegrini, M. Epigenetic differences between shoots and roots in Arabidopsis reveals tissue-specific regulation. Epigenetics 2014, 9, 236–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seymour, D.K.; Koenig, D.; Hagmann, J.; Becker, C.; Weigel, D. Evolution of DNA Methylation Patterns in the Brassicaceae is Driven by Differences in Genome Organization. PLoS Genetics 2014, 10, e1004785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Y.; Rong, T.; Cao, M. Analysis of DNA methylation in different maize tissues. J. Genet. Genom. 2008, 35, 41–48. [Google Scholar] [CrossRef]
- Zemach, A.; Kim, M.Y.; Silva, P.; Rodrigues, J.A.; Dotson, B.; Brooks, M.D.; Zilberman, D. Local DNA hypomethylation activates genes in rice endosperm. Proc. Natl. Acad. Sci. USA 2010, 107, 18729–18734. [Google Scholar] [CrossRef] [Green Version]
- Hsieh, T.F.; Ibarra, C.A.; Silva, P.; Zemach, A.; Eshed-Williams, L.; Fischer, R.L.; Zilberman, D. Genome-wide demethylation of Arabidopsis endosperm. Science 2009, 324, 1451–1454. [Google Scholar] [CrossRef] [Green Version]
- Gahlaut, V.; Zinta, G.; Jaiswal, V.; Kumar, S. Quantitative Epigenetics: A New Avenue for Crop Improvement. Epigenomes 2020, 4, 25. [Google Scholar] [CrossRef]
- Pan, Y.; Wane, W.; Zhao, X.; Zhu, L.; Fu, B.; Li, Z. DNA methylation alterations of rice in response to cold stress. Omics J. 2011, 4, 364–369. [Google Scholar]
- Zhang, W.; Wang, N.; Yang, J.; Guo, H.; Liu, Z.; Zheng, X.; Li, S.; Xiang, F. The salt-induced transcription factor GmMYB84 confers salinity tolerance in soybean. Plant Sci. 2020, 291, 110326. [Google Scholar] [CrossRef]
- Qian, Y.; Hu, W.; Liao, J.; Zhang, J.; Ren, Q. The Dynamics of DNA methylation in the maize (Zea mays L.) inbred line B73 response to heat stress at the seedling stage. Biochem. Biophys. Res. Commun. 2019, 512, 742–749. [Google Scholar] [CrossRef]
- Gardiner, L.J.; Joynson, R.; Omony, J.; Rusholme-Pilcher, R.; Olohan, L.; Lang, D.; Bai, C.; Hawkesford, M.; Salt, D.; Spannagl, M.; et al. Hidden variation in polyploid wheat drives local adaptation. Genome Res. 2018, 28, 1319–1332. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.; Bhushan, B.; Gaikwad, K.; Yadav, O.P.; Kumar, S.; Rai, R.D. Induced defence responses of contrasting bread wheat genotypes under differential salt stress imposition. Indian J. Biochem. Biophys. 2015, 52, 75–85. [Google Scholar]
- Lämke, J.; Bäurle, I. Epigenetic and chromatin-based mechanisms in environmental stress adaptation and stress memory in plants. Genome Biol. 2017, 18, 124. [Google Scholar] [CrossRef]
- Kumar, G.; Rattan, U.K.; Singh, A.K. Chilling-mediated DNA methylation changes during dormancy and its release reveal the importance of epigenetic regulation during winter dormancy in Apple (Malus x domestica Borkh.). PLoS ONE 2016, 11, e0149934. [Google Scholar] [CrossRef] [Green Version]
- Bäurle, I. Plant Heat Adaptation: Priming in response to heat stress. F1000Research 2016, 5, 694. [Google Scholar] [CrossRef]
- Liu, T.; Li, Y.; Duan, W.; Huang, F.; Hou, X. Cold acclimation alters DNA methylation patterns and confers tolerance to heat and increases growth rate in Brassica rapa. J. Exp. Bot. 2017, 68, 1213–1224. [Google Scholar] [CrossRef] [Green Version]
- Kawakatsu, T.; Huang, S.C.; Jupe, F.; Sasaki, E.; Schmitz, R.J.; Urich, M.A.; Castanon, R.; Nery, J.R.; Barragan, C.; He, Y.; et al. Epigenomic diversity in global collection of Arabidopsis thaliana accession. Cell 2016, 166, 492–505. [Google Scholar] [CrossRef] [Green Version]
- Eichten, S.R.; Briskine, R.; Song, J.; Li, Q.; Swanson-Wagner, R.; Hermanson, P.J.; Waters, A.J.; Starr, E.; West, P.T.; Tiffin, P.; et al. Epigenetic and Genetic Influences on DNA Methylation Variation in Maize Populations. Plant Cell 2013, 25, 2783–2797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, Y.; Zhang, J.; Liu, Y.; Liu, S.; Liu, Z.; Duan, Z.; Wang, Z.; Zhu, B.; Guo, Y.L.; Tian, Z. DNA methylation footprints during soybean domestication and improvement. Genome Biol. 2018, 19, 128. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Xie, L.; Zhang, Q.; Ouyang, W.; Deng, L.; Guan, P.; Ma, M.; Li, Y.; Zhang, Y.; Xiao, Q.; et al. Integrative analysis of reference epigenomes in 20 rice varieties. Nat. Commun. 2020, 11, 2658. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, R.J.; He, Y.; Valdés-López, O.; Khan, S.M.; Joshi, T.; Urich, M.A.; Nery, J.R.; Diers, B.; Xu, D.; Stacey, G.; et al. Epigenome-wide inheritance of cytosine methylation variants in a recombinant inbred population. Genome Res. 2013, 23, 1663–1674. [Google Scholar] [CrossRef] [Green Version]
- Takata, M.; Kishima, Y.; Sano, Y. DNA Methylation Polymorphisms in Rice and Wild Rice Strains: Detection of Epigenetic Markers. Breed. Sci. 2005, 55, 57–63. [Google Scholar] [CrossRef] [Green Version]
- Schmitz, R.J.; Ecker, J.R. Epigenetic and epigenomic variation in Arabidopsis thaliana. Trends Plant Sci. 2012, 17, 149–154. [Google Scholar] [CrossRef] [Green Version]
- Ong-Abdullah, M.; Ordway, J.M.; Jiang, N.; Ooi, S.E.; Kok, S.Y.; Sarpan, N.; Azimi, N.; Hashim, A.T.; Ishak, Z.; Rosli, S.K.; et al. Loss of Karma transposon methylation underlies the mantled somaclonal variant of oil palm. Nature 2015, 525, 533–537. [Google Scholar] [CrossRef] [Green Version]
- Cocciolone, S.M.; Chopra, S.; Flint-Garcia, S.A.; McMullen, M.D.; Peterson, T. Tissue-specific patterns of a maize Myb transcription factor are epigenetically regulated. Plant J. 2001, 27, 467–478. [Google Scholar] [CrossRef] [Green Version]
- Quadrana, L.; Almeida, J.; Asís, R.; Duffy, T.; Dominguez, P.G.; Bermúdez, L.; Conti, G.; Corrêa da Silva, J.V.; Peralta, I.E.; Colot, V.; et al. Natural occurring epialleles determine vitamin e accumulation in tomato fruits. Nat. Commun. 2014, 5, 4027. [Google Scholar] [CrossRef] [Green Version]
- Hofmeister, B.T.; Lee, K.; Rohr, N.A.; Hall, D.W.; Schmitz, R.J. Stable inheritance of DNA methylation allows creation of epigenotype maps and the study of epiallele inheritance patterns in the absence of genetic variation. Genome Biol. 2017, 18, 155. [Google Scholar] [CrossRef]
- Cortijo, S.; Wardenaar, R.; Colomé-Tatché, M.; Gilly, A.; Etcheverry, M.; Labadie, K.; Caillieux, E.; Hospital, F.; Aury, J.M.; Wincker, P.; et al. Mapping the Epigenetic Basis of Complex Traits. Science 2014, 343, 1145–1148. [Google Scholar] [CrossRef]
- Furci, L.; Jain, R.; Stassen, J.; Berkowitz, O.; Whelan, J.; Roquis, D.; Baillet, V.; Colot, V.; Johannes, F.; Ton, J. Identification and characterisation of hypomethylated DNA loci controlling quantitative resistance in Arabidopsis. eLife 2019, 8, e40655. [Google Scholar] [CrossRef]
- Can, S.N.; Nunn, A.; Galanti, D.; Langenberger, D.; Becker, C.; Volmer, K.; Heer, K.; Opgenoorth, L.; Fernandez-Pozo, N.; Rensing, S.A. The EpiDiverse Plant Epigenome-Wide Association Studies (EWAS) Pipeline. Epigenomes 2021, 5, 12. [Google Scholar] [CrossRef]
- McCartney, D.L.; Hillary, R.F.; Stevenson, A.J.; Ritchie, S.J.; Walker, R.M.; Zhang, Q.; Morris, S.W.; Bermingham, M.L.; Campbell, A.; Murray, A.D.; et al. Epigenetic prediction of complex traits and death. Genome Biol. 2018, 19, 136. [Google Scholar] [CrossRef] [Green Version]
- Johannes, F.; Porcher, E.; Teixeira, F.K.; Saliba-Colombani, V.; Simon, M.; Agier, N.; Bulski, A.; Albuisson, J.; Heredia, F.; Audigier, P.; et al. Assessing the impact of transgenerational epigenetic variation on complex traits. PLoS Genet. 2009, 5, e1000530. [Google Scholar] [CrossRef]
- Hu, Y.; Morota, G.; Rosa, G.J.; Gianola, D. Prediction of plant height in arabidopsis thaliana using DNA methylation data. Genetics 2015, 201, 779–793. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Chen, G.; Hermanson, P.J.; Xu, Q.; Sun, C.; Chen, W.; Kan, Q.; Li, M.; Crisp, P.A.; Yan, J.; et al. Population-level analysis reveals the widespread occurrence and phenotypic consequence of DNA methylation variation not tagged by genetic variation in maize. Genome Biol. 2019, 20, 243. [Google Scholar] [CrossRef]
- López, C.M.R.; Wilkinson, M.J. Epi-fingerprinting and epi-interventions for improved crop production and food quality. Front. Plant Sci. 2015, 6, 397. [Google Scholar]
- Hauben, M.; Haesendonckx, B.; Standaert, E.; Van Der Kelen, K.; Azmi, A.; Akpo, H.; Breusegem, F.V.; Guisez, Y.; Bots, M.; Lambert, B.; et al. Energy use efficiency is characterized by an epigenetic component that can be directed through artificial selection to increase yield. Proc. Natl. Acad. Sci. USA 2009, 106, 20109–20114. [Google Scholar] [CrossRef] [Green Version]
- Tricker, P.J.; López, C.M.R.; Gibbings, G.; Hadley, P.; Wilkinson, M.J. Transgenerational, dynamic methylation of stomata genes in response to low relative humidity. Int. J. Mol. Sci. 2013, 14, 6674–6689. [Google Scholar] [CrossRef] [Green Version]
- Sun, H.; Guo, Z.; Gao, L.; Zhao, G.; Zhang, W.; Zhou, R.; Wu, Y.; Wang, H.; An, H.; Jia, J. DNA methylation pattern of Photoperiod-B1 is associated with photoperiod insensitivity in wheat (Triticum aestivum). New Phytol. 2014, 204, 682–692. [Google Scholar] [CrossRef]
- Kakoulidou, I.; Avramidou, E.V.; Baránek, M.; Brunel-Muguet, S.; Farrona, S.; Johannes, F.; Kaiserli, E.; Liberman-Lazarovich, M.; Martinelli, F.; Mladenov, V.; et al. Epigenetics for crop improvement in times of global change. Biology 2021, 10, 766. [Google Scholar] [CrossRef]
- Tricker, P.J. Transgenerational inheritance or resetting of stress-induced epigenetic modifications: Two sides of the same coin. Front. Plant Sci. 2015, 6, 699. [Google Scholar] [CrossRef] [Green Version]
- Varotto, S.; Tani, E.; Abraham, E.; Krugman, T.; Kapazoglou, A.; Melzer, R.; Radanović, A.; Miladinović, D. Epigenetics: Possible applications in climate-smart crop breeding. J. Exp. Bot. 2020, 71, 5223–5236. [Google Scholar] [CrossRef]
- Latutrie, M.; Gourcilleau, D.; Pujol, B. Epigenetic variation for agronomic improvement: An opportunity for vegetatively propagated crops. Am. J. Bot. 2019, 106, 1281–1284. [Google Scholar] [CrossRef]
- Sarkar, D.; Kar, S.K.; Chattopadhyay, A.; Shikha Rakshit, A.; Tripathi, V.K.; Dubey, P.; Abhilash, P. Low input sustainable agriculture: A viable climate-smart option for boosting food production in a warming world. Ecol. Indic. 2020, 115, 106412. [Google Scholar] [CrossRef]
- Xu, G.; Lyu, J.; Li, Q.; Liu, H.; Wang, D.; Zhang, M.; Springer, N.M.; Ross-Ibarra, J.; Yang, J. Evolutionary and functional genomics of DNA methylation in maize domestication and improvement. Nat. Commun. 2020, 11, 5539. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).