Abstract
Brassica napus L. provides high-quality edible oil and clean energy for humans. For a long time, rapeseed breeders have tried to breed improved varieties through traditional breeding strategies. However, B. napus is an allotetraploid species containing many repetitive sequences. It is very inefficient to change traits through traditional genetic methods such as hybridization and random mutagenesis due to gene redundancy. Today, the burgeoning CRISPR/Cas9 technology has been applied in polyploid rapeseed for gene function research and targeted genetic improvement because of its unique advantages of high efficiency and simplicity. This review summarizes current reports about the application of CRISPR/Cas9 system for gene function research and genetic improvement in rapeseed, involving important agronomic traits such as yield, oil content, and fatty acid composition. The application status of emerging precise genome editing technology in plants and several potential limitations and technical bottlenecks in rapeseed gene editing is discussed, which will provide confidence for researchers in rapeseed gene function research and genetic improvement through genome editing technology.
1. Introduction
Genome editing technologies are ideal tools to perform knockouts, chromosomal recombination, and site-directed insertion/substitution at specific sites of genes and chromosomal regions [1]. Recently, genome editing-mediated DNA modification based on site-directed nucleases (SDNs), including zinc finger nucleases (ZFNs), transcriptional activator-like effector nucleases (TALENs), and clustered regularly interspaced short palindromic repeats-associated protein (CRISPR/Cas), have been used in crop improvement by specific and accurate gene modification [2]. ZFNs and TALENs, two systems based on sequence-specific endonucleases including a specific DNA-binding domain and a cleavage domain, have been successfully used in rice [3], maize [3], wheat [4], and tomato [5]. However, the large-scale application of both in plants is limited as they both require a complex construction method [6,7]. Lately, the CRISPR/Cas technology, the forward genome editing technology, is rapidly replacing the application of ZFNs and TALENs in plants because of its convenience and high accuracy [8]. The CRISPR/Cas system was developed from the clustered regularly interspaced short palindromic repeats (CRISPR) adaptive immune system for eliminating invading viral and plasmid DNA in bacteria and archaea, which depend on the endonuclease action of CRISPR-associated (Cas) proteins with sequence specificity guided by CRISPR RNAs (crRNAs) [9]. In comparison with other SDNs, the CRISPR/Cas systems are more efficient for genome editing because the specificity of editing is determined by nucleotide complementarity of the guide RNA to a specific sequence [10]. The CRISPR/Cas tools can shorten the time of identifying gene function and developing new cultivars in the crop improvement field. To date, the CRISPR/Cas techniques have been successfully used in more than fifty plant species, including the oilseed crop rapeseed [11].
Rapeseed (Brassica napus L., AACC, 4n = 38), as one of the most important oil crops grown worldwide, is an allotetraploid species that originated from the hybridization of the diploid parents Brassica rapa (AA, 2n = 20) and Brassica. oleracea (CC, 2n = 18) 7500 years ago, and possesses a relatively complex genome containing many repetitive sequences [12]. Multiple copies of genes in B. napus exhibiting high sequence similarity interferes with gene function study. For rapeseed, knocking out all homologous genes is essential for obtaining a reliable phenotype [13]. In recent years, an efficient and universal CRISPR/Cas system has aroused lots of attention in multi-copy gene knockout due to the ability to introduce mutations at multiple sites concurrently [14,15]. In addition, due to the short domestication history and single ancestral sources [16], B. napus has very limited genetic diversity, which is one of the major hindrances for rapeseed improvement. In addition to genetic source from wild relatives, genetics and genomics approaches such as distant hybridization and transgenic technologies have extensively been applied to improve agronomic traits in rapeseed. Mutagenesis, created by random mutagens such as X-rays, ethyl methanesulfonate (EMS), and T-DNA insertion, is also another effective way to expand rapeseed germplasm for functional genomics research and breeding applications [17,18]. Although heritable and stable, mutagenesis from these methods is random and may result in unexpected mutations, requiring intensive screening which is time-consuming and laborious—especially for polyploid rapeseed [19]. Genome editing technologies, which can modify DNA at a specific site and limit unexcepted mutation, are perfect choices to achieve higher efficiency and accuracy in genetic modification than conventional breeding [20]. As the second most important oil crop, achieving further improvement of agronomical traits to meet the demand of the future has always been the main direction of rapeseed breeding. CRISPR/Cas9, as the most adequately developed and popular genome editing technology, has been successfully used for rapeseed gene modification and germplasm creation. This review summarizes the research status and future development direction of CRISPR/Cas9 in rapeseed gene function verification and genetic improvement, hoping to provide a reference for rapeseed breeding researchers.
2. CRISPR/Cas9: A Magnificent Tool for Plant Genome Editing
ZFNs and TALENs are two SDNs-mediated genome editing technologies that have been successfully applied in plants. Although able to create double-stranded breaks (DSBs) at a specific genomic site [21], ZFNs are not easily designed and constructed in plants. In addition, ZFNs are high in cost and can be affected by imprecise DNA sequence recognition [22,23]. In comparison, TALENs technology performs a lower off-target probability and a higher target-binding specificity [24]. However, the construction of TALENs is also tedious and complicated due to the complex tandem repeat feature in the DNA binding domains of the TALEN protein [1]. Therefore, the application of both ZFNs and TALENs has been limited due to the complexity, poorer precision, and lower cutting efficiency of targets. A simple, reliable, efficient, and economical method has become the urgent demand for precise plant genome modification [2].
The CRISPR/Cas system is a prokaryotic adaptive immune system against invading genetic elements, like invasive phages and plasmids [25]. This system is categorized as Type I, Type II, and Type III according to phylogeny, sequence, locus organization, and contents, among which the Type II system is the most studied due to its ability to produce DSBs in the target DNA [2]. Cas9 is derived from the type II prokaryotic CRISPR adaptive immune system of Streptococcus pyogenes [26]. This protein consists of separate RucV and HNH nuclease domains, which respectively cleaves one of the target DNA strands together and produces a blunt-ended DSB at the same time in vivo [26,27]. Unlike the type I and type III systems that utilize a large multi-Cas protein complex for CRISPR RNA (crRNA) binding and target sequence degradation [28], type II systems only require a single multi-functional Cas9 protein to recognize double-stranded DNA substrates and cleave each strand with a distinct nuclease domain (HNH or RuvC) [29]. In the endogenous CRISPR/Cas9 system, the Cas9 protein and the single guide RNA (gRNA or sgRNA) are two indispensable components for target cutting [30]. The gRNA comprises crRNA and trans-activating crRNA (tracrRNA), which contributes to crRNA maturation and Cas9 complex formation [30]. The crRNA contains a spacer which consists of a ~20 nt fragment and is complementary to a specific site of target genes, followed by a protospacer adjacent motif (PAM) in the target genes of interest [31]. Based on both the PAM sequence in the target DNA and RNA–DNA complementarity base pairing between the guide RNA sequence and the complementary target DNA sequence, Cas9 nuclease induces DSBs at ~3 bp upstream of the PAM motif [26]. Unlike ZFNs and TALENs, the Cas9/sgRNA nuclease complex can achieve precise specificity for its target site by the hybridization of the sgRNA to the target [26]. The DSBs are subsequently repaired either by the error-prone nonhomologous end-joining (NHEJ) pathway, which causes nucleotide insertions or deletions, or by the homology-directed repair (HDR) pathway directing DNA sequence substitutions in the sites [32]. The CRISPR/Cas9 system can improve the specificity of editing without complex protein engineering of DNA-recognition domains for each DNA target site to be modified [33]. Importantly, multiplex editing can be achieved by designing multiple sgRNAs [34], which makes CRISPR/Cas9 technology simpler and more efficient in creating mutations at the DNA targets sites than ZFNs and TALENs.
From the first use for plant gene editing in 2013 to now, CRISPR/Cas9 technology has been widely applied in 41 food crop species such as rice, wheat, tomato, etc., 15 industrial crops such as cichorium intybus, coffee, dandelion, etc., 8 ornamental crops such as lily, lotus, rose, etc., 6 oil crops such as rapeseed, soybean, sunflower, etc., 1 fiber crop (cotton), and 1 feed crop (alfalfa) [31,35,36]. These reports show that CRISPR/Cas9 is playing an important role in plant functional genomics research and crop improvement.
3. CRISPR/Cas9-Mediated Genomic Modification Accelerates Rapeseed Improvement
Rapeseed is an oilseed crop with high economic value and wide application [37]. Presently, improving agronomic traits by modifying the rapeseed genome through molecular genetics and genomics methods is the urgent demand for rapeseed farming and production. The novel genome editing technology CRISPR/Cas9 for convenient and accurate genomic modification can shorten the time to identify gene function and produce new breeding material. In 2017, the CRISPR/Cas9 system was first applied in rapeseed to target two BnALC homologous genes for site-directed mutagenesis to avoid seed loss by increasing shatter resistance [38]. One transgenic T1 plant with four alc mutant alleles using a single target sequence was obtained and was stably inherited to the offspring without off-target effect. First application of CRISPR/Cas9 in rapeseed offers new opportunities for its gene function research and breeding. In rapeseed, with the wide application of the CRISPR/Cas9 system, a general CRISPR/Cas9-based gene-editing procedure which contains multiple programs as illustrated in Figure 1 has been formed, including the selection of genes for editing according to target traits, the selection of single or multiple copy knockout, designing and synthesis of sgRNAs, the construction of CRISPR/Cas9 plasmids, transformation, the screening of target gene-edited rapeseed, and phenotypic analysis of mutants.
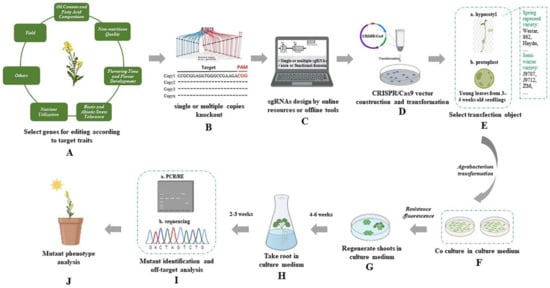
Figure 1.
The general workflow of CRISPR/Cas9-based gene editing in rapeseed. (A) Select genes for editing according to target traits. The traits that B. napus genes edited by CRISPR/Cas9 are involved in are shown here; (B) perform single or multiple copy knockout according to function differentiation-related information among homologous copies; (C) design sgRNAs for selected target genes using online and offline tools (sgRNAs are usually designed in exon or functional domain, which can lead to a greater probability of functional mutation); (D) CRISPR/Cas9 vector with the sgRNA coding sequence construction and transformation; (E) select genetic transformation object (the representative receptor materials successfully used for transformation for CRISPR/Cas9 vector are shown on the right); (F) deliver CRISPR/Cas9 vector by co-culturing Agrobacterium with transfection object in the culture medium; (G,H) differentiate callus and regenerate plants by inducing reagents in the culture medium, and identify positive plants by resistance or fluorescence; (I) identify target and/or off-target editing by (a) polymerase chain reaction restricted enzyme (PCR-RE) and/or (b) sequencing; (J) build target gene-edited rapeseed lines for phenotyping.
Over the last five years, CRISPR/Cas9-mediated genomic modification in rapeseed has been focused on important agronomical trait improvements. Here, we summarize the related research and expect to provide references and point out the direction for further application.
3.1. Yield
Seed size, seed numbers per silique, and silique numbers per plant are closely related to the yield of rapeseed [39]. ENHANCER3 OF DA1 (EOD3/CYP78A6) is reported to regulate silique length and seed size in Arabidopsis and wheat [40,41]. All four homologous copies of BnaEOD3 have been knocked out to increase yield by CRISPR/Cas9 in rapeseed, and the seed weight per plant of the quadrable mutants increased by 13.9% on average compared with wild type [42]. Moreover, other genes in the CYP78A subfamily have been identified that can play a role in seed development in many plants [43,44,45,46,47]. For example, KLUH/CYP78A5 was reported to control seed size in Arabidopsis, wheat, soybean, and tomato [46,47,48,49]. Like CYP78A6 and CYP78A9 in Arabidopsis, CYP78A3 in wheat promotes cell expansion in the integument of the embryo [40,41,43,45]. In rice, CYP78A11 and CYP78A13 affect the seed size by regulating stability between the embryo and endosperm [44,50,51]. In sweet cherry, PaCYP78A9 promotes fruit size [52]. Therefore, these genes may be considered as attractive targets for improving seed yield-related agronomic traits in B. napus genome engineering. The CLV signaling pathway has been reported to control yield-related traits in maize, rice, and tomato [53,54,55,56,57,58], and the CLAVATA3 (CLV3) and CLAVATA1 (CLV1) genes in the pathway regulate the multilocular trait in B. rapa and B. juncea, respectively [59,60]. Researchers evaluated the function of CLV pathway genes by knockout CLV3 and its receptors CLV1 and CLV2 using CRISPR/Cas9 and obtained mutations of both copies of each BnCLV gene in variety J9707 [61]. Double homozygous knockout of BnaA04.CLV3 and BnaC04.CLV3 showed heritable multilocular silique phenotype and provided excellent starting materials for high-yield breeding of rapeseed.
In rapeseed breeding, improving plant architecture is a major challenge. The branch number is one of the main factors influencing rapeseed plant architecture and is directly correlated to the yield. Semi-dwarf germplasm with increased branching was obtained for rapeseed breeding by the CRISPR/Cas9 system to edit two homologs of more axillary growth 1 (MAX1) that could repress the vegetative growth of axillary buds [62]. This study is the first report that CRISPR/Cas9 system has been applied in improving plant architecture in rapeseed and provides ideal germplasm resources for high-yield rapeseed breeding. In Arabidopsis, BREVIPEDICELLUS (BP) encodes a knotted1-like homeobox gene which controls the regulation of pedicel bending and leaf morphogenesis [63,64]. Similarly, Fan, et al. [65] found that the down-regulation of two close homologs of BP, namely BnaA03.BP and BnaC03.BP, could produce more compact plants by decreasing the branch angle, and constructed CRISPR/Cas9 vectors to knock them out. As expected, the homozygous mutations plants in the BnaA03.BP gene showed semi-dwarf, slightly drooping siliques and erect axillary buds. Also, the bnac03.bp homozygous mutants showed similar phenotypes to bnaa03.bp but were shorter in height. Strikingly, the homozygous mutants which simultaneously knocked out both BnaBP genes displayed extremely dwarf, smaller branch angles, and severely drooping and short siliques with sterility.
The yield loss was often caused by pod shatter occurring at the mature period in rapeseed, which accounts for about 20% of the total yield, up to 50% under adverse weather conditions [66]. To avoid production loss, rapeseed is usually harvested before full maturity, however, which causes chlorophyll-contaminated oil extracted from the immature seed and makes its quality lower [67]. Besides, as pod shatter plays a key role in affecting mechanized harvesting in rapeseed, enhancing pod-shattering resistance can extend the mechanical operation period and reduce the yield loss [68]. In Arabidopsis, the development of specialized silique tissues essential for fruit dehiscence has been revealed to be controlled by the basic regulators SHATTERPROOF1 (SHP1), SHATTERPROOF2(SHP2), INDEHISCENT (IND), ALCATRAZ (ALC), and JAGGED (JAG) [69,70]. For improving pod-shattering resistance of rapeseed, similar work has been done in editing BnALC [38], BnJAG.A08 [70], BnIND [71] and BnSHP1 and BnSHP2 homologs [72] with CRISPR/Cas9. The BnA03.ind/BnC03.ind mutants, bnjag.a08 mutants, and the mutants combining mutagenesis at five homeologs of BnSHP1 and BnSHP2 (BnSHP1A09, BnSHP1A04, BnSHP1C04-A, BnSHP2C04-B, and BnSHP2A05) showed significantly improved pod-shattering resistance, which provided good germplasm resources for pod-shattering resistance breeding.
3.2. Oil Content and Fatty Acid Composition
Increasing oil content and optimizing the fatty acid (FA) composition have always been two key goals for rapeseed breeding. Lysophosphatidic acid acyltransferase (LPAT), which catalyzes FAs into triacylglycerol (TAG), is an important enzyme in the Kennedy pathway, and promotes further oil production in the form of TAG [73]. Zhang, et al. [74] knocked out all seven BnLPAT2 homologous copies and four BnLPAT5 homologous copies using CRISPR/Cas9 to analyze their respective roles in the oil biosynthesis of rapeseed. The knockout mutants of different copies of BnLPAT2 and BnLPAT5 showed decreased oil content in varying degrees, with a 32% decrease in Bnlpat2 lines, 29% in Bnlpat5 lines, and 39% in Bnlpat2/Bnlpat5 double mutant lines. This report confirmed that the oil content could be altered by all the copies of the BnLPAT, and knockout of all copies displayed an enlarged size of oil bodies and decreased oil content. As same as LPAT, Glycerol-3-phosphate acyltransferase (GPAT), and diacylglycerol acyltransferase (DGAT) also control TAG biosynthesis [73]. Therefore, functional characterization of GPAT and DGAT in rapeseed is a promising work for the future.
For decades, yellow-seeded rapeseed has been preferred due to its relevant higher oil content, lower pigmentation, and fiber content compared with black-seeded counterparts [75]. Accumulating evidence shows that transparent testa 8 (TT8) regulates the accumulation of flavonoids in various crops [76,77,78,79,80]. The allelic variation of B. napus transparent testa 2 (BnTT2) on the C genome has been identified to participate in seed color formation and FA biosynthesis in rapeseed [81]. Using the CRISPR/Cas9 technology, yellow-seeded mutants were created by knockout BnTT8 [82] and BnTT2 homologs [83], which showed increased seed oil and altered FA composition. Like Arabidopsis, the formation of seed color in Brassica species is due to the deposition of an oxidized form of flavonoids, proanthocyanidins, in the endothelial layer of the inner integument of the seed coat [84]. In Arabidopsis, the MYB-bHLH-WD40 (MBW) complex plays an important role in the flavonoid pathway, including MYB5-TT8-TTG1, TT2-TT8-TTG1, TT2-GL3-TTG1, and TT2-EGL3-TTG1 [85]. Moreover, PAP2/MYB90 and PAP1/MYB75 were also determined as components of the MBW complex [86]. To date, only BnTT8 and BnTT2 yellow-seeded mutants have been created. It is very promising to create more yellow-seeded mutants by knocking out key genes in the flavonoid pathway.
Fatty acid composition is the main factor that determines the nutritional quality of vegetable oils, and optimizing fatty acid composition has always been the main goals in rapeseed improvement [18]. In order to develop new rapeseed variations with superior FA profile, the CRISPR/cas9 technology was applied to target multiple copies of the BnaFAD2 gene that could affect three major FA compositions (oleic, linoleic, and linolenic acid) and proportions in oilseed crops [87]. The oleic acid content of double mutants Bnafad2.A5/Bnafad2.C5 increased up to 80% comparison to 66.43% in wild type and increased significantly over the Bnafad2.A5 and Bnafad2.C5 single mutants, while the content of linoleic acid and linolenic acid decreased correspondingly. Furthermore, mutations at BnaFAD2.A5 changed the C18 fatty acid profile more significantly than BnaFAD2.C5.
3.3. Non-Nutritious Quality
Many nutrients remain in the seedcake after oil extraction in many oil crops, including rapeseed. Since the existence of anti-nutritional factors makes the seedcake taste bad and have low edible value, the rapeseed seedcake is currently only used as animal feed. Phytic acid (PA), also known as inositol hexaphosphate, is the main source of phosphorus in plants, accounting for 65–90% of total seed phosphorus in various plants from cereals to oilseeds [88]. Due to the lack of phytase, humans are unable to metabolize PA. When a large amount of PA is ingested, the absorption of minerals and proteins is reduced because PA can form indigestible complexes by binding with them [89]. To reduce the PA content in rapeseed, researchers have tried to used CRISPR/Cas9 to knock out the inositol tetrakisphosphate kinase (ITPK) gene that encodes the enzyme that catalyzes the penultimate step of phytate synthesis [90]. The homozygous triple mutant of BnITPK showed a 35% reduction in PA, while there was no change in plant performance [91].
Glucosinolates (GSLs) are also among the anti-nutritional factors, and eliminating or reducing the GSLs content in the rapeseed seed is necessary to improve feed and food value [92]. In Brassica species, GSLs are transported by the glucosinolate transporters (GTRs) from the vegetative tissues to seeds [92]. Recent research reported CRISPR/Cas9 gene editing for the knockout of BnGTR1 and BnGTR2 through an optimized protoplast transfection and regeneration system in rapeseed [93]. Regenerated plants with targeted mutations in BnGTR1 and BnGTR2, blocking glucosinolate transport, were obtained with a high mutation frequency in rapeseed. Low seed glucosinolate germplasms that have been created by editing BnaA06.GTR2, BnaC03.GTR2, BnaA02.GTR2 and BnaC02.GTR2, which have a higher functional similarity with GTR2 than the other two homologs in rapeseed, are not suitable for breeding due to their poor performance in other traits [94]. This may be because the edited genes are involved in the formation of other traits.
3.4. Flowering Time and Flower Development
Flowering time not only affects the yield in rapeseed, but also influences the sowing date of next rotation crops [95], and proper flowering time is an improved target in rapeseed breeding. TERMINAL FLOWER 1 (TFL1) is a member of the phosphatidylethanoamine-binding protein (PEBP) family, which can control flowering time in higher plants [96]. By CRISPR/Cas9, researchers have knocked out four different homologous copies of TFL1 in B. napus and found that mutation of only BnaC03.TFL1 enhanced earlier flowering [96]. The other copies, BnaA10.TFL1, BnaC03.TFL1 and BnaC09.TFL1, affected plant architecture. The Arabidopsis histone 3 lysine 36 (H3K36) methyltransferase SDG8 is a pleiotropic gene, which participates in the biological processes of several plants, including flowering time [97,98]. Two homologs of SDG8, namely, BnaSDG8.A and BnaSDG8.C in rapeseed were knocked out simultaneously using CRISPR/Cas9 to characterize and investigate their effects on floral transition [99]. Compared with the control, the knockout double mutants Bnasdg8.A/Bnasdg8.C showed an early flowering phenotype.
The seed yield of many Brassica crops is often hindered by abnormal flower organs, especially under abiotic stress [100]. However, the molecular mechanism of these abnormal floral organs is still not clear. As a class A gene in the ABCE model in the plant, APETALA2 (AP2) regulates floral organ development [101]. All four B. napus AP2 homologous genes have been targeted by the CRISPR/Cas9 system, and the quadruple mutant showed carpels sepals, missing petals, and a reduced number of stamens [102].
Rapeseed flower tourism is popular in many places around the world, and the ornamental value of rapeseed has attracted more and more attention in China. Therefore, different flower colors have become one of the goals in rapeseed genetic breeding. Carotenoids provide flowers and fruits with different colors, such as yellow, orange, and red, and these colors attract animals for pollination and seed dispersal [103]. The yellow flower color of B. napus is caused by carotenoids and the nuclear-encoded plastid enzyme zeaxanthin epoxidase (ZEP) is involved in carotenoid biosynthesis [104,105]. Both BnaA09.ZEP and BnaC09.ZEP were targeted by CRISPR/Cas9 technology using the yellow-flowered variety Westar as receptor material [104]. BnaA09.zep/BnaC09.zep double mutants exhibiting orange flowers with greatly enhanced lutein content and sharply reduced violaxanthin content were obtained. This study provides a useful orange-flowered germplasm resource for developing ornamental B. napus varieties. Carotenoid biosynthesis, as well as the related genes and enzymes, have been well-reported in many plants [106,107]. For example, some genes related to carotenoid metabolism in the petals of O. fragrans, including OfPSY, OfPDS, OfZDS, OfLCYE, OfLCYB, OfHYB, OfZEP, OfNCED, OfCCD1, and OfCCD4, have been identified [108,109,110,111]. Hitherto, the underlying molecular basis of the color formation in rapeseed flowers is not very clear [112]. Studying the function of carotenoid biosynthesis-related genes in rapeseed using CRISPR/Cas9 technology is important for breeding varieties with high ornamental value, as well as for learning more about the mechanism underlying flower coloration.
3.5. Biotic and Abiotic Stress Tolerance
Verticillium longisporum (Vl43) is a hemibiotrophic fungal pathogen of B. napus that causes yield reduction by impairing plant growth and resulting in premature senescence [113]. The knockout of AtCRT1a (calreticulin), a host susceptibility gene, caused a sharp decline in the sensitivity of plants to Vl43 in Arabidopsis [114]. The CRISPR/Cas9 system has been introduced to knockout CRT1a in B. napus to obtain Vl43-resistant materials. The double BnaC09.CRT1a/BnaA09.CRT1a mutants displayed reduced susceptibility to Vl43, while the BnaC09.CRT1a copy mutants showed a weaker reduced-susceptibility phenotype than the BnaA09. CRT1a [114].
Sclerotinia sclerotiorum induces sclerotinia stem rot in B. napus, usually leading to a serious yield loss of 5% to 100% [115]. Previous reports have shown WRKY11 and WRKY70 controls salicylic acid (SA) and jasmonic acid (JA) -induced disease resistance response to pathogens in Arabidopsis [116,117,118,119]. In 2018, researchers revealed that the plants with CRISPR/Cas9-mediated mutations of three homologous BnWRKY70 genes were more resistant to S. sclerotiorum in rapeseed [120], which provides theoretical guidance and germplasm resources for rapeseed with high resistance against S. sclerotinia.
Like other large-scale crops, rapeseed was often affected by weeds during the planting process. Weeds can be effectively controlled by herbicides to keep the yield and quality, and breeding herbicide-tolerant varieties is a profitable way to deal with weed threats. Glyphosate is very effective in controlling extensive weeds and causes little harm to the environment [121]. However, it is rarely used directly in rapeseed fields due to its toxicity to rapeseed. Studies have shown that mutations at the glyphosate-binding site of enolpyruvylshikimate-3-phosphate synthase (EPSPS) genes confer resistance to glyphosate in different plants [122,123,124]. By precise gene replacement of BnaC04EPSPS, induced by CRISPR/Cas9, the researchers have produced glyphosate-tolerant B. napus plants [125]. Acetolactate synthase (ALS) is considered as an important enzyme for branched-chain amino acid biosynthesis and as an ideal target of several important herbicides [126]. An efficient cytidine-deaminase-mediated base editor (CBE, C•G to T•A) system for the production of herbicide-resistant rapeseed by precisely editing BnALS1 genes has been reported [127].
Rapeseed is very sensitive to water deficits in the whole process of growth and development [128]. Therefore, it is of great significance to investigate the physiological and molecular mechanisms of rapeseed response to drought stress for drought tolerance breeding. There has been evidence that DELLA proteins can improve drought tolerance [129,130]. Using CRISPR/Cas9 to edit BnaRGA (REPRESSOR OF ga1-3) genes that code for DELLA proteins [131], a mutant BnaA6.rga-D displaying improved drought tolerance was obtained [132], which provided desirable drought-resistant germplasms for rapeseed breeding.
3.6. Nutrient Utilization
Promoting the efficiency of nutrition resource utilization will decrease the quantity of fertilizer to save rapeseed production costs and reduce water eutrophication. Boron (B) plays key roles in cell wall formation, pollen tube growth, membrane integrity, nitrogen fixation, sugar transport, and plant metabolism [133]. Usually, the available B concentration is very low in soils [134]. B deficiency inhibits the growth of plant apical and flower development, and finally leads to declined seed yield [133]. B. napus is hypersensitive to B deficiency and has the typical phenomenon of ‘flowering without seed setting’ under B deficiency [135]. In B. napus, CRISPR/Cas9-mediated knockout lines of BnaA9.WRKY47 which showed strong binding activity with conserved sequences containing a W box in the promoters of the B transport-related genes BnaNIP5;1s and BnaBOR1s had increased sensitivity to low B and lower contents of B than wild-type plants [136]. Combined with overexpression of BnaA9.WRKY47, it was demonstrated that BnaA9.WRKY47 contributed to improving the adaptability to B deficiency in rapeseed.
3.7. Others
Self-incompatibility (SI) is a genetic mechanism that prevents inbreeding in hermaphrodite angiosperms via the rejection of self-pollen, and SI lines are conducive to the utilization of heterosis [137]. SI is determined by the S locus, which harbors genes encoding the stigma papilla cell-specific S-receptor kinase (SRK) and the pollen surface-localized ligand of SRK/S-locus cysteine-rich protein/S-locus protein 11 (SCR/SP11) in cruciferous plants [138,139]. Despite being generated from two SI species, Brassica rapa and Brassica oleracea, B. napus is self-compatible (SC). The plasma membrane-tethered M-locus protein kinase (MLPK) has been reported to interact with the activated SRK [140]. Four paralogue MLPK genes in B. napus, BnaA3.MLPK, BnaC3.MLPK, BnaA4.MLPK, and BnaC4.MLPK, were identified and all four paralogous genes were simultaneously knocked out to reveal their function in controlling the SI response of rapeseed by CRISPR/Cas9 [141]. The quadruple Bnamlpk mutant has abundant pollen germination, elongated pollen tube, and a large number of seeds after self-pollination compared with the control. Similar work using CRISPR/Cas9 was done by Dou, et al. [142], creating new SI B. napus lines by targeting BnS6-Smi2, which specifically inhibits BnSCR7 transcription.
Extensive leaf shape diversity, including entire, serrated, and lobed leaves, exist in B. napus. The lobed leaves in rapeseed have been confirmed as a desirable trait in high-density planting and hybrid production [143]. Lobed leaves can improve heat transfer and the leaf lobes serve as a very good indicator character in hybrid production because they can be easily identified even in the early stages [144,145,146]. In B. napus, leaf lobing is regulated by an incompletely dominant locus at the distal end of chromosome A10 [147]. The gene expression differences between HY and its near-isogenic line (NIL) strongly suggest that the lobed leaf shape is positively related to enhanced A LATE MERISTEM IDENTITY1 (LMI1)-like gene (BnLMI1) transcript levels [144]. To test whether BnA10.LMI1 plays a role in rapeseed leaf lobe development process, the knockout mutations of BnA10.LMI1 gene were generated in both HY (the lobed-leaf parent) and J9707 (serrated leaf) genetic backgrounds by CRISPR/Cas9 technology [144]. In the HY background, the homozygous and heterozygous BnA10.LMI1 mutants had unlobed-leaf and intermediate leaf shapes respectively. In contrast, mutations at one or both BnA10.LMI1 alleles in the J9707 background exhibited similar phenotypes to the control in leaf shape, which indicated that the serrated leaf was a recessive trait and was obviously consistent with the incompletely dominant nature of the lobed-leaf trait. The phenotype of CRISPR/Cas9 mediated mutants indicates that BnA10.LMI1 promotes the formation of leaf lobes in rapeseed (Table 1).

Table 1.
List of gene-editing research by using CRISPR/Cas9 technology on rapeseed.
4. Conclusions and Future Perspectives
4.1. Advantages of CRISPR/Cas9 Application in Polyploid Rapeseed
Several potential limitations still worry researchers, such as multi-copy gene knockout and off-target effects. B. napus is an allotetraploid species, and most genes in B. napus are multiple-copy genes with redundant functions [12]. Due to gene redundancy, it is very inefficient to change traits through random mutations. It is often necessary to edit multiple genes to improve one trait. Fortunately, different single-guide RNAs (sgRNAs) can guide the Cas9 protein to specific sites, which offer an opportunity to achieve multiple gene editing by expressing Cas9 along with the multiple sgRNAs [148,149]. CRISPR/Cas9 technology has obvious advantages in polyploid rapeseed because multiple mutations can be induced in one step. In rapeseed, the most common strategy is to introduce mutations at multiple sites or genes concurrently by stacking multiple independent sgRNA-expressing cassettes. At present, there are more and more studies on multi-copy gene knockout in B. napus using CRISPR/Cas9, which shows that the prospect of CRISPR/Cas9 in the study of multi-copy gene function is encouraging in B. napus. The CRISPR/Cas9 system is an efficient tool and widely used for crop improvement either by a single or multiplex genome editing approach [150]. Analyzing the functions of homologous genes and gene family members with high sequence similarity frequently require multiple gene mutants. Based on many research facts are listed in this review, different sgRNAs connected in tandem show a huge advantage in multiple gene editing, due to their ability to efficiently generate mutants with multiple gene mutations [151]. The specificity of CRISPR/Cas9 technology for targeted gene editing in plants is also a worrying issue. Generally, to ensure that mutation is as specific as possible, potential off-target sites should be checked. Researchers can adopt some measures to avoid off-target mutations as much as possible. For example, combined with other CRISPR/Cas9 online design tools, BLAST searches against the Brassica genome using the full 23 nucleotide sequence (protospacer plus PAM) as a query can allow the selection of guides with no, or a minimal number of, predicted off-targets [152]. Recent studies report that off-target mutations can be tackled by designing precise sgRNA [153]. Therefore, it is essential to design sgRNAs with minimal off-target activities to avoid unexpected outcomes. Currently, to avoid off-target effects as much as possible, several free online prediction tools, such as CRISPOR [154] and CCTop [155], have been used to help researchers design sgRNA. Also, several machine learning-based methods for identifying sgRNAs with high on-target activity have been developed and applied to sgRNA activity prediction in agronomy [156,157]. The development of these methods helps to identify the activity of designed sgRNA in crops and avoid unexpected results. Many groups listed in this review have reported that they did not identify any off-target activity at potential off-target sites. This is consistent with the conclusion of Matres, et al. [158]: when gRNAs are carefully designed, off-target editing is negligible, and the frequency of occurrence is much lower than naturally occurring diversity in plants. For rapeseed breeders, the off-target mutation is not the main constraint, as any undesirable mutations will be low frequency, predictable, and testable [152]. In summary, CRISPR/Cas9-mediated genome editing technology has been proven to simultaneously alter multiple homologous genes without any off-target editing and generates mutations that are stable and heritable by offspring.
4.2. Identification of CRISPR Mutants and Detection of Mutation Types in Rapeseed
Amplification of single copies by polymerase chain reaction (PCR) using paralog-specific primers is a common method for identifying multi-copy mutants, followed by sequencing. For complex mutations, the amplicons need to be subcloned into the T-vector, followed by monoclonal sequencing. Mutation analysis work usually requires a large investment in sequencing costs and analysis time for lots of samples by Sanger sequencing. In addition, complex chimeric mutations caused by genome editing are difficult to decode. The Hi-TOM method developed by Liu, et al. [159] is gradually being more widely used because of its simplicity, rapidity, and high throughput. In this method, only two rounds of PCR are needed to complete the construction of a multi-sample mixed sequencing library. After obtaining the sequencing data, the resulting sequencing data are uploaded to the Hi-TOM online tool (http://www.hi-tom.net/hi-tom/) to obtain the detailed mutation sequence of each locus of each sample and the corresponding genotype information. At present, the Hi-TOM method has been applied in the identification of rapeseed mutants [42,82].
4.3. CRISPR Transformation Receptor Restriction Needs to Be Further Broken
So far, the application of CRISPR/Cas9 in rapeseed has mainly relied on stable transformation by Agrobacterium tumefaciens to deliver the CRISPR vector. However, several excellent B. napus varieties are usually not easy to transform due to a lack of the traits suitable for culture and regeneration [160]. At present, the reported varieties for Agrobacterium tumefaciens-mediated genetic transformation include spring rapeseed varieties and semi-winter varieties, among which the spring varieties include Westar, 862, and Haydn, etc., and the semi-winter varieties include J9707, J9712, and ZS6, etc. Also, there is a problem that more efficient transformation techniques are required for recalcitrant commercial rapeseed varieties [161]. One of the ways to help alleviate the bottleneck is to use morphogenic genes. For maize, this bottleneck has been mitigated by using the morphogenic genes WUS2 and BBM [162]. Over the past three decades, basic research has provided us with a detailed understanding of the genes that control morphogenesis. These insights will continue to provide inspiration for testing morphogenetic genes and, together with new methods to control expression, will lead to continuous improvements that expand the range of different rapeseed varieties suitable for transformation [163]. Moreover, the tissue culture procedures are often technically demanding, time-consuming and laborious. Therefore, developing no-tissue-culture-required delivery methods, such as nanoparticles or virus delivery, will contribute to further extending the application of CRISPR/Cas9 in rapeseed.
4.4. Application of CRISPR/Cas9 in High-Throughput Gene Editing
The precision, coverage, and flexibility provided by CRISPR/Cas9 are playing an important role in genome editing in rapeseed improvement. In recent years, CRISPR technology has been developed rapidly and has shown unparalleled advantages in high-throughput gene editing. The high efficiency of CRISPR/Cas9 technology is very suitable for high-throughput gene editing in various organisms and cell types [164]. Since the targeting specificity of CRISPR/Cas9 is conferred by a 20bp sgRNA, array-based synthesis of oligonucleotide libraries on a large scale can be easily generated. Such powerful genome-scale CRISPR/Cas9 mutagenesis systems have been successfully used for rice and maize research [165,166]. For example, Lu et al. (2017) used the CRISPR/Cas9 system to perform genome-scale mutagenesis in rice and generated a library of targeted loss-of-function mutants, which provided a useful resource for rice research and breeding. Furthermore, Liu et al. (2020) successfully targeted 743 candidate genes related to traits relevant for agronomy and nutrition by integrating multiplexed CRISPR/Cas9-based high-throughput targeted mutagenesis with genetic mapping and genomic approaches, which provided the guidance and reference for further optimizing experiments on high-throughput CRISPR in plants. In general, their research proves that it is feasible for the powerful genome-scale CRISPR/Cas9 mutagenesis systems to be applied to rapeseed. Utilizing this system to achieve high-throughput targeted gene editing and screen large-scale mutant libraries will be a good way to identify novel genes that can improve target traits in rapeseed varieties.
4.5. Application Prospect of CRISPR/Cas Precise Genome Editing in Rapeseed Improvement
Current progress in CRISPR technology has offered more and more opportunities for gene function study and genetic improvement that have not been seen before [167,168]. Clustered regularly interspaced short palindromic repeats interference (CRISPRi) and CRISPR-mediated gene activation (CRISPRa), which both derive from the CRISPR/Cas9 technology, can be used in gene functional studies. Both CRISPRi and CRISPRa can repress or activate multiple target genes simultaneously with no detectable off-target activities [169,170]. Furthmore, the CRISPR/Cas system has been applied to live cell chromatin imaging [171,172,173,174] and manipulation of chromatin topology [175,176]. In general, CRISPRi/a technologies lead to transient changes in gene expression [170,177,178]. Epigenetics has been an attractive target for crop improvement as it is a key factor for controlling biological pathways. DNA methylation, histone modification, and non-coding RNAs are all main epigenetic factors that contribute to regulating gene expression. CRISPR provides a genomic targeting system capable of interfacing with many aspects of the epigenome. CRISPR/Cas-based genome and epigenome editing has been used to improve drought stress tolerance in Arabidopsis by fusing dCas9 protein with a histone acetyltransferase (AtHA T1) and using this fused CRISPR/dCas9 system to target the abscisic acid (ABA)-responsive element-binding protein1 (AREB1)/ABRE-binding factor2 (ABF2) [179]. Two powerful emerging technologies, base editing and prime editing, have been applied in plants and have greatly enhanced the effectiveness of gene editing for crop improvement [180,181]. Desired changes can be installed by both base editors (BEs) and prime editors (PEs) without the donor DNA and a DSB introduction in the genome [182]. BEs and PEs generate edited plants with nucleobase precision as the incidence of DSB is usually low during the editing process [182]. BEs install C•G-to-T•A and A•T-to-G•C transitions [183,184,185] and have been successfully used in plants [186,187]. Several studies reported by Wu, et al. [127] suggest BEs have become an efficient tool for precise genetic modification of important agronomic traits in rapeseed. However, the base editing technologies only can generate the four transition mutations [188] and are constrained by the PAM motif and the editing window [189]. Fortunately, the recent breakthrough of prime editing based on the CRISPER/Cas system can overcome these limitations [190]. Prime editing has enabled search-and-replace editing instead of single base substitutions, conferring precision genome editing by installing small insertions, deletions, point mutations, and combination edits [191]. Prime editing was first applied in human cells [191] and has since been quickly developed for use in plants including rice [192,193,194,195,196], wheat [193], maize [197], and tomato [198] by multiple research groups around the word. Compared to other gene-editing technologies, prime editing can reduce the risk of unwanted off-target activities and poses few restrictions on the edited sequence [199]. With continuous development and progress, the CRISPR technology will likely further revolutionize basic research and precision breeding in rapeseed.
4.6. Government Regulation and the Future of Gene-Edited Rapeseed
Although the exogenous DNA could be excluded from the gene-edited crops by progeny separation, it is still difficult to commercialize gene-edited crops in many areas, such as the European Union and New Zealand, due to their following a ‘process basis’ that leads to expensive and time-consuming genetically modified (GM) safety tests. Recently, China’s agriculture ministry released preliminary guidelines which will be followed in the safety evaluation of gene-edited plants without foreign genes. Since the release of the new regulations, Chinese researchers have been very excited and eager to submit applications to use their gene-edited crops [200]. The preliminary guidelines state that gene-edited plants without foreign genes can directly apply for a production application safety certificate after an intermediate test when target traits do not increase environmental safety and food safety risks. In China, the process for receiving a biosafety certificate for gene-edited crops under the new guidelines shortens the approval time to one to two years in comparison to the six years needed for a GM crop to get biosafety approval [200]. The new guidelines open the door to the commercialization of gene-edited crops and provide new opportunities for Chinese breeders to develop gene-edited crops with superior traits.
At present, the CRISPR/Cas9 system has not been explored fully yet for trait improvement in rapeseed. One of the main reasons is that most studies have relied on stable transformation by Agrobacterium tumefaciens, which leads to randomness of gene insertion and is subject to GMO regulations. Developing transgene-free genome editing approaches in rapeseed are also required, which could provide a promising means for developing rapeseed varieties with reduced regulatory limitations. In this direction, novel delivery methods that do not introduce exogenous DNA may be able to avoid GM regulation. Recently, researchers have developed an optimized protoplast transient transfection method in rapeseed, which is an efficient solution for delivering CRISPR complexes [93]. This optimized protoplast regeneration protocol will provide important guidance for other rapeseed researchers. In addition, transgenic cotton was successfully produced by transforming pollen with magnetic nanoparticles and then pollinating plants with this magnetofected pollen [201]. Several other nanoparticles, including carbon nanotubes [202,203], DNA origami, and DNA nanostructures [204] have been successfully investigated for unassisted delivery of exogenous DNA. If delivery by nanomaterials without tissue culturing could be successfully applied in CRISPR/Cas9 for rapeseed genome editing, it would be a shortcut to creating non-GM rapeseed while avoiding strict GM regulations. With further development and remaining challenges gradually resolved, the CRISPR/Cas9 technology will produce more excellent rapeseed germplasm resources and create great economic value.
Author Contributions
Conceptualization, Q.T. and H.C.; writing—original draft preparation, Q.T.; writing parts of the manuscript—review & editing, Q.T., B.L., Y.F., W.Z., J.H. and H.C. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by the National Natural Science Foundation of China (32001583 and U2004149), the Key Research Plan Project of Shaanxi Province (2020ZDLNY04-01) and the Key Scientific Research Projects of Colleges and Universities in Henan Province (K20A180031).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Zhang, Y.; Ma, X.; Xie, X.; Liu, Y.-G. CRISPR/Cas9-Based Genome Editing in Plants. Prog. Mol. Biol. Transl. Sci. 2017, 149, 133–150. [Google Scholar] [CrossRef] [PubMed]
- Gupta, D.; Bhattacharjee, O.; Mandal, D.; Sen, M.K.; Dey, D.; Dasgupta, A.; Kazi, T.A.; Gupta, R.; Sinharoy, S.; Acharya, K.; et al. CRISPR-Cas9 system: A new-fangled dawn in gene editing. Life Sci. 2019, 232, 116636. [Google Scholar] [CrossRef] [PubMed]
- Shan, Q.; Wang, Y.; Chen, K.; Liang, Z.; Li, J.; Zhang, Y.; Zhang, K.; Liu, J.; Voytas, D.F.; Zheng, X.; et al. Rapid and efficient gene modification in rice and Brachypodium using TALENs. Mol. Plant 2013, 6, 1365–1368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Cheng, X.; Shan, Q.; Zhang, Y.; Liu, J.; Gao, C.; Qiu, J.-L. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 2014, 32, 947–951. [Google Scholar] [CrossRef]
- Čermák, T.; Baltes, N.J.; Čegan, R.; Zhang, Y.; Voytas, D.F. High-frequency, precise modification of the tomato genome. Genome Biol. 2015, 16, 232. [Google Scholar] [CrossRef] [Green Version]
- Gaj, T.; Gersbach, C.A.; Barbas, C.F., III. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013, 31, 397–405. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Rudis, M.R.; Cheplick, M.H.; Millwood, R.J.; Yang, J.-P.; Ondzighi-Assoume, C.A.; Montgomery, G.A.; Burris, K.P.; Mazarei, M.; Chesnut, J.D.; et al. Lipofection-mediated genome editing using DNA-free delivery of the Cas9/gRNA ribonucleoprotein into plant cells. Plant Cell Rep. 2020, 39, 245–257. [Google Scholar] [CrossRef]
- Woo, J.W.; Kim, J.; Kwon, S.I.; Corvalán, C.; Cho, S.W.; Kim, H.; Kim, S.-G.; Kim, S.-T.; Choe, S.; Kim, J.-S. DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat. Biotechnol. 2015, 33, 1162–1164. [Google Scholar] [CrossRef]
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar] [CrossRef]
- Lino, C.A.; Harper, J.C.; Carney, J.P.; Timlin, J.A. Delivering CRISPR: A review of the challenges and approaches. Drug Deliv. 2018, 25, 1234–1257. [Google Scholar] [CrossRef] [Green Version]
- Metje-Sprink, J.; Menz, J.; Modrzejewski, D.; Sprink, T. DNA-free genome editing: Past, present and future. Front. Plant Sci. 2018, 9, 1957. [Google Scholar] [CrossRef] [PubMed]
- Chalhoub, B.; Denoeud, F.; Liu, S.; Parkin, I.; Tang, H.; Wang, X.; Chiquet, J.; Belcram, H.; Tong, C.; Samans, B. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 2014, 345, 950–953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wells, R.; Trick, M.; Soumpourou, E.; Clissold, L.; Morgan, C.; Werner, P.; Gibbard, C.; Clarke, M.; Jennaway, R.; Bancroft, I. The control of seed oil polyunsaturate content in the polyploid crop species Brassica napus. Mol. Breed. 2014, 33, 349–362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, W.; Zhou, H.; Bi, H.; Fromm, M.; Yang, B.; Weeks, D.P. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 2013, 41, e188. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-P.; Xing, H.-L.; Dong, L.; Zhang, H.-Y.; Han, C.-Y.; Wang, X.-C.; Chen, Q.-J. Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 2015, 16, 144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beilstein, M.A.; Al-Shehbaz, I.A.; Kellogg, E. Brassicaceae phylogeny and trichome evolution. Am. J. Bot. 2006, 93, 607–619. [Google Scholar] [CrossRef]
- Amosova, A.V.; Zoshchuk, S.A.; Volovik, V.T.; Shirokova, A.V.; Horuzhiy, N.E.; Mozgova, G.V.; Yurkevich, O.Y.; Artyukhova, M.A.; Lemesh, V.A.; Samatadze, T.E.; et al. Phenotypic, biochemical and genomic variability in generations of the rapeseed (Brassica napus L.) mutant lines obtained via chemical mutagenesis. PLoS ONE 2019, 14, e0221699. [Google Scholar] [CrossRef]
- Tang, S.; Liu, D.; Lu, S.; Yu, L.; Li, Y.; Lin, S.; Li, L.; Du, Z.; Liu, X.; Ma, W.; et al. Development and screening of EMS mutants with altered seed oil content or fatty acid composition in Brassica napus. Plant J. 2020, 104, 1410–1422. [Google Scholar] [CrossRef]
- Parry, M.A.J.; Madgwick, P.J.; Bayon, C.; Tearall, K.; Hernandez-Lopez, A.; Baudo, M.; Rakszegi, M.; Hamada, W.; Al-Yassin, A.; Ouabbou, H.; et al. Mutation discovery for crop improvement. J. Exp. Bot. 2009, 60, 2817–2825. [Google Scholar] [CrossRef] [Green Version]
- Chen, K.; Wang, Y.; Zhang, R.; Zhang, H.; Gao, C. CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 2019, 70, 667–697. [Google Scholar] [CrossRef]
- Farooq, R.; Hussain, K.; Nazir, S.; Javed, M.R.; Masood, N. CRISPR/Cas9; A robust technology for producing genetically engineered plants. Cell. Mol. Biol. 2018, 64, 31–38. [Google Scholar] [CrossRef]
- Ramirez, C.; Foley, J.E.; Wright, D.A.; Müller-Lerch, F.; Rahman, S.H.; Cornu, T.I.; Winfrey, R.J.; Sander, J.D.; Fu, F.; Townsend, J.A.; et al. Unexpected failure rates for modular assembly of engineered zinc fingers. Nat. Methods 2008, 5, 374–375. [Google Scholar] [CrossRef] [PubMed]
- Sanjana, N.E.; Cong, L.; Zhou, Y.; Cunniff, M.M.; Feng, G.G.; Zhang, F. A transcription activator-like effector toolbox for genome engineering. Nat. Protoc. 2012, 7, 171–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joung, J.K.; Sander, J.D. TALENs: A widely applicable technology for targeted genome editing. Nat. Rev. Mol. Cell Biol. 2013, 14, 49–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altenbuchner, J. Editing of the Bacillus subtilis genome by the CRISPR-Cas9 system. Appl. Environ. Microbiol. 2016, 82, 5421–5427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Villion, M.; Moineau, S. The double-edged sword of CRISPR-Cas systems. Cell Res. 2013, 23, 15–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van der Oost, J.; Westra, E.R.; Jackson, R.N.; Wiedenheft, B. Unravelling the structural and mechanistic basis of CRISPR–Cas systems. Nat. Rev. Microbiol. 2014, 12, 479–492. [Google Scholar] [CrossRef]
- Makarova, K.S.; Haft, D.H.; Barrangou, R.; Brouns, S.J.J.; Charpentier, E.; Horvath, P.; Moineau, S.; Mojica, F.J.M.; Wolf, Y.I.; Yakunin, A.F.; et al. Evolution and classification of the CRISPR–Cas systems. Nat. Rev. Microbiol. 2011, 9, 467–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hryhorowicz, M.; Lipiński, D.; Zeyland, J.; Słomski, R. CRISPR/Cas9 immune system as a tool for genome engineering. Arch. Immunol. Ther. Exp. 2016, 65, 233–240. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.; Yang, F.; Zhang, J.; Liu, H.; Rahman, S.; Islam, S.; Ma, W.; She, M. Application of CRISPR/Cas9 in crop quality improvement. Int. J. Mol. Sci. 2021, 22, 4206. [Google Scholar] [CrossRef] [PubMed]
- Symington, L.S.; Gautier, J. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 2011, 45, 247–271. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.H.; Davis, K.M.; Liu, D.R. Chemical biology approaches to genome editing: Understanding, controlling, and delivering programmable nucleases. Cell Chem. Biol. 2016, 23, 57–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y.; et al. A Robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef] [PubMed]
- Agnès, R.; Pauline, C.; Wendy, H.J.E.T.i.L.S. Use of CRISPR systems in plant genome editing: Toward new opportunities in agriculture. Emerg. Top. Life Sci. 2017, 1, 169–182. [Google Scholar]
- Li, W.; Teng, F.; Li, T.; Zhou, Q. Simultaneous generation and germline transmission of multiple gene mutations in rat using CRISPR-Cas systems. Nat. Biotechnol. 2013, 31, 684–686. [Google Scholar] [CrossRef]
- Xiao, Y.G.; Sun, Q.B.; Kang, X.J.; Chen, C.B.; Ni, M. SHORT HYPOCOTYL UNDER BLUE1 or HAIKU2 mixepression alters canola and Arabidopsis seed development. New Phytol. 2016, 209, 636–649. [Google Scholar] [CrossRef]
- Braatz, J.; Harloff, H.J.; Mascher, M.; Stein, N.; Himmelbach, A.; Jung, C. CRISPR-Cas9 targeted mutagenesis leads to simultaneous modification of different homoeologous gene copies in polyploid oilseed rape (Brassica napus). Plant Physiol. 2017, 174, 935–942. [Google Scholar] [CrossRef] [Green Version]
- Li, N.; Shi, J.Q.; Wang, X.F.; Liu, G.H.; Wang, H.Z. A combined linkage and regional association mapping validation and fine mapping of two major pleiotropic QTLs for seed weight and silique length in rapeseed (Brassica napus L.). BMC Plant Biol. 2014, 14, 114. [Google Scholar] [CrossRef] [Green Version]
- Fang, W.; Wang, Z.; Cui, R.; Li, J.; Li, Y. Maternal control of seed size by EOD3/CYP78A6 in Arabidopsis thaliana. Plant J. 2012, 70, 929–939. [Google Scholar] [CrossRef]
- Ma, M.; Wang, Q.; Li, Z.J.; Cheng, H.H.; Liu, X.; Song, W.; Appels, R.; Zhao, H. Expression ofTaCYP78A3, a gene encoding cytochrome P450 CYP78A3 protein in wheat (Triticum aestivum L.), affects seed size. Plant J. 2015, 83, 312–325. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.H.U.; Hu, L.; Zhu, M.; Zhai, Y.; Khan, S.U.; Ahmar, S.; Amoo, O.; Zhang, K.; Fan, C.; Zhou, Y. Targeted mutagenesis of EOD3 gene in Brassica napus L. regulates seed production. J. Cell. Physiol. 2021, 236, 1996–2007. [Google Scholar] [CrossRef] [PubMed]
- Sotelo-Silveira, M.; Cucinotta, M.; Chauvin, A.-L.; Montes, R.A.C.; Colombo, L.; Marsch-Martínez, N.; de Folter, S. Cytochrome P450CYP78A9 is involved in Arabidopsis reproductive development. Plant Physiol. 2013, 162, 779–799. [Google Scholar] [CrossRef] [Green Version]
- Nagasawa, N.; Hibara, K.-I.; Heppard, E.P.; Velden, K.A.V.; Luck, S.; Beatty, M.; Nagato, Y.; Sakai, H. GIANT EMBRYO encodes CYP78A13, required for proper size balance between embryo and endosperm in rice. Plant J. 2013, 75, 592–605. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Meyerowitz, E.M. Overexpression of a gene encoding a cytochrome P450, CYP78A9, induces large and seedless fruit in Arabidopsis. Plant Cell 2000, 12, 1541–1550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakrabarti, M.; Zhang, N.; Sauvage, C.; Muños, S.; Blanca, J.; Cañizares, J.; Diez, M.J.; Schneider, R.; Mazourek, M.; McClead, J.; et al. A cytochrome P450 regulates a domestication trait in cultivated tomato. Proc. Natl. Acad. Sci. USA 2013, 110, 17125–17130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adamski, N.M.; Anastasiou, E.; Eriksson, S.; O’Neill, C.M.; Lenhard, M. Local maternal control of seed size by KLUH/CYP78A5 -dependent growth signaling. Proc. Natl. Acad. Sci. USA 2009, 106, 20115–20120. [Google Scholar] [CrossRef] [Green Version]
- Ma, M.; Zhao, H.; Li, Z.; Hu, S.; Song, W.; Liu, X. TaCYP78A5 regulates seed size in wheat (Triticum aestivum). J. Exp. Bot. 2016, 67, 1397–1410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, B.; Dai, A.; Wei, H.; Yang, S.; Wang, B.; Jiang, N.; Feng, X. Arabidopsis KLU homologue GmCYP78A72 regulates seed size in soybean. Plant Mol. Biol. 2016, 90, 33–47. [Google Scholar] [CrossRef]
- Xu, F.; Fang, J.; Ou, S.; Gao, S.; Zhang, F.; Du, L.; Xiao, Y.; Wang, H.; Sun, X.; Chu, J.; et al. Variations in CYP78A13 coding region influence grain size and yield in rice. Plant Cell Environ. 2015, 38, 800–811. [Google Scholar] [CrossRef]
- Miyoshi, K.; Ahn, B.-O.; Kawakatsu, T.; Ito, Y.; Itoh, J.-I.; Nagato, Y.; Kurata, N. PLASTOCHRON1, a timekeeper of leaf initiation in rice, encodes cytochrome P450. Proc. Natl. Acad. Sci. USA 2004, 101, 875–880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, X.; Liu, C.; Song, L.; Li, Y.; Li, M. PaCYP78A9, a cytochrome P450, regulates fruit size in sweet cherry (Prunus avium L.). Front. Plant Sci. 2017, 8, 2076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taguchi-Shiobara, F.; Yuan, Z.; Hake, S.; Jackson, D. The fasciated ear2 gene encodes a leucine-rich repeat receptor-like protein that regulates shoot meristem proliferation in maize. Genes Dev. 2001, 15, 2755–2766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzaki, T. The gene FLORAL ORGAN NUMBER1 regulates floral meristem size in rice and encodes a leucine-rich repeat receptor kinase orthologous to Arabidopsis CLAVATA1. Development 2004, 131, 5649–5657. [Google Scholar] [CrossRef] [Green Version]
- Bommert, P.; Lunde, C.; Nardmann, J.; Vollbrecht, E.; Running, M.; Jackson, D.; Hake, S.; Werr, W. Thick tassel dwarf1 encodes a putative maize ortholog of the Arabidopsis CLAVATA1 leucine-rich repeat receptor-like kinase. Development 2005, 132, 1235–1245. [Google Scholar] [CrossRef] [Green Version]
- Chu, H.; Qian, Q.; Liang, W.; Yin, C.; Tan, H.; Yao, X.; Yuan, Z.; Yang, J.; Huang, H.; Luo, D.; et al. The FLORAL ORGAN NUMBER4 gene encoding a putative ortholog of Arabidopsis CLAVATA3 regulates apical meristem size in rice. Plant Physiol. 2006, 142, 1039–1052. [Google Scholar] [CrossRef] [Green Version]
- Bommert, P.; Nagasawa, N.S.; Jackson, D. Quantitative variation in maize kernel row number is controlled by the FASCIATED EAR2 locus. Nat. Genet. 2013, 45, 334–337. [Google Scholar] [CrossRef]
- Xu, C.; Liberatore, K.L.; MacAlister, C.A.; Huang, Z.; Chu, Y.-H.; Jiang, K.; Brooks, C.; Ogawa-Ohnishi, M.; Xiong, G.; Pauly, M.; et al. A cascade of arabinosyltransferases controls shoot meristem size in tomato. Nat. Genet. 2015, 47, 784–792. [Google Scholar] [CrossRef]
- Fan, C.; Wu, Y.; Yang, Q.; Yang, Y.; Meng, Q.; Zhang, K.; Li, J.; Wang, J.; Zhou, Y. A novel single-nucleotide mutation in a CLAVATA3 gene homolog controls a multilocular silique trait in Brassica rapa L. Mol. Plant 2014, 7, 1788–1792. [Google Scholar] [CrossRef] [Green Version]
- Xu, P.; Cao, S.Q.; Hu, K.N.; Wang, X.H.; Huang, W.; Wang, G.; Lv, Z.W.; Liu, Z.S.; Wen, J.; Yi, B.; et al. Trilocular phenotype in Brassica juncea L. resulted from interruption of CLAVATA1 gene homologue (BjMc1) transcription. Sci. Rep. 2017, 7, 3498. [Google Scholar] [CrossRef]
- Yang, Y.; Zhu, K.; Li, H.; Han, S.; Meng, Q.; Khan, S.U.; Fan, C.; Xie, K.; Zhou, Y. Precise editing of CLAVATA genes in Brassica napus L. regulates multilocular silique development. Plant Biotechnol. J. 2018, 16, 1322–1335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, M.; Zhang, L.; Tang, M.; Liu, J.; Liu, H.; Yang, H.; Fan, S.; Terzaghi, W.; Wang, H.; Hua, W. Knockout of two Bna MAX 1 homologs by CRISPR /Cas9-targeted mutagenesis improves plant architecture and increases yield in rapeseed (Brassica napus L.). Plant Biotechnol. J. 2020, 18, 644–654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lincoln, C.; Long, J.; Yamaguchi, J.; Hake, S.S.J.T.P.C.O. A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell 1994, 6, 1859–1876. [Google Scholar] [PubMed] [Green Version]
- Venglat, S.P.; Dumonceaux, T.; Rozwadowski, K.; Parnell, L.; Babic, V.; Keller, W.; Martienssen, R.; Selvaraj, G.; Datla, R. The homeobox gene BREVIPEDICELLUS is a key regulator of inflorescence architecture in Arabidopsis. Proc. Natl. Acad. Sci. USA 2002, 99, 4730–4735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, S.; Zhang, L.; Tang, M.; Cai, Y.; Liu, J.; Liu, H.; Liu, J.; Terzaghi, W.; Wang, H.; Hua, W.; et al. CRISPR/Cas9-targeted mutagenesis of the BnaA03.BP gene confers semi-dwarf and compact architecture to rapeseed (Brassica napus. L). Plant Biotechnol. J. 2021, 19, 2383–2385. [Google Scholar] [CrossRef]
- Hu, Q.; Hua, W.; Yin, Y.; Zhang, X.K.; Liu, L.J.; Shi, J.Q.; Zhao, Y.G.; Qin, L.; Chen, C.; Wang, H. Rapeseed research and production in China. Crop J. 2017, 5, 127–135. [Google Scholar] [CrossRef] [Green Version]
- Hu, Z.Y.; Yang, H.L.; Zhang, L.; Wang, X.F.; Liu, G.H.; Wang, H.Z.; Hua, W. A large replum-valve joint area is associated with increased resistance to pod shattering in rapeseed. J. Plant Res. 2015, 128, 813–819. [Google Scholar] [CrossRef]
- Kuai, J.; Sun, Y.; Liu, T.; Zhang, P.; Zhou, M.; Wu, J.; Zhou, G. Physiological mechanisms behind differences in pod shattering resistance in rapeseed (Brassica napus L.) varieties. PLoS ONE 2016, 11, e0157341. [Google Scholar] [CrossRef] [Green Version]
- Ballester, P.; Ferrándiz, C. Shattering fruits: Variations on a dehiscent theme. Curr. Opin. Plant Biol. 2017, 35, 68–75. [Google Scholar] [CrossRef]
- Zaman, Q.; Chu, W.; Hao, M.; Shi, Y.; Sun, M.; Sang, S.-F.; Mei, D.; Cheng, H.; Liu, J.; Li, C.; et al. CRISPR/Cas9-mediated multiplex genome editing of JAGGED gene in Brassica napus L. Biomolecules 2019, 9, 725. [Google Scholar] [CrossRef] [Green Version]
- Zhai, Y.; Cai, S.; Hu, L.; Yang, Y.; Amoo, O.; Fan, C.; Zhou, Y. CRISPR/Cas9-mediated genome editing reveals differences in the contribution of INDEHISCENT homologues to pod shatter resistance in Brassica napus L. Theor. Appl. Genet. 2019, 132, 2111–2123. [Google Scholar] [CrossRef] [PubMed]
- Zaman, Q.U.; Wen, C.; Yuqin, S.; Mengyu, H.; Desheng, M.; Jacqueline, B.; Baohong, Z.; Chao, L.; Qiong, H. Characterization of SHATTERPROOF homoeologs and CRISPR-Cas9-mediated genome editing enhances pod-shattering resistance in Brassica napus L. CRISPR J. 2021, 4, 360–370. [Google Scholar] [CrossRef] [PubMed]
- Chapman, K.D.; Ohlrogge, J.B. Compartmentation of triacylglycerol accumulation in plants. J. Biol. Chem. 2012, 287, 2288–2294. [Google Scholar] [CrossRef] [Green Version]
- Zhang, K.; Nie, L.; Cheng, Q.; Yin, Y.; Chen, K.; Qi, F.; Zou, D.; Liu, H.; Zhao, W.; Wang, B.; et al. Effective editing for lysophosphatidic acid acyltransferase 2/5 in allotetraploid rapeseed (Brassica napus L.) using CRISPR-Cas9 system. Biotechnol. Biofuels 2019, 12, 225. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.J.; Zhu, S.; Yuan, Y.; Wang, Y.; Zeng, L.; Batley, J.; Wang, Y.-P. Transcriptomic comparison between developing seeds of yellow- and black-seeded Brassica napus reveals that genes influence seed quality. BMC Plant Biol. 2019, 19, 203. [Google Scholar] [CrossRef]
- Li, P.; Chen, B.; Zhang, G.; Chen, L.; Dong, Q.; Wen, J.; Mysore, K.; Zhao, J. Regulation of anthocyanin and proanthocyanidin biosynthesis by Medicago truncatula bHLH transcription factor MtTT8. New Phytol. 2016, 210, 905–921. [Google Scholar] [CrossRef]
- Escaray, F.J.; Passeri, V.; Perea-García, A.; Antonelli, C.J.; Damiani, F.; Ruiz, O.A.; Paolocci, F. The R2R3-MYB TT2b and the bHLH TT8 genes are the major regulators of proanthocyanidin biosynthesis in the leaves of Lotus species. Planta 2017, 246, 243–261. [Google Scholar] [CrossRef]
- Lim, S.-H.; Kim, D.-H.; Kim, J.K.; Lee, J.-Y.; Ha, S.-H. A radish basic helix-loop-helix transcription factor, RsTT8 acts a positive regulator for anthocyanin biosynthesis. Front. Plant Sci. 2017, 8, 1917. [Google Scholar] [CrossRef]
- Nemesio-Gorriz, M.; Blair, P.B.; Dalman, K.; Hammerbacher, A.; Arnerup, J.; Stenlid, J.; Mukhtar, S.M.; Elfstrand, M. Identification of Norway spruce MYB-bHLH-WDR transcription factor complex members linked to regulation of the flavonoid pathway. Front. Plant Sci. 2017, 8, 305. [Google Scholar] [CrossRef] [Green Version]
- Li, C.H.; Qiu, J.; Huang, S.R.; Yin, J.M.; Yang, G.S. AaMYB3 interacts with AabHLH1 to regulate proanthocyanidin accumulation in Anthurium andraeanum (Hort.)—Another strategy to modulate pigmentation. Hortic. Res. 2019, 6, 14. [Google Scholar] [CrossRef] [Green Version]
- Zhou, L.; Li, Y.; Hussain, N.; Li, Z.; Wu, D.; Jiang, L. Allelic variation of BnaC.TT2.a and its association with seed coat color and fatty acids in rapeseed (Brassica napus L.). PLoS ONE 2016, 11, e0146661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhai, Y.; Yu, K.; Cai, S.; Hu, L.; Amoo, O.; Xu, L.; Yang, Y.; Ma, B.; Jiao, Y.; Zhang, C.; et al. Targeted mutagenesis of BnTT8 homologs controls yellow seed coat development for effective oil production in Brassica napus L. Plant Biotechnol. J. 2020, 18, 1153–1168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, T.; Chen, X.; Guo, T.; Rong, H.; Chen, Z.; Sun, Q.; Batley, J.; Jiang, J.; Wang, Y. Targeted knockout of BnTT2 homologues for yellow-seeded Brassica napus with reduced flavonoids and improved fatty acid composition. J. Agric. Food Chem. 2020, 68, 5676–5690. [Google Scholar] [CrossRef] [PubMed]
- Lepiniec, L.; Debeaujon, I.; Routaboul, J.-M.; Baudry, A.; Pourcel, L.; Nesi, N.; Caboche, M. Genetics and biochemistry of seed flavonoids. Annu. Rev. Plant Biol. 2006, 57, 405–430. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Grain, D.; Bobet, S.; Le Gourrierec, J.; Thévenin, J.; Kelemen, Z.; Lepiniec, L.; Dubos, C. Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB–bHLH–WDR complexes and their targets in Arabidopsis seed. New Phytol. 2014, 202, 132–144. [Google Scholar] [CrossRef]
- Appelhagen, I.; Jahns, O.; Bartelniewoehner, L.; Sagasser, M.; Weisshaar, B.; Stracke, R. Leucoanthocyanidin dioxygenase in Arabidopsis thaliana: Characterization of mutant alleles and regulation by MYB–BHLH–TTG1 transcription factor complexes. Gene 2011, 484, 61–68. [Google Scholar] [CrossRef]
- Huang, H.B.; Cui, T.T.; Zhang, L.L.; Yang, Q.-Y.; Yang, Y.; Xie, K.B.; Fan, C.C.; Zhou, Y.M. Modifications of fatty acid profile through targeted mutation at BnaFAD2 gene with CRISPR/Cas9-mediated gene editing in Brassica napus. Theor. Appl. Genet. 2020, 133, 2401–2411. [Google Scholar] [CrossRef]
- Overturf, K.; Raboy, V.; Cheng, Z.J.; Hardy, R.W. Mineral availability from barley low phytic acid grains in rainbow trout (Oncorhynchus mykiss) diets. Aquac. Nutr. 2003, 9, 239–246. [Google Scholar] [CrossRef]
- Shi, J.; Wang, H.; Schellin, K.; Li, B.; Faller, M.; Stoop, J.M.; Meeley, R.B.; Ertl, D.; Ranch, J.P.; Glassman, K. Embryo-specific silencing of a transporter reduces phytic acid content of maize and soybean seeds. Nat. Biotechnol. 2007, 25, 930–937. [Google Scholar] [CrossRef]
- Sun, Y.; Thompson, M.; Lin, G.; Butler, H.; Gao, Z.; Thornburgh, S.; Yau, K.; Smith, D.A.; Shukla, V.K. Inositol 1,3,4,5,6-pentakisphosphate 2-kinase from maize: Molecular and biochemical characterization. Plant Physiol. 2007, 144, 1278–1291. [Google Scholar] [CrossRef] [Green Version]
- Sashidhar, N.; Harloff, H.J.; Potgieter, L.; Jung, C. Gene editing of three BnITPK genes in tetraploid oilseed rape leads to significant reduction of phytic acid in seeds. Plant Biotechnol. J. 2020, 18, 2241–2250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nour-Eldin, H.H.; Andersen, T.G.; Burow, M.; Madsen, S.R.; Jørgensen, M.E.; Olsen, C.E.; Dreyer, I.; Hedrich, R.; Geiger, D.; Halkier, B.A. NRT/PTR transporters are essential for translocation of glucosinolate defence compounds to seeds. Nature 2012, 488, 531–534. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Sandgrind, S.; Moss, O.; Guan, R.; Ivarson, E.; Wang, E.S.; Kanagarajan, S.; Zhu, L.-H. Efficient protoplast regeneration protocol and CRISPR/Cas9-mediated editing of glucosinolate transporter (GTR) genes in rapeseed (Brassica napus L.). Front. Plant Sci. 2021, 12, 680859. [Google Scholar] [CrossRef] [PubMed]
- Tan, Z.; Xie, Z.; Dai, L.; Zhang, Y.; Zhao, H.; Tang, S.; Wan, L.; Yao, X.; Guo, L.; Hong, D. Genome- and transcriptome-wide association studies reveal the genetic basis and the breeding history of seed glucosinolate content in Brassica napus. Plant Biotechnol. J. 2022, 20, 211–225. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Long, Y.; Zhang, L.B.; Dalton-Morgan, J.; Batley, J.; Yu, L.J.; Meng, J.L.; Li, M.T. Genome wide analysis of flowering time trait in multiple environments via high-throughput genotyping technique in Brassica napus L. PLoS ONE 2015, 10, e0119425. [Google Scholar] [CrossRef] [Green Version]
- Sriboon, S.; Li, H.; Guo, C.; Senkhamwong, T.; Dai, C.; Liu, K. Knock-out of TERMINAL FLOWER 1 genes altered flowering time and plant architecture in Brassica napus. BMC Genet. 2020, 21, 52. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Zhao, Z.; Dong, A.; Soubigou-Taconnat, L.; Renou, J.-P.; Steinmetz, A.; Shen, W.-H. Di- and tri- but not monomethylation on histone H3 lysine 36 marks active transcription of genes involved in flowering time regulation and other processes in Arabidopsis thaliana. Mol. Cell. Biol. 2008, 28, 1348–1360. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Z.; Yu, Y.; Meyer, D.; Wu, C.; Shen, W.-H. Prevention of early flowering by expression of FLOWERING LOCUS C requires methylation of histone H3 K36. Nat. Cell Biol. 2005, 7, 1256–1260. [Google Scholar] [CrossRef]
- Jiang, L.; Li, D.; Jin, L.; Ruan, Y.; Shen, W.-H.; Liu, C. Histone lysine methyltransferases BnaSDG8.A and BnaSDG8.C are involved in the floral transition in Brassica napus. Plant J. 2018, 95, 672–685. [Google Scholar] [CrossRef]
- Zhou, Y.T.; Wang, H.Y.; Zhou, L.; Wang, M.P.; Li, H.P.; Wang, M.L.; Zhao, Y. Analyses of the floral organ morphogenesis and the differentially expressed genes of an apetalous flower mutant in Brassica napus. Plant Cell Rep. 2008, 27, 9–20. [Google Scholar] [CrossRef]
- Jofuku, K.D.; Boer, B.; Okamuro, M.J.P.C. Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 1994, 6, 1211–1225. [Google Scholar] [PubMed] [Green Version]
- Zhang, Y.; Huang, S.; Wang, X.; Liu, J.; Guo, X.; Mu, J.; Tian, J.; Wang, X. Defective APETALA2 Genes Lead to Sepal Modification in Brassica crops. Front. Plant Sci. 2018, 9, 367. [Google Scholar] [CrossRef] [PubMed]
- Kevan, P.G. Floral colors through the insect eye: What they are and what they mean. In The Handbook of Experimental Pollination Biology; Van Nostrand Reinhold Company: New York, NY, USA, 1983. [Google Scholar]
- Liu, Y.; Ye, S.; Yuan, G.; Ma, X.; Heng, S.; Yi, B.; Ma, C.; Shen, J.; Tu, J.; Fu, T.; et al. Gene silencing of BnaA09.ZEP and BnaC09.ZEP confers orange color in Brassica napus flowers. Plant J. 2020, 104, 932–949. [Google Scholar] [CrossRef] [PubMed]
- Nikolov, L.A. Brassicaceae flowers: Diversity amid uniformity. J. Exp. Bot. 2019, 70, 2623–2635. [Google Scholar] [CrossRef]
- Nisar, N.; Li, L.; Lu, S.; Khin, N.C.; Pogson, B.J. Carotenoid metabolism in plants. Mol. Plant 2015, 8, 68–82. [Google Scholar] [CrossRef] [Green Version]
- Yuan, H.; Zhang, J.; Nageswaran, D.; Li, L. Carotenoid metabolism and regulation in horticultural crops. Hortic. Res. 2015, 2, 15036. [Google Scholar] [CrossRef] [Green Version]
- Huang, F.-C.; Molnár, P.; Schwab, W. Cloning and functional characterization of carotenoid cleavage dioxygenase 4 genes. J. Exp. Bot. 2009, 60, 3011–3022. [Google Scholar] [CrossRef] [Green Version]
- Baldermann, S.; Kato, M.; Kurosawa, M.; Kurobayashi, Y.; Fujita, A.; Fleischmann, P.; Watanabe, N. Functional characterization of a carotenoid cleavage dioxygenase 1 and its relation to the carotenoid accumulation and volatile emission during the floral development of Osmanthus fragrans Lour. J. Exp. Bot. 2010, 61, 2967–2977. [Google Scholar] [CrossRef] [Green Version]
- Han, Y.; Li, L.; Dong, M.; Yuan, W.; Shang, F. cDNA cloning of the phytoene synthase (PSY) and expression analysis of PSY and carotenoid cleavage dioxygenase genes in Osmanthus fragrans. Biologia 2013, 68, 258–263. [Google Scholar] [CrossRef] [Green Version]
- Han, Y.J.; Wang, X.H.; Chen, W.C.; Dong, M.F.; Yuan, W.J.; Liu, X.; Shang, F.D. Differential expression of carotenoid-related genes determines diversified carotenoid coloration in flower petal of Osmanthus fragrans. Tree Genet. Genomes 2014, 10, 329–338. [Google Scholar] [CrossRef]
- Yin, N.-W.; Wang, S.-X.; Jia, L.-D.; Zhu, M.-C.; Yang, J.; Zhou, B.-J.; Yin, J.-M.; Lu, K.; Wang, R.; Li, J.-N.; et al. Identification and characterization of major constituents in different-colored rapeseed petals by UPLC–HESI-MS/MS. J. Agric. Food Chem. 2019, 67, 11053–11065. [Google Scholar] [CrossRef] [PubMed]
- Lopisso, D.T.; Knüfer, J.; Koopmann, B.; Von Tiedemann, A. The vascular pathogen Verticillium longisporum does not affect water relations and plant responses to drought stress of its host, Brassica napus. Phytopathology 2017, 107, 444–454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pröbsting, M.; Schenke, D.; Hossain, R.; Häder, C.; Thurau, T.; Wighardt, L.; Schuster, A.; Zhou, Z.; Ye, W.; Rietz, S.; et al. Loss of function of CRT1a (calreticulin) reduces plant susceptibility to Verticillium longisporum in both Arabidopsis thaliana and oilseed rape (Brassica napus). Plant Biotechnol. J. 2020, 18, 2328–2344. [Google Scholar] [CrossRef] [PubMed]
- Saharan, G.S.; Mehta, N. Sclerotinia Diseases of Crop Plants: Biology, Ecology and Disease Management; Springer: Berlin/Heidelberg, Germany, 2008. [Google Scholar]
- Jiang, C.-H.; Huang, Z.-Y.; Xie, P.; Gu, C.; Li, K.; Wang, D.-C.; Yu, Y.-Y.; Fan, Z.-H.; Wang, C.-J.; Wang, Y.-P.; et al. Transcription factors WRKY70 and WRKY11 served as regulators in rhizobacterium Bacillus cereus AR156-induced systemic resistance to Pseudomonas syringae pv. tomato DC3000 in Arabidopsis. J. Exp. Bot. 2016, 67, 157–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Journot-Catalino, N.; Somssich, I.E.; Roby, D.; Kroj, T. The transcription factors WRKY11 and WRKY17 act as negative regulators of basal resistance in Arabidopsis thaliana. Plant Cell 2006, 18, 3289–3302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Brader, G.; Palva, E.T. The WRKY70 transcription factor: A node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 2004, 16, 319–331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shim, J.S.; Jung, C.; Lee, S.; Min, K.; Lee, Y.-W.; Choi, Y.; Lee, J.S.; Song, J.T.; Kim, J.-K.; Choi, Y.D. AtMYB44 regulates WRKY70 expression and modulates antagonistic interaction between salicylic acid and jasmonic acid signaling. Plant J. 2013, 73, 483–495. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Lin, L.; Liu, D.; Wu, D.; Fang, Y.; Wu, J.; Wang, Y. CRISPR/Cas9-mediated multiplex genome editing of the BnWRKY11 and BnWRKY70 genes in Brassica napus L. Int. J. Mol. Sci. 2018, 19, 2716. [Google Scholar] [CrossRef] [Green Version]
- Duke, S.O.; Powles, S.B. Glyphosate: A once-in-a-century herbicide. Pest Manag. Sci. 2008, 64, 319–325. [Google Scholar] [CrossRef]
- García, M.J.; Palma-Bautista, C.; Vazquez-Garcia, J.G.; Rojano-Delgado, A.M.; Osuna, M.D.; Torra, J.; De Prado, R. Multiple mutations in the EPSPS and ALS genes of Amaranthus hybridus underlie resistance to glyphosate and ALS inhibitors. Sci. Rep. 2020, 10, 17681. [Google Scholar] [CrossRef]
- Li, J.; Peng, Q.; Han, H.; Nyporko, A.; Kulynych, T.; Yu, Q.; Powles, S.; Nyporko, A.; Kulynych, T. Glyphosate resistance in Tridax procumbens via a novel EPSPS Thr-102-Ser substitution. J. Agric. Food Chem. 2018, 66, 7880–7888. [Google Scholar] [CrossRef]
- Yu, Q.; Jalaludin, A.; Han, H.; Chen, M.; Sammons, R.D.; Powles, S.B. Evolution of a double amino acid substitution in the 5-enolpyruvylshikimate-3-phosphate synthase in Eleusine indica conferring high-level glyphosate resistance. Plant Physiol. 2015, 167, 1440–1447. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Wan, L.; Xin, Q.; Zhang, X.; Song, Y.; Wang, P.; Hong, D.; Fan, Z.; Yang, G. Optimizing glyphosate tolerance in rapeseed by CRISPR/Cas9-based geminiviral donor DNA replicon system with Csy4-based single-guide RNA processing. J. Exp. Bot. 2021, 72, 4796–4808. [Google Scholar] [CrossRef] [PubMed]
- Powles, S.B.; Yu, Q. Evolution in action: Plants resistant to herbicides. Annu. Rev. Plant Biol. 2010, 61, 317–347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.; Chen, C.; Xian, G.; Liu, D.; Lin, L.; Yin, S.; Sun, Q.; Fang, Y.; Zhang, H.; Wang, Y. Engineering herbicide-resistant oilseed rape by CRISPR/Cas9-mediated cytosine base-editing. Plant Biotechnol. J. 2020, 18, 1857–1859. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Monroe, J.G.; Suhail, Y.; Villiers, F.; Mullen, J.; Pater, D.; Hauser, F.; Jeon, B.W.; Bader, J.S.; Kwak, J.M.; et al. Molecular and systems approaches towards drought-tolerant canola crops. New Phytol. 2016, 210, 1169–1189. [Google Scholar] [CrossRef] [Green Version]
- Nir, I.; Shohat, H.; Panizel, I.; Olszewski, N.; Aharoni, A.; Weiss, D. The tomato DELLA protein PROCERA acts in guard cells to promote stomatal closure. Plant Cell 2017, 29, 3186–3197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Liu, L.; Cheng, C.; Ren, Z.; Xu, S.; Li, X. GAI functions in the plant response to dehydration stress in Arabidopsis thaliana. Int. J. Mol. Sci. 2020, 21, 819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, H.; Wu, J.-J.; Tang, T.; Liu, K.-D.; Dai, C. CRISPR/Cas9-mediated genome editing efficiently creates specific mutations at multiple loci using one sgRNA in Brassica napus. Sci. Rep. 2017, 7, 7489. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Yan, G.; Duan, Z.; Wang, Z.; Kang, C.; Guo, L.; Liu, K.; Tu, J.; Shen, J.; Yi, B.; et al. Roles of the Brassica napus DELLA protein BnaA6.RGA, in modulating drought tolerance by interacting with the ABA signaling component BnaA10.ABF2. Front. Plant Sci. 2020, 11, 577. [Google Scholar] [CrossRef]
- Marschner, P. Marschner’s Mineral Nutrition of Higher Plants, 3rd ed.; Academic Press: San Diego, CA, USA, 2012. [Google Scholar]
- Shorrocks, V.M. The occurrence and correction of boron deficiency. Plant Soil 1997, 193, 121–148. [Google Scholar] [CrossRef]
- Xu, F.S.; Wang, Y.H.; Ying, W.H.; Meng, J.L. Inheritance of boron nutrition efficiency in Brassica napus. J. Plant Nutr. 2002, 25, 901–912. [Google Scholar] [CrossRef]
- Feng, Y.N.; Cui, R.; Wang, S.L.; He, M.L.; Hua, Y.P.; Shi, L.; Ye, X.S.; Xu, F.S. Transcription factor BnaA9.WRKY47 contributes to the adaptation of Brassica napus to low boron stress by upregulating the boric acid channel gene BnaA3.NIP5;1. Plant Biotechnol. J. 2020, 18, 1241–1254. [Google Scholar] [CrossRef] [Green Version]
- De Nettancourt, D. Incompatibility and Incongruity in Wild and Cultivated Plants; Springer: Berlin/Heidelberg, Germany, 2001. [Google Scholar]
- Schopfer, C.R.; Nasrallah, M.E.; Nasrallah, J.B. The male determinant of self-incompatibility in Brassica. Science 1999, 286, 1697–1700. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, G.; Kai, N.; Hirose, T.; Fukui, K.; Nishio, T.; Takayama, S.; Isogai, A.; Watanabe, M.; Hinata, K. Genomic organization of the S locus: Identification and characterization of genes in SLG/SRK region of S9 haplotype of Brassica campestris (syn. rapa). Genetics 1999, 153, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Kakita, M.; Murase, K.; Iwano, M.; Matsumoto, T.; Watanabe, M.; Shiba, H.; Isogai, A.; Takayama, S. Two distinct forms of M-locus protein kinase localize to the plasma membrane and interact directly with S-locus receptor kinase to transduce self-incompatibility signaling in Brassica rapa. Plant Cell 2007, 19, 3961–3973. [Google Scholar] [CrossRef] [Green Version]
- Chen, F.; Yang, Y.; Li, B.; Liu, Z.; Khan, F.; Zhang, T.; Zhou, G.; Tu, J.; Shen, J.; Yi, B.; et al. Functional analysis of M-locus protein kinase revealed a novel regulatory mechanism of self-incompatibility in Brassica napus L. Int. J. Mol. Sci. 2019, 20, 3303. [Google Scholar] [CrossRef] [Green Version]
- Dou, S.; Zhang, T.; Tu, J.; Shen, J.; Yi, B.; Wen, J.; Fu, T.; Dai, C.; Ma, C. Generation of novel self-incompatible Brassica napus by CRISPR/Cas9. Plant Biotechnol. J. 2021, 19, 875–877. [Google Scholar] [CrossRef]
- Hui-Ming, P.U.; Shou-Zhong, F.U.; Cun-Kou, Q.I.; Zhang, J.F.; Yi-Mei, W.U.; Gao, J.Q.; Chen, X.J. Inheritance of divided leaf trait of rapeseed (Brassica napus) and application in hybrid breeding. Chin. J. Oil Crop Sci. 2001, 23, 60–62. [Google Scholar]
- Hu, L.; Zhang, H.; Yang, Q.; Meng, Q.; Han, S.; Nwafor, C.C.; Khan, M.H.U.; Fan, C.; Zhou, Y. Promoter variations in a homeobox gene, BnA10.LMI1, determine lobed leaves in rapeseed (Brassica napus L.). Theor. Appl. Genet. 2018, 131, 2699–2708. [Google Scholar] [CrossRef]
- Vogel, S. Leaves in the lowest and highest winds: Temperature, force and shape. New Phytol. 2009, 183, 13–26. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.-H.; Zhang, J.; Liu, D.; Stiller, W.; Liu, D.; Zhang, Z.; Llewellyn, D.; Wilson, I. Integrated mapping and characterization of the gene underlying the okra leaf trait in Gossypium hirsutum L. J. Exp. Bot. 2016, 67, 763–774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ni, X.Y.; Huang, J.X.; Ali, B.; Zhou, W.J.; Zhao, J.Y. Genetic analysis and fine mapping of the LOBED-LEAF 1 (BnLL1) gene in rapeseed (Brassica napus L.). Euphytica 2015, 204, 29–38. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.L.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.B.; Jiang, W.Y.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [Green Version]
- Lawrenson, T.; Shorinola, O.; Stacey, N.; Li, C.D.; Østergaard, L.; Patron, N.; Uauy, C.; Harwood, W. Induction of targeted, heritable mutations in barley and Brassica oleracea using RNA-guided Cas9 nuclease. Genome Biol. 2015, 16, 258–270. [Google Scholar] [CrossRef] [Green Version]
- Lohani, N.; Jain, D.; Singh, M.B.; Bhalla, P.L. Engineering multiple abiotic stress tolerance in canola, Brassica napus. Front. Plant Sci. 2020, 11, 3. [Google Scholar] [CrossRef]
- Xiong, X.; Liu, W.; Jiang, J.; Xu, L.; Huang, L.; Cao, J. Efficient genome editing of Brassica campestris based on the CRISPR/Cas9 system. Mol. Genet. Genom. 2019, 294, 1251–1261. [Google Scholar] [CrossRef]
- Lawrenson, T.; Hundleby, P.; Harwood, W. Creating targeted gene knockouts in Brassica oleracea using CRISPR/Cas9. Methods Mol. Biol. 2019, 1917, 155–170. [Google Scholar] [CrossRef]
- Jacinto, F.V.; Link, W.; Ferreira, B.I. CRISPR/Cas9-mediated genome editing: From basic research to translational medicine. J. Cell. Mol. Med. 2020, 24, 3766–3778. [Google Scholar] [CrossRef] [Green Version]
- Concordet, J.-P.; Haeussler, M. CRISPOR: Intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 2018, 46, W242–W245. [Google Scholar] [CrossRef]
- Stemmer, M.; Thumberger, T.; Keyer, M.D.S.; Wittbrodt, J.; Mateo, J.L. CCTop: An intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PLoS ONE 2015, 10, e0124633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niu, M.; Lin, Y.; Zou, Q. sgRNACNN: Identifying sgRNA on-target activity in four crops using ensembles of convolutional neural networks. Plant Mol. Biol. 2021, 105, 483–495. [Google Scholar] [CrossRef] [PubMed]
- Hesami, M.; Najafabadi, M.Y.; Adamek, K.; Torkamaneh, D.; Jones, A.M.P. Synergizing off-target predictions for in silico insights of CENH3 knockout in cannabis through CRISPR/Cas. Molecules 2021, 26, 2053. [Google Scholar] [CrossRef] [PubMed]
- Matres, J.M.; Hilscher, J.; Datta, A.; Armario-Nájera, V.; Baysal, C.; He, W.; Huang, X.; Zhu, C.; Valizadeh-Kamran, R.; Trijatmiko, K.R.; et al. Genome editing in cereal crops: An overview. Transgenic Res. 2021, 30, 461–498. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, C.; Jiao, X.; Zhang, H.; Song, L.; Li, Y.; Gao, C.; Wang, K. Hi-TOM: A platform for high-throughput tracking of mutations induced by CRISPR/Cas systems. Sci. China Life Sci. 2019, 62, 1–7. [Google Scholar] [CrossRef]
- Hamada, H.; Linghu, Q.; Nagira, Y.; Miki, R.; Taoka, N.; Imai, R. An in planta biolistic method for stable wheat transformation. Sci. Rep. 2017, 7, 11443. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Bhalla, P.L. In vitro shoot regeneration from commercial cultivars of Australian canola (Brassica napus L.). Aust. J. Agric. Res. 2004, 55, 753–756. [Google Scholar] [CrossRef]
- Ananiev, E.V.; Wu, C.; Chamberlin, M.A.; Svitashev, S.; Schwartz, C.; Gordon-Kamm, W.; Tingey, S. Artificial chromosome formation in maize (Zea mays L.). Chromosoma 2009, 118, 157–177. [Google Scholar] [CrossRef] [Green Version]
- Gordon-Kamm, B.; Sardesai, N.; Arling, M.; Lowe, K.; Hoerster, G.; Betts, S.; Jones, T. Using morphogenic genes to improve recovery and regeneration of transgenic plants. Plants 2019, 8, 38. [Google Scholar] [CrossRef] [Green Version]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.-J.; Jian, L.M.; Xu, J.T.; Zhang, Q.H.; Zhang, M.L.; Jin, M.L.; Peng, Y.; Yan, J.L.; Han, B.Z.; Liu, J.; et al. High-throughput CRISPR/Cas9 mutagenesis streamlines trait gene identification in maize. Plant Cell 2020, 32, 1397–1413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Y.M.; Ye, X.; Guo, R.M.; Huang, J.; Wang, W.; Tang, J.Y.; Tan, L.T.; Zhu, J.-K.; Chu, C.C.; Qian, Y.W. Genome-wide targeted mutagenesis in rice using the CRISPR/Cas9 system. Mol. Plant 2017, 10, 1242–1245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, C. Genome engineering for crop improvement and future agriculture. Cell 2021, 184, 1621–1635. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Malzahn, A.A.; Sretenovic, S.; Qi, Y. The emerging and uncultivated potential of CRISPR technology in plant science. Nat. Plants 2019, 5, 778–794. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, L.A.; Larson, M.H.; Morsut, L.; Liu, Z.; Brar, G.A.; Torres, S.E.; Stern-Ginossar, N.; Brandman, O.; Whitehead, E.H.; Doudna, J.A.; et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 2013, 154, 442–451. [Google Scholar] [CrossRef] [Green Version]
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 2013, 152, 1173–1183. [Google Scholar] [CrossRef] [Green Version]
- Khosravi, S.; Ishii, T.; Dreissig, S.; Houben, A. Application and prospects of CRISPR/Cas9-based methods to trace defined genomic sequences in living and fixed plant cells. Chromosom. Res. 2020, 28, 7–17. [Google Scholar] [CrossRef]
- Wu, X.; Mao, S.; Ying, Y.; Krueger, C.J.; Chen, A.K. Progress and challenges for live-cell imaging of genomic loci using CRISPR-based platforms. Genom. Proteom. Bioinform. 2019, 17, 119–128. [Google Scholar] [CrossRef]
- Fu, Y.; Rocha, P.; Luo, V.M.; Raviram, R.; Deng, Y.; Mazzoni, E.O.; Skok, J.A. CRISPR-dCas9 and sgRNA scaffolds enable dual-colour live imaging of satellite sequences and repeat-enriched individual loci. Nat. Commun. 2016, 7, 11707. [Google Scholar] [CrossRef]
- Ma, H.; Naseri, A.; Reyes-Gutierrez, P.; Wolfe, S.A.; Zhang, S.; Pederson, T. Multicolor CRISPR labeling of chromosomal loci in human cells. Proc. Natl. Acad. Sci. USA 2015, 112, 3002–3007. [Google Scholar] [CrossRef] [Green Version]
- Yim, Y.Y.; Teague, C.D.; Nestler, E.J. In vivo locus-specific editing of the neuroepigenome. Nat. Rev. Neurosci. 2020, 21, 471–484. [Google Scholar] [CrossRef] [PubMed]
- Adli, M. The CRISPR tool kit for genome editing and beyond. Nat. Commun. 2018, 9, 1911. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, D.C.; Gjaltema, R.A.F.; Jilderda, L.J.; Jellema, P.; Dokter-Fokkens, J.; Ruiters, M.H.J.; Rots, M.G. Writing of H3K4Me3 overcomes epigenetic silencing in a sustained but context-dependent manner. Nat. Commun. 2016, 7, 12284. [Google Scholar] [CrossRef]
- Amabile, A.; Migliara, A.; Capasso, P.; Biffi, M.; Cittaro, D.; Naldini, L.; Lombardo, A. Inheritable silencing of endogenous genes by hit-and-run targeted epigenetic editing. Cell 2016, 167, 219–232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paixão, J.F.R.; Gillet, F.-X.; Ribeiro, T.P.; Bournaud, C.; Lourenço-Tessutti, I.T.; Noriega, D.D.; De Melo, B.P.; De Almeida-Engler, J.; Grossi-De-Sa, M.F. Improved drought stress tolerance in Arabidopsis by CRISPR/dCas9 fusion with a Histone AcetylTransferase. Sci. Rep. 2019, 9, 8080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nadakuduti, S.S.; Enciso-Rodríguez, F. Advances in genome editing with CRISPR systems and transformation technologies for plant DNA manipulation. Front. Plant Sci. 2020, 11, 637159. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Cheng, X.; Li, R.; Yao, J.; Li, Z.; Cheng, Y. Advances in application of genome editing in tomato and recent development of genome editing technology. Theor. Appl. Genet. 2021, 134, 2727–2747. [Google Scholar] [CrossRef]
- Molla, K.A.; Sretenovic, S.; Bansal, K.C.; Qi, Y.P. Precise plant genome editing using base editors and prime editors. Nat. Plants 2021, 7, 1166–1187. [Google Scholar] [CrossRef]
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016, 533, 420–424. [Google Scholar] [CrossRef] [Green Version]
- Nishida, K.; Arazoe, T.; Yachie, N.; Banno, S.; Kakimoto, M.; Tabata, M.; Mochizuki, M.; Miyabe, A.; Araki, M.; Hara, K.Y.; et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 2016, 353, aaf8729. [Google Scholar] [CrossRef]
- Gaudelli, N.M.; Komor, A.C.; Rees, H.A.; Packer, M.S.; Badran, A.H.; Bryson, D.I.; Liu, D.R. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 2017, 551, 464–471. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Y.; Sun, Y.W.; Du, J.L.; Zhao, Y.D.; Xia, L.Q. Generation of targeted point mutations in rice by a modified CRISPR/Cas9 system. Mol. Plant 2017, 10, 526–529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, B.; Yan, F.; Kuang, Y.J.; Li, N.; Zhang, D.W.; Zhou, X.P.; Lin, H.H.; Zhou, H.B. Improved base editor for efficiently inducing genetic variations in rice with CRISPR/Cas9-guided hyperactive hAID mutant. Mol. Plant 2018, 11, 623–626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajput, M.; Choudhary, K.; Kumar, M.; Vivekanand, V.; Chawade, A.; Ortiz, R.; Pareek, N. RNA interference and CRISPR/Cas gene editing for crop improvement: Paradigm shift towards sustainable agriculture. Plants 2021, 10, 1914. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, H.; Chen, J.; Yan, L.; Xia, L. Toward precision genome editing in crop plants. Mol. Plant 2020, 13, 811–813. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Koblan, L.W.; Liu, D.R. Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 2020, 38, 824–844. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef]
- Tang, X.; Sretenovic, S.; Ren, Q.R.; Jia, X.Y.; Li, M.K.; Fan, T.T.; Yin, D.S.; Xiang, S.Y.; Guo, Y.H.; Liu, L.; et al. Plant prime editors enable precise gene editing in rice cells. Mol. Plant 2020, 13, 667–670. [Google Scholar] [CrossRef]
- Lin, Q.; Zong, Y.; Xue, C.; Wang, S.; Jin, S.; Zhu, Z.; Wang, Y.; Anzalone, A.V.; Raguram, A.; Doman, J.L.; et al. Prime genome editing in rice and wheat. Nat. Biotechnol. 2020, 38, 582–585. [Google Scholar] [CrossRef]
- Li, H.Y.; Li, J.Y.; Chen, J.L.; Yan, L.; Xia, L.Q. Precise modifications of both exogenous and endogenous genes in rice by prime editing. Mol. Plant 2020, 13, 671–674. [Google Scholar] [CrossRef]
- Hua, K.; Jiang, Y.W.; Tao, X.P.; Zhu, J.K. Precision genome engineering in rice using prime editing system. Plant Biotechnol. J. 2020, 18, 2167–2169. [Google Scholar] [CrossRef] [PubMed]
- Butt, H.; Rao, G.S.; Sedeek, K.; Aman, R.; Kamel, R.; Mahfouz, M. Engineering herbicide resistance via prime editing in rice. Plant Biotechnol. J. 2020, 18, 2370–2372. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.-Y.; Chai, Y.-P.; Lu, M.-H.; Han, X.-L.; Lin, Q.P.; Zhang, Y.; Zhang, Q.; Zhou, Y.; Wang, X.-C.; Gao, C.X.; et al. Prime editing efficiently generates W542L and S621I double mutations in two ALS genes in maize. Genome Biol. 2020, 21, 257. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.M.; Tian, Y.F.; Shen, R.D.; Yao, Q.; Zhong, D.T.; Zhang, X.N.; Zhu, J.K. Precise genome modification in tomato using an improved prime editing system. Plant Biotechnol. J. 2021, 19, 415–417. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Zhang, L.; Zhang, K.; Ran, Y. Progresses, challenges, and prospects of genome editing in soybean (Glycine max). Front. Plant Sci. 2020, 11, 571138. [Google Scholar] [CrossRef] [PubMed]
- Mallapaty, S. China’s approval of gene-edited crops energizes researchers. Nature 2022, 602, 559–560. [Google Scholar] [CrossRef]
- Zhao, X.; Meng, Z.G.; Wang, Y.; Chen, W.J.; Sun, C.J.; Cui, B.; Cui, J.H.; Yu, M.L.; Zeng, Z.H.; Guo, S.D.; et al. Pollen magnetofection for genetic modification with magnetic nanoparticles as gene carriers. Nat. Plants 2017, 3, 956–964. [Google Scholar] [CrossRef]
- Kwak, S.-Y.; Lew, T.T.S.; Sweeney, C.J.; Koman, V.B.; Wong, M.H.; Bohmert-Tatarev, K.; Snell, K.D.; Seo, J.S.; Chua, N.-H.; Strano, M.S. Chloroplast-selective gene delivery and expression in planta using chitosan-complexed single-walled carbon nanotube carriers. Nat. Nanotechnol. 2019, 14, 447–455. [Google Scholar] [CrossRef]
- Demirer, G.S.; Zhang, H.; Matos, J.L.; Goh, N.S.; Cunningham, F.J.; Sung, Y.; Chang, R.; Aditham, A.J.; Chio, L.; Cho, M.-J.; et al. High aspect ratio nanomaterials enable delivery of functional genetic material without DNA integration in mature plants. Nat. Nanotechnol. 2019, 14, 456–464. [Google Scholar] [CrossRef]
- Zhang, H.; Demirer, G.S.; Zhang, H.; Ye, T.; Goh, N.S.; Aditham, A.J.; Cunningham, F.J.; Fan, C.; Landry, M.P. DNA nanostructures coordinate gene silencing in mature plants. Proc. Natl. Acad. Sci. USA 2019, 116, 7543–7548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).