Genetic Engineering Technologies for Improving Crop Yield and Quality
Abstract
1. Introduction
2. Genetic Engineering Technology
2.1. Transgenic Technology
2.2. Gene Editing Technology
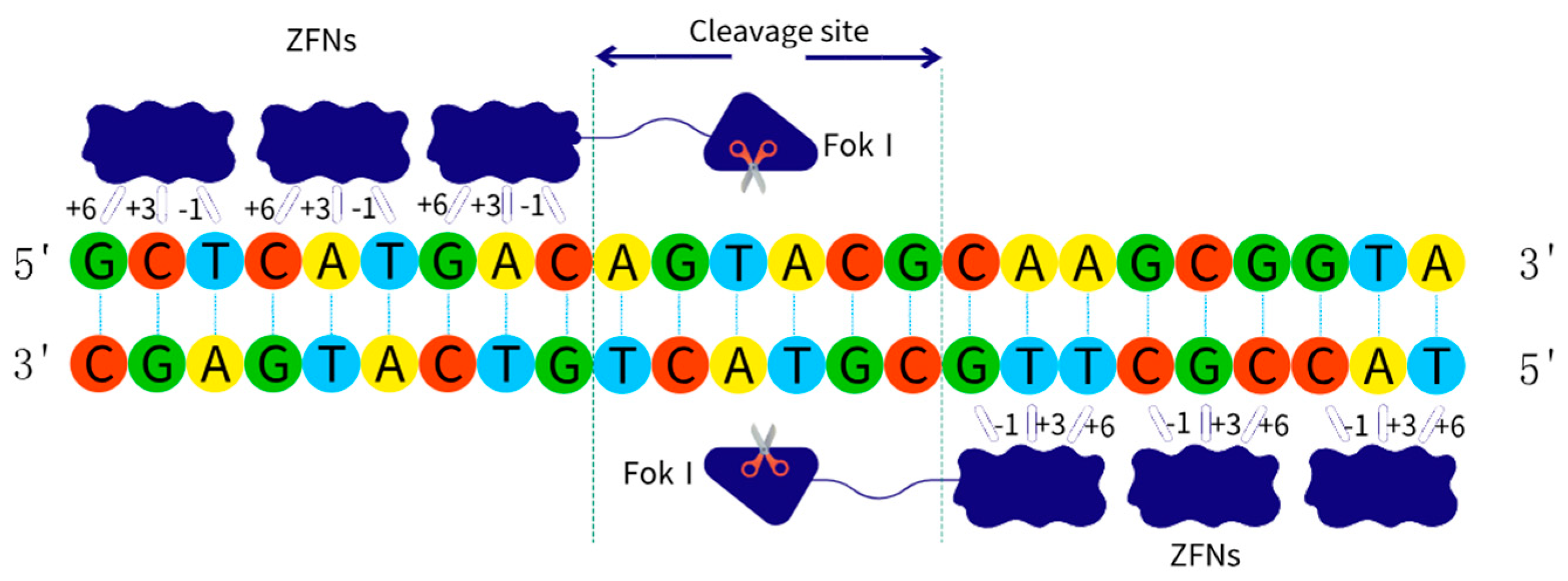
2.2.1. ZFNs
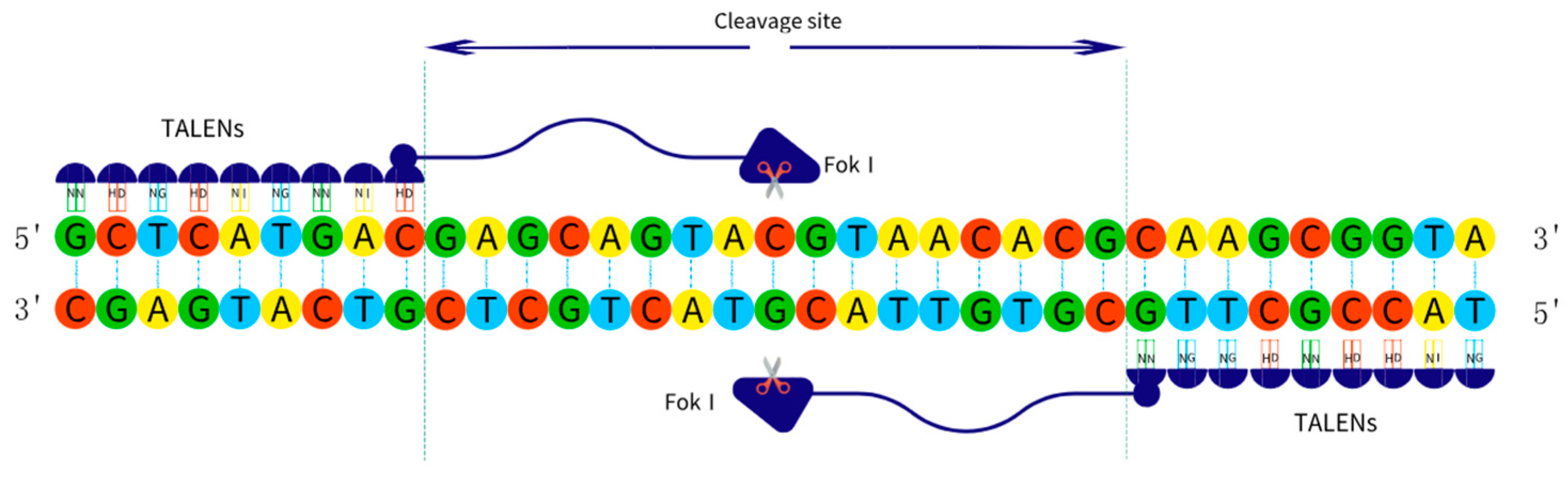
2.2.2. TALENs
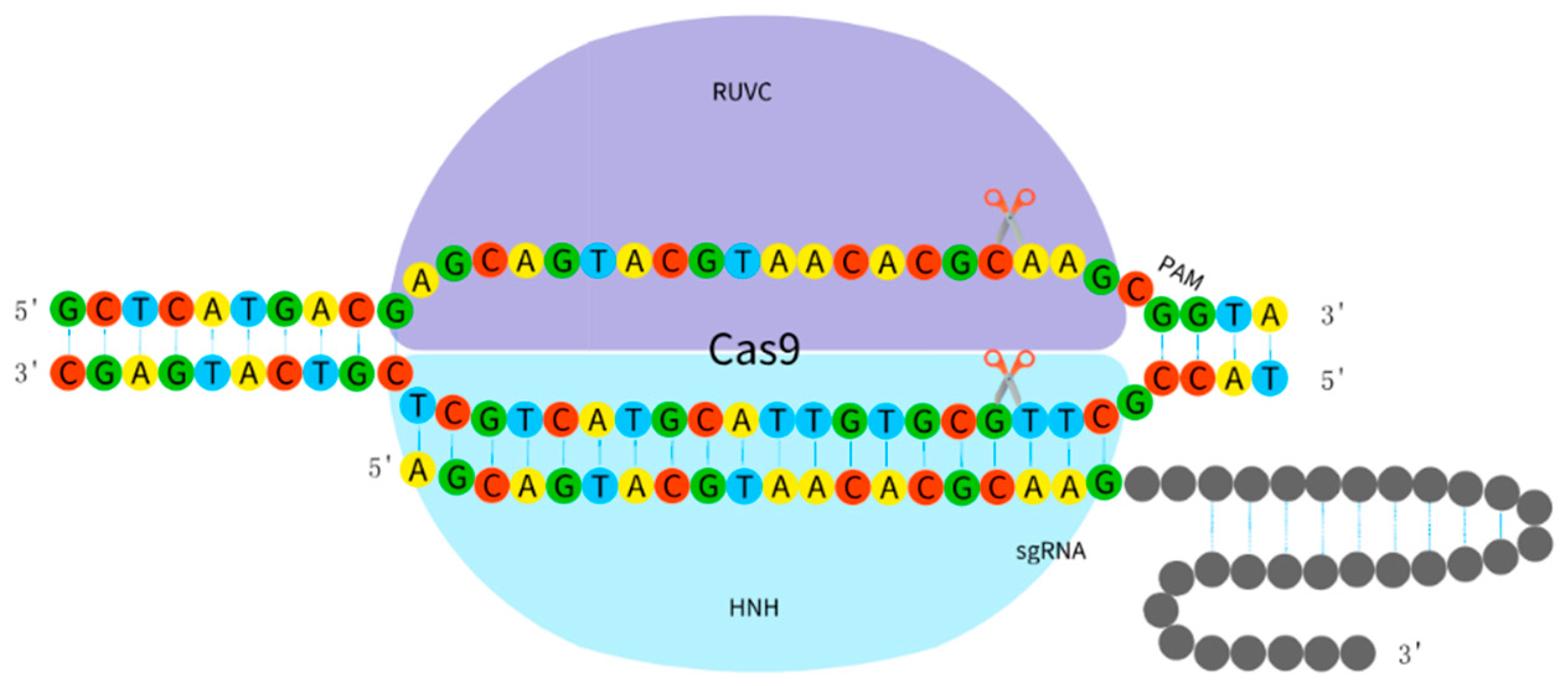
2.2.3. CRISPR/Cas9 System
3. Effects of Gene Editing Technology
3.1. Gene Knockout
3.2. Gene Knock-In
3.3. Gene Regulation
4. Comparison of Transgenic Technology and Gene Editing Technology
5. Optimization of Genetic Transformation and Regeneration Efficiency
6. Application of Genetic Engineering Technology in Improving Crop Yield and Quality
6.1. Transgenic Technology
6.2. DNA Recombination Technology
6.3. RNA Interference Technology
6.4. Gene Editing Technology
7. Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tian, Z.; Wang, J.; Li, J.; Han, B. Designing future crops: Challenges and strategies for sustainable agriculture. Plant J. Cell Mol. Biol. 2021, 105, 1165–1178. [Google Scholar] [CrossRef]
- Zhu, Q.; Yu, S.; Zeng, D.; Liu, H.; Wang, H.; Yang, Z.; Xie, X.; Shen, R.; Tan, J.; Li, H.; et al. Development of “purple endosperm rice” by engineering anthocyanin biosynthesis in the endosperm with a high-efficiency transgene stacking system. Mol. Plant 2017, 10, 918–929. [Google Scholar] [CrossRef] [PubMed]
- Cucina, M.; Regni, L. New advances on nutrients recovery from agro-industrial and livestockwastes for sustainable farming. Agronomy 2021, 11, 2308. [Google Scholar] [CrossRef]
- Lanigan, T.M.; Kopera, H.C.; Saunders, T.L. Principles of genetic engineering. Genes 2020, 11, 291. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Estrella, L.; Depicker, A.; Montagu, M.V.; Schell, J. Expression of chimaeric genes transferred into plant cells using a Ti-plasmid-derived vector. Nature 1983, 303, 209–213. [Google Scholar] [CrossRef]
- Mackelprang, R.; Lemaux, P.G. Genetic engineering and editing of plants: An analysis of new and persisting questions. Annu. Rev. Plant Biol. 2020, 71, 659–687. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Liu, Z.; Zhang, S.; Shen, S.; Wang, D.; Wang, J. The progress of plant transgenic technology. Chin. Agric. Sci. Bull. 2014, 30, 234–240. [Google Scholar]
- Gelvin, S.B. Agrobacterium-mediated plant transformation: The biology behind the “gene-jockeying” tool. Microbiol. Mol. Biol. Rev. MMBR 2003, 67, 16–37. [Google Scholar] [CrossRef]
- Helenius, E.; Boije, M.; Niklander-Teeri, V.; Palva, E.T.; Teeri, T.H. Gene delivery into intact plants using the heliosTM gene gun. Plant Mol. Biol. Rep. 2000, 18, 278–288. [Google Scholar] [CrossRef]
- Ribeiro, T.P.; Lourenco-Tessutti, I.T.; de Melo, B.P.; Morgante, C.V.; Filho, A.S.; Lins, C.B.J.; Ferreira, G.F.; Mello, G.N.; Macedo, L.L.P.; Lucena, W.A.; et al. Improved cotton transformation protocol mediated by Agrobacterium and biolistic combined-methods. Planta 2021, 254, 20. [Google Scholar] [CrossRef]
- Travella, S.; Ross, S.M.; Harden, J.; Everett, C.; Snape, J.W.; Harwood, W.A. A comparison of transgenic barley lines produced by particle bombardment and Agrobacterium-mediated techniques. Plant Cell Rep. 2005, 23, 780–789. [Google Scholar] [CrossRef]
- Gurusaravanan, P.; Vinoth, S.; Jayabalan, N. An improved Agrobacterium-mediated transformation method for cotton (Gossypium hirsutum L. ‘KC3’) assisted by microinjection and sonication. In Vitro Cell. Dev. Biol.-Plant 2020, 56, 111–121. [Google Scholar] [CrossRef]
- Yan, Y.; Zhu, X.; Yu, Y.; Li, C.; Zhang, Z.; Wang, F. Nanotechnology strategies for plant genetic engineering. Adv. Mater. 2021, 34, e2106945. [Google Scholar] [CrossRef]
- Kim, H.; Kim, J.S. A guide to genome engineering with programmable nucleases. Nat. Rev. Genet. 2014, 15, 321–334. [Google Scholar] [CrossRef]
- Moscou, M.J.; Bogdanove, A.J. A simple cipher governs DNA recognition by TAL effectors. Science 2009, 326, 1501. [Google Scholar] [CrossRef]
- Nishimasu, H.; Ran, F.A.; Hsu, P.D.; Konermann, S.; Shehata, S.I.; Dohmae, N.; Ishitani, R.; Zhang, F.; Nureki, O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 2014, 156, 935–949. [Google Scholar] [CrossRef]
- Lieber, M.R.; Ma, Y.; Pannicke, U.; Schwarz, K. Mechanism and regulation of human non-homologous DNA end-joining. Nat. Rev. Mol. Cell Biol. 2003, 4, 712–720. [Google Scholar] [CrossRef]
- Murray, J.M.; Carr, A.M. Integrating DNA damage repair with the cell cycle. Curr. Opin. Cell Biol. 2018, 52, 120–125. [Google Scholar] [CrossRef]
- Begemann, M.B.; Gray, B.N.; January, E.; Gordon, G.C.; He, Y.; Liu, H.; Wu, X.; Brutnell, T.P.; Mockler, T.C.; Oufattole, M. Precise insertion and guided editing of higher plant genomes using Cpf1 CRISPR nucleases. Sci. Rep. 2017, 7, 11606. [Google Scholar] [CrossRef]
- Wang, M.; Lu, Y.; Botella, J.R.; Mao, Y.; Hua, K.; Zhu, J. Gene targeting by homology-directed repair in rice using a geminivirus-based CRISPR/Cas9 system. Mol. Plant 2017, 10, 1007–1010. [Google Scholar] [CrossRef]
- Carroll, D. Genome engineering with Zinc-Finger nucleases. Genetics 2011, 188, 773–782. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, S.A.; Nekludova, L.; Pabo, C.O. DNA recognition by cys2his2 Zinc Finger proteins. Annu. Rev. Biophys. Biomol. Struct. 2000, 29, 183–212. [Google Scholar] [CrossRef]
- Li, L.; Wu, L.P.; Chandrasegaran, S. Functional domainsin FokI restriction endonuclease. Proc. Natl. Acad. Sci. USA 1992, 8, 4275–4279. [Google Scholar] [CrossRef]
- Shukla, V.K.; Doyon, Y.; Miller, J.C.; DeKelver, R.C.; Moehle, E.A.; Worden, S.E.; Mitchell, J.C.; Arnold, N.L.; Gopalan, S.; Meng, X.; et al. Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature 2009, 459, 437–441. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.-J.; Nogoy, F.M.; Lee, S.-K.; Cho, Y.-G.; Kang, K.-K. Application of ZFN for Site Directed Mutagenesis of Rice SSIVa Gene. Biotechnol. Bioprocess Eng. 2018, 23, 108–115. [Google Scholar] [CrossRef]
- Khan, S.H. Genome-editing technologies: Concept, pros, and cons of various genome-editing techniques and bioethical concerns for clinical application. Mol. Ther. Nucleic Acids 2019, 16, 326–334. [Google Scholar] [CrossRef] [PubMed]
- Boch, J.; Bonas, U. Xanthomonas AvrBs3 family-type III effectors: Discovery and function. Annu. Rev. Phytopathol. 2010, 48, 419–436. [Google Scholar] [CrossRef] [PubMed]
- Boch, J.; Scholze, H.; Schornack, S.; Landgraf, A.; Hahn, S.; Kay, S.; Lahaye, T.; Nickstadt, A.; Bonas, U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 2009, 326, 1509–1512. [Google Scholar] [CrossRef]
- Chandrasekaran, A.P.; Song, M.; Ramakrishna, S. Genome editing: A robust technology for human stem cells. Cell. Mol. Life Sci. CMLS 2017, 74, 3335–3346. [Google Scholar] [CrossRef]
- Mussolino, C.; Morbitzer, R.; Lutge, F.; Dannemann, N.; Lahaye, T.; Cathomen, T. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011, 39, 9283–9293. [Google Scholar] [CrossRef]
- Briggs, A.W.; Rios, X.; Chari, R.; Yang, L.; Zhang, F.; Mali, P.; Church, G.M. Iterative capped assembly: Rapid and scalable synthesis of repeat-module DNA such as TAL effectors from individual monomers. Nucleic Acids Res. 2012, 40, e117. [Google Scholar] [CrossRef]
- Li, T.; Liu, B.; Chen, C.Y.; Yang, B. TALEN-Mediated Homologous Recombination Produces Site-Directed DNA Base Change and Herbicide-Resistant Rice. J. Genet. Genom. Yi Chuan Xue Bao 2016, 43, 297–305. [Google Scholar] [CrossRef]
- Liang, Z.; Zhang, K.; Chen, K.; Gao, C. Targeted Mutagenesis in Zea mays Using TALENs and the CRISPR/Cas System. J. Genet. Genom. 2014, 41, 63–68. [Google Scholar] [CrossRef]
- Luo, M.; Li, H.; Chakraborty, S.; Morbitzer, R.; Rinaldo, A.; Upadhyaya, N.; Bhatt, D.; Louis, S.; Richardson, T.; Lahaye, T.; et al. Efficient TALEN-mediated gene editing in wheat. Plant Biotechnol. J. 2019, 17, 2026–2028. [Google Scholar] [CrossRef]
- Cox, D.B.; Platt, R.J.; Zhang, F. Therapeutic genome editing: Prospects and challenges. Nat. Med. 2015, 21, 121–131. [Google Scholar] [CrossRef]
- Marraffini, L.A.; Sontheimer, E.J. Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature 2010, 463, 568–571. [Google Scholar] [CrossRef]
- Gohil, N.; Bhattacharjee, G.; Lam, N.L.; Perli, S.D.; Singh, V. CRISPR-Cas systems: Challenges and future prospects. Prog. Mol. Biol. Transl. Sci. 2021, 180, 141–151. [Google Scholar] [CrossRef]
- Sapranauskas, R.; Gasiunas, G.; Fremaux, C.; Barrangou, R.; Horvath, P.; Siksnys, V. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011, 39, 9275–9282. [Google Scholar] [CrossRef]
- Yusa, K.; Rashid, S.T.; Strick-Marchand, H.; Varela, I.; Liu, P.Q.; Paschon, D.E.; Miranda, E.; Ordonez, A.; Hannan, N.R.; Rouhani, F.J.; et al. Targeted gene correction of alpha1-antitrypsin deficiency in induced pluripotent stem cells. Nature 2011, 478, 391–394. [Google Scholar] [CrossRef]
- Fu, Y.; Sander, J.D.; Reyon, D.; Cascio, V.M.; Joung, J.K. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 2014, 32, 279–284. [Google Scholar] [CrossRef]
- Guilinger, J.P.; Thompson, D.B.; Liu, D.R. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat. Biotechnol. 2014, 32, 577–582. [Google Scholar] [CrossRef] [PubMed]
- Zuo, E.; Sun, Y.; Yuan, T.; He, B.; Zhou, C.; Ying, W.; Liu, J.; Wei, W.; Zeng, R.; Li, Y.; et al. A rationally engineered cytosine base editor retains high on-target activity while reducing both DNA and RNA off-target effects. Nat. Methods 2020, 17, 600–604. [Google Scholar] [CrossRef]
- Neha, D.; Momin, M.; Khan, T.; Gharat, S.; Ningthoujam, R.S.; Omri, A. Metallic nanoparticles as drug delivery system for the treatment of cancer. Expert Opin. Drug Deliv. 2021, 18, 1261–1290. [Google Scholar] [CrossRef] [PubMed]
- Walton, R.T.; Christie, K.A.; Whittaker, M.N.; Kleinstiver, B.P. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science 2020, 368, 6488. [Google Scholar] [CrossRef] [PubMed]
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016, 533, 420–424. [Google Scholar] [CrossRef] [PubMed]
- Gaudelli, N.M.; Komor, A.C.; Rees, H.A.; Packer, M.S.; Badran, A.H.; Bryson, D.I.; Liu, D.R. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature 2017, 551, 464–471. [Google Scholar] [CrossRef] [PubMed]
- Kurt, I.C.; Zhou, R.; Iyer, S.; Garcia, S.P.; Miller, B.R.; Langner, L.M.; Grunewald, J.; Joung, J.K. CRISPR C-to-G base editors for inducing targeted DNA transversions in human cells. Nat. Biotechnol. 2021, 39, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Newby, G.A.; Liu, D.R. In vivo somatic cell base editing and prime editing. Mol. Ther. J. Am. Soc. Gene Ther. 2021, 29, 3107–3124. [Google Scholar] [CrossRef]
- Zhao, D.; Li, J.; Li, S.; Xin, X.; Hu, M.; Price, M.A.; Rosser, S.J.; Bi, C.; Zhang, X. Glycosylase base editors enable C-to-A and C-to-G base changes. Nat. Biotechnol. 2021, 39, 35–40. [Google Scholar] [CrossRef]
- Hua, K.; Tao, X.; Yuan, F.; Wang, D.; Zhu, J.K. Precise A.T to G.C Base Editing in the Rice Genome. Mol. Plant 2018, 11, 627–630. [Google Scholar] [CrossRef]
- Zong, Y.; Song, Q.; Li, C.; Jin, S.; Zhang, D.; Wang, Y.; Qiu, J.L.; Gao, C. Efficient C-to-T base editing in plants using a fusion of nCas9 and human APOBEC3A. Nat. Biotechnol. 2018, 36, 950–953. [Google Scholar] [CrossRef]
- Zeng, D.; Liu, T.; Tan, J.; Zhang, Y.; Zheng, Z.; Wang, B.; Zhou, D.; Xie, X.; Guo, M.; Liu, Y.-G.; et al. PhieCBEs: Plant High-Efficiency Cytidine Base Editors with Expanded Target Range. Mol. Plant 2020, 13, 1666–1669. [Google Scholar] [CrossRef]
- Xu, R.; Kong, F.; Qin, R.; Li, J.; Liu, X.; Wei, P. Development of an efficient plant dual cytosine and adenine editor. J. Integr. Plant Biol. 2021, 63, 1600–1605. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef]
- Alfatih, A.; Wu, J.; Jan, S.U.; Zhang, Z.S.; Xia, J.Q.; Xiang, C.B. Loss of rice PARAQUAT TOLERANCE 3 confers enhanced resistance to abiotic stresses and increases grain yield in field. Plant Cell Environ. 2020, 43, 2743–2754. [Google Scholar] [CrossRef]
- Cai, Y.; Wang, N.; Wang, S.; Wang, H. CRISPR/Cas9 mediated homology recombination insertion knockout of human SH2B3 gene. Chin. J. Cell Biol. 2018, 40, 252–259. [Google Scholar] [CrossRef]
- Komatsu, A.; Ohtake, M.; Shimatani, Z.; Nishida, K. Production of herbicide-sensitive strain to prevent volunteer rice infestation using a CRISPR-Cas9 cytidine deaminase fusion. Front. Plant Sci. 2020, 11, 925. [Google Scholar] [CrossRef]
- Lin, Q.; Zong, Y.; Xue, C.; Wang, S.; Jin, S.; Zhu, Z.; Wang, Y.; Anzalone, A.V.; Raguram, A.; Doman, J.L.; et al. Prime genome editing in rice and wheat. Nat. Biotechnol. 2020, 38, 582–585. [Google Scholar] [CrossRef]
- Shen, L.; Hua, Y.; Fu, Y.; Li, J.; Liu, Q.; Jiao, X.; Xin, G.; Wang, J.; Wang, X.; Yan, C.; et al. Rapid generation of genetic diversity by multiplex CRISPR/Cas9 genome editing in rice. Sci. China Life Sci. 2017, 60, 506–515. [Google Scholar] [CrossRef]
- Duan, K.; Cheng, Y.; Ji, J.; Wang, C.; Wei, Y.; Wang, Y. Large chromosomal segment deletions by CRISPR/LbCpf1-mediated multiplex gene editing in soybean. J. Integr. Plant Biol. 2021, 63, 1620–1631. [Google Scholar] [CrossRef]
- Jiang, T.; Zhang, X.; Weng, Z.; Xue, W. Deletion and replacement of long genomic sequences using prime editing. Nat. Biotechnol. 2021, 40, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Chen, W.; Suiter, C.C.; Lee, C.; Chardon, F.M.; Yang, W.; Leith, A.; Daza, R.M.; Martin, B.; Shendure, J. Precise genomic deletions using paired prime editing. Nat. Biotechnol. 2021, 40, 218–226. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Tian, Y.; Shen, R.; Yao, Q.; Wang, M.; Chen, M.; Dong, J.; Zhang, T.; Li, F.; Lei, M.; et al. Targeted, efficient sequence insertion and replacement in rice. Nat. Biotechnol. 2020, 38, 1402–1407. [Google Scholar] [CrossRef]
- Lin, Q.; Jin, S.; Zong, Y.; Yu, H.; Zhu, Z.; Liu, G.; Kou, L.; Wang, Y.; Qiu, J.L.; Li, J.; et al. High-efficiency prime editing with optimized, paired pegRNAs in plants. Nat. Biotechnol. 2021, 39, 923–927. [Google Scholar] [CrossRef] [PubMed]
- Anzalone, A.V.; Gao, X.D.; Podracky, C.J.; Nelson, A.T.; Koblan, L.W.; Raguram, A.; Levy, J.M.; Mercer, J.A.M.; Liu, D.R. Programmable deletion, replacement, integration and inversion of large DNA sequences with twin prime editing. Nat. Biotechnol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 2013, 152, 1173–1183. [Google Scholar] [CrossRef] [PubMed]
- Lowder, L.G.; Zhang, D.; Baltes, N.J.; Paul, J.W.; Tang, X.; Zheng, X.; Voytas, D.F.; Hsieh, T.F.; Zhang, Y.; Qi, Y. A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol. 2015, 169, 971–985. [Google Scholar] [CrossRef] [PubMed]
- Lowder, L.G.; Zhou, J.; Zhang, Y.; Malzahn, A.; Zhong, Z.; Hsieh, T.F.; Voytas, D.F.; Zhang, Y.; Qi, Y. Robust transcriptional activation in plants using multiplexed CRISPR-Act2.0 and mTALE-Act systems. Mol. Plant 2018, 11, 245–256. [Google Scholar] [CrossRef]
- Moradpour, M.; Abdulah, S.N.A. CRISPR/dCas9 platforms in plants: Strategies and applications beyond genome editing. Plant Biotechnol. J. 2020, 18, 32–44. [Google Scholar] [CrossRef]
- Shen, S.; Lu, Z.; Jin, J.; Cai, Y. Genetic modes of epigenetic modification and its research progress. Chin. Sci. Bull. 2016, 61, 3878–3886. [Google Scholar] [CrossRef][Green Version]
- Gallego-Bartolome, J.; Gardiner, J.; Liu, W.; Papikian, A.; Ghoshal, B.; Kuo, H.Y.; Zhao, J.M.; Segal, D.J.; Jacobsen, S.E. Targeted DNA demethylation of the Arabidopsis genome using the human TET1 catalytic domain. Proc. Natl. Acad. Sci. USA 2018, 115, E2125–E2134. [Google Scholar] [CrossRef]
- Papikian, A.; Liu, W.; Gallego-Bartolome, J.; Jacobsen, S.E. Site-specific manipulation of Arabidopsis loci using CRISPR-Cas9 SunTag systems. Nat. Commun. 2019, 10, 729. [Google Scholar] [CrossRef]
- Ding, Y.; Shao, Y.; Xu, Y.; Chen, F. Screening citrinin mutants from the transformants library of monascus ruber M-7 by Agrobacterium-mediated DNA transfer. Microbiol. China 2006, 4, 52–57. [Google Scholar]
- Wu, K.; Wu, Y.; Zhang, C.; Fu, Y.; Liu, Z.; Zhang, X. Simultaneous silencing of two different Arabidopsis genes with a novel virus-induced gene silencing vector. Plant. Methods 2021, 17, 6. [Google Scholar] [CrossRef]
- Iwase, A.; Mita, K.; Nonaka, S.; Ikeuchi, M.; Koizuka, C.; Ohnuma, M.; Ezura, H.; Imamura, J.; Sugimoto, K. WIND1-based acquisition of regeneration competency in Arabidopsis and rapeseed. J. Plant Res. 2015, 128, 389–397. [Google Scholar] [CrossRef]
- Iwase, A.; Mita, K.; Favero, D.S.; Mitsuda, N.; Sasaki, R.; Kobayshi, M.; Takebayashi, Y.; Kojima, M.; Kusano, M.; Oikawa, A.; et al. WIND1 induces dynamic metabolomic reprogramming during regeneration in Brassica napus. Dev. Biol. 2018, 442, 40–52. [Google Scholar] [CrossRef]
- Lowe, K.; Wu, E.; Wang, N.; Hoerster, G.; Hastings, C.; Cho, M.J.; Scelonge, C.; Lenderts, B.; Chamberlin, M.; Cushatt, J.; et al. Morphogenic Regulators Baby boom and Wuschel Improve Monocot Transformation. Plant Cell 2016, 28, 1998–2015. [Google Scholar] [CrossRef] [PubMed]
- Debernardi, J.M.; Tricoli, D.M.; Ercoli, M.F.; Hayta, S.; Ronald, P.; Palatnik, J.F.; Dubcovsky, J. A GRF-GIF chimeric protein improves the regeneration efficiency of transgenic plants. Nat. Biotechnol. 2020, 38, 1274–1279. [Google Scholar] [CrossRef]
- Feng, Q.; Xiao, L.; He, Y.; Liu, M.; Wang, J.; Tian, S.; Zhang, X.; Yuan, L. Highly efficient, genotype-independent transformation and gene editing in watermelon (Citrullus lanatus) using a chimeric ClGRF4-GIF1 gene. J. Integr. Plant Biol. 2021, 63, 2038–2042. [Google Scholar] [CrossRef]
- Kong, J.; Martin-Ortigosa, S.; Finer, J.; Orchard, N.; Gunadi, A.; Batts, L.A.; Thakare, D.; Rush, B.; Schmitz, O.; Stuiver, M.; et al. Overexpression of the Transcription Factor GROWTH-REGULATING FACTOR5 Improves Transformation of Dicot and Monocot Species. Front. Plant Sci. 2020, 11, 572319. [Google Scholar] [CrossRef]
- Wang, K.; Shi, L.; Liang, X.; Zhao, P.; Wang, W.; Liu, J.; Chang, Y.; Hiei, Y.; Yanagihara, C.; Du, L.; et al. The gene TaWOX5 overcomes genotype dependency in wheat genetic transformation. Nat. Plants 2022, 8, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, M.; Li, Y.; Zhang, Q.; Lindsey, K.; Daniell, H.; Jin, S.; Zhang, X. Multi-omics analyses reveal epigenomics basis for cotton somatic embryogenesis through successive regeneration acclimation process. Plant Biotechnol. J. 2019, 17, 435–450. [Google Scholar] [CrossRef] [PubMed]
- Ellison, E.E.; Nagalakshmi, U.; Gamo, M.E.; Huang, P.J.; Dinesh-Kumar, S.; Voytas, D.F. Multiplexed heritable gene editing using RNA viruses and mobile single guide RNAs. Nat. Plants 2020, 6, 620–624. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Hu, J.; Sun, Y.; Li, B.; Zhang, D.; Li, W.; Liu, J.; Li, D.; Gao, C.; Zhang, Y.; et al. Highly efficient heritable genome editing in wheat using an RNA virus and bypassing tissue culture. Mol. Plant 2021, 14, 1787–1798. [Google Scholar] [CrossRef]
- Wang, H.; Liao, S.; Li, M.; Wei, J.; Zhu, B.; Gu, L.; Li, L.; Du, X. TmNAS3 from Triticum monococum directly regulated by TmbHLH47 increases Fe content of wheat grain. Gene 2021, 811, 146096. [Google Scholar] [CrossRef]
- Fischer, S.E.J. RNA interference and MicroRNA-mediated silencing. Curr. Protoc. Mol. Biol. 2015, 112, 2611–2615. [Google Scholar] [CrossRef]
- Zhang, X.; Mogel, K.J.H.V.; Lor, V.S.; Hirsch, C.N.; Vries, B.D.; Kaeppler, H.F.; Tracy, W.F.; Kaeppler, S.M. Maize sugary enhancer1 (se1) is a gene affecting endosperm starch metabolism. Proc. Natl. Acad. Sci. USA 2019, 116, 20776–20785. [Google Scholar] [CrossRef]
- Wang, H.; Li, Y.; Chern, M.; Zhu, Y.; Zhang, L.; Lu, J.; Li, X.; Dang, W.; Ma, X.; Yang, Z.; et al. Suppression of rice miR168 improves yield, flowering time and immunity. Nat. Plants 2021, 7, 129–136. [Google Scholar] [CrossRef]
- Lu, K.; Wu, B.; Wang, J.; Zhu, W.; Nie, H.; Qian, J.; Huang, W.; Fang, Z. Blocking amino acid transporter OsAAP3 improves grain yield by promoting outgrowth buds and increasing tiller number in rice. Plant Biotechnol. J. 2018, 16, 1710–1722. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, R.; Gao, J.; Song, G.; Li, J.; Li, W.; Qi, Y.; Li, Y.; Li, G. CRISPR/Cas9-mediated genome editing for wheat grain quality improvement. Plant Biotechnol. J. 2021, 19, 1684–1686. [Google Scholar] [CrossRef]
- Perez, L.; Soto, E.; Farre, G.; Juanos, J.; Villorbina, G.; Bassie, L.; Medina, V.; Serrato, A.J.; Sahrawy, M.; Rojas, J.A.; et al. CRISPR/Cas9 mutations in the rice Waxy/GBSSI gene induce allele-specific and zygosity-dependent feedback effects on endosperm starch biosynthesis. Plant Cell Rep. 2019, 38, 417–433. [Google Scholar] [CrossRef]
- Zeng, D.; Liu, T.; Ma, X.; Wang, B.; Zheng, Z.; Zhang, Y.; Xie, X.; Yang, B.; Zhao, Z.; Zhu, Q.; et al. Quantitative regulation of Waxy expression by CRISPR/Cas9-based promoter and 5′UTR-intron editing improves grain quality in rice. Plant Biotechnol. J. 2020, 18, 2385–2387. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, X.; Zheng, X.; Wang, W.; Yin, X.; Liu, H.; Ma, C.; Niu, X.; Zhu, J.; Wang, F. Creation of aromatic maize by CRISPR/Cas. J. Integr. Plant Biol. 2021, 63, 1664–1670. [Google Scholar] [CrossRef]
- Xu, R.; Yang, Y.; Qin, R.; Li, H.; Qiu, C.; Li, L.; Wei, P.; Yang, J. Rapid improvement of grain weight via highly efficient CRISPR/Cas9-mediated multiplex genome editing in rice. J. Genet. Genom. Yi Chuan Xue Bao 2016, 43, 529–532. [Google Scholar] [CrossRef]
- Miao, C.; Xiao, L.; Hua, K.; Zou, C.; Zhao, Y.; Bressan, R.A.; Zhu, J. Mutations in a subfamily of abscisic acid receptor genes promote rice growth and productivity. Proc. Natl. Acad. Sci. USA 2018, 115, 6058–6063. [Google Scholar] [CrossRef]
- Huang, X.; Su, F.; Huang, S.; Mei, F.; Niu, X.; Ma, C.; Zhang, H.; Zhu, X.; Zhu, J.; Zhang, J. Novel Wx alleles generated by base editing for improvement of rice grain quality. J. Integr. Plant biology 2021, 63, 1632–1638. [Google Scholar] [CrossRef]
- Liu, T.; Zeng, D.; Zheng, Z.; Lin, Z.; Xue, Y.; Li, T.; Xie, X.; Ma, G.; Liu, Y.G.; Zhu, Q. The ScCas9++ variant expands the CRISPR toolbox for genome editing in plants. J. Integr. Plant Biol. 2021, 63, 1611–1619. [Google Scholar] [CrossRef]
- Zhu, X.; Rong, W.; Wang, K.; Guo, W.; Zhou, M.; Wu, J.; Ye, X.; Wei, X.; Zhang, Z. Overexpression of TaSTT3b-2B improves resistance to sharp eyespot and increases grain weight in wheat. Plant Biotechnol. J. 2021, 1–17. [Google Scholar] [CrossRef]
- Fanelli, A.; Rancour, D.M.; Sullivan, M.; Karlen, S.D.; Ralph, J.; Riano-Pachon, D.M.; Vicentini, R.; Silva, T.D.F.; Ferraz, A.; Hatfield, R.D.; et al. Overexpression of a sugarcane BAHD acyltransferase alters hydroxycinnamate content in maize cell wall. Front. Plant Sci. 2021, 12, 626168. [Google Scholar] [CrossRef]
- Sun, X.; Xue, X.; Wang, X.; Zhang, C.; Zheng, D.; Song, W.; Zhao, J.; Wei, J.; Wu, Z.; Zhang, Z. Natural variation of ZmCGT1 is responsible for isoorientin accumulation in maize silk. Plant J. Cell Mol. Biol. 2022, 109, 64–76. [Google Scholar] [CrossRef]
- Zeng, J.; Wang, X.; Miao, Y.; Wang, C.; Zang, M.; Chen, X.; Li, M.; Li, X.; Wang, Q.; Li, K.; et al. Metabolic engineering of wheat provitamin a by simultaneously overexpressing CrtB and silencing carotenoid hydroxylase (TaHYD). J. Agric. Food Chem. 2015, 63, 9083–9092. [Google Scholar] [CrossRef]
- Wang, W.; Tian, B.; Pan, Q.; Chen, Y.; He, F.; Bai, G.; Akhunova, A.; Trick, H.N.; Akhunov, E. Expanding the range of editable targets in the wheat genome using the variants of the Cas12a and Cas9 nucleases. Plant Biotechnol. J. 2021, 19, 2428–2441. [Google Scholar] [CrossRef]
- Hu, J.; Huang, L.; Chen, G.; Liu, H.; Zhang, Y.; Zhang, R.; Zhang, S.; Liu, J.; Hu, Q.; Hu, F.; et al. The elite alleles of OsSPL4 regulate grain size and increase grain yield in rice. Rice 2021, 14, 90. [Google Scholar] [CrossRef]
- Zheng, S.; Ye, C.; Lu, J.; Liufu, J.; Lin, L.; Dong, Z.; Li, J.; Zhuang, C. Improving the rice photosynthetic efficiency and yield by editing OsHXK1 via CRISPR/Cas9 system. Int. J. Mol. Sci. 2021, 22, 9554. [Google Scholar] [CrossRef]
- Hui, S.; Li, H.; Mawia, A.M.; Zhou, L.; Cai, J.; Ahmad, S.; Lai, C.; Wang, J.; Jiao, G.; Xie, L.; et al. Production of aromatic three-line hybrid rice using novel alleles of BADH2. Plant Biotechnol. J. 2022, 20, 59–74. [Google Scholar] [CrossRef] [PubMed]
- Ning, Q.; Jian, Y.; Du, Y.; Li, Y.; Shen, X.; Jia, H.; Zhao, R.; Zhan, J.; Yang, F.; Jackson, D.; et al. An ethylene biosynthesis enzyme controls quantitative variation in maize ear length and kernel yield. Nat. Commun. 2021, 12, 5832. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, P.; Jakimo, N.; Jacobson, J.M. Minimal PAM specificity of a highly similar SpCas9 ortholog. Sci. Adv. 2018, 4, eaau0766. [Google Scholar] [CrossRef] [PubMed]
- Ma, G.; Kuang, Y.; Lu, Z.; Li, X.; Xu, Z.; Ren, B.; Zhou, X.; Zhou, H. CRISPR/Sc++-mediated genome editing in rice. J. Integr. Plant Biol. 2021, 63, 1606–1610. [Google Scholar] [CrossRef]
- Liu, M.S.; Gong, S.; Yu, H.H.; Jung, K.; Johnson, K.A.; Taylor, D.W. Engineered CRISPR/Cas9 enzymes improve discrimination by slowing DNA cleavage to allow release of off-target DNA. Nat. Commun. 2020, 11, 3576. [Google Scholar] [CrossRef]
- Zhong, Z.; Sretenovic, S.; Ren, Q.; Yang, L.; Bao, Y.; Qi, C.; Yuan, M.; He, Y.; Liu, S.; Liu, X.; et al. Improving plant genome editing with high-fidelity xCas9 and non-canonical PAM-Targeting Cas9-NG. Mol. Plant 2019, 12, 1027–1036. [Google Scholar] [CrossRef]
- Qin, R.; Li, J.; Liu, X.; Xu, R.; Yang, J.; Wei, P. SpCas9-NG self-targets the sgRNA sequence in plant genome editing. Nat. Plants 2020, 6, 197–201. [Google Scholar] [CrossRef]
- Li, S.; Li, J.; He, Y.; Xu, M.; Zhang, J.; Du, W.; Zhao, Y.; Xia, L. Precise gene replacement in rice by RNA transcript-templated homologous recombination. Nat. Biotechnol. 2019, 37, 445–450. [Google Scholar] [CrossRef]
| Serial No. | Parameter | Transgenic Technology | Gene Editing Technology | Reference |
|---|---|---|---|---|
| 1 | Whether it is targeted | No targeting | Targeting using proteins or nucleic acids | [53,58,60] |
| 2 | Is it possible to edit without exogenous DNA | No | Yes | [55,57] |
| 3 | The efficiency of target genome modification | Low | High | [2,14,73] |
| 4 | Probability of causing gene silencing | High | Low | [11] |
| 5 | Genetic stability | Low | High | [11] |
| 6 | Types of modifications that can be generated | Single | Diverse | [10,53,58,60,74] |
| Genetic Engineering Technology | Target | Crop | Crop Benefit | References |
|---|---|---|---|---|
| Transgenic technology | Knock in TmNAS3 | Wheat | Increase grain size and Fe content | [85] |
| Transgenic technology | Overexpress TaSTT3b-2B | Wheat | Increase grain weight for higher yield | [98] |
| Transgenic technology | Knock in ScAT10 | Maize | Increase the ratio of p-coumaric acid/ferulic acid | [99] |
| Transgenic technology | Overexpress ZmCGT1 | Maize | Increase the isoorientin content in maize silk | [100] |
| DNA recombinant technology | Knock in eight genes related to anthocyanin synthesis | Rice | Increase the rice anthocyanins in the endosperm | [2] |
| RNA silencing technology | Silence microRNA168 | Rice | Increase the number of ears for higher yield | [88] |
| RNA silencing technology | Silence Se1 | Maize | Increase soluble sugar content | [87] |
| RNA silencing technology combined with transgenic technology | Overexpress CrtB and silence TaHYD | Wheat | Increase the β-carotene content in wheat endosperm | [101] |
| CRISPR/Cas12a single site editing technology | Edit TaGW7-B1 | Wheat | Increase grain weight for higher yield | [102] |
| CRISPR/Cas9 single site editing technology | Knock out OsAAP3 | Rice | Increase tiller number for higher yield | [89] |
| CRISPR/Cas9 single site editing technology | Edit OsSPL4 | Rice | Generate two new high-quality alleles for higher yield | [103] |
| CRISPR/Cas9 single site editing technology | Edit CREs of Wx | Rice | Reduce amylose content appropriately | [92] |
| CRISPR/Cas9 single site editing technology | Knock out OsHXK | Rice | Increase the rate of photosynthesis for higher yield | [104] |
| CRISPR/Cas9 single site editing technology | Edit OsBADH2 | Rice | Produce moderate aroma | [105] |
| CRISPR/Cas9 single site editing technology | Knock out Ppo | Wheat | Inhibite dough browning | [90] |
| CRISPR/Cas9 single site editing technology | Edit ZmACO2 | Maize | Increase the number of grains per ear for higher yield | [106] |
| CRISPR/Cas9 multiple sites editing technology | Knock out GW2, GW5, and TGW6 | Rice | Increase the volume and the weight of grains for higher yield | [94] |
| CRISPR/Cas9 multiple sites editing technology | Knock out PYL1, PYL4, and PYL6 | Rice | Increase yield | [95] |
| CRISPR/Cas9 multiple sites editing technology | Knock out ZmBADH2a and ZmBADH2b | Maize | Produce popcorn aroma | [93] |
| ABEmax-nCas9NG, Anc689CEBmax-nCas9NG | Substitute base of Wx | Rice | Reduce amylose content appropriately | [96] |
| PevoCDA1-ScCas9n++ | Substitute base of OsWx | Rice | Reduce amylose content appropriately | [97] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, R.; Yang, X.; Rao, Y. Genetic Engineering Technologies for Improving Crop Yield and Quality. Agronomy 2022, 12, 759. https://doi.org/10.3390/agronomy12040759
Ye R, Yang X, Rao Y. Genetic Engineering Technologies for Improving Crop Yield and Quality. Agronomy. 2022; 12(4):759. https://doi.org/10.3390/agronomy12040759
Chicago/Turabian StyleYe, Runle, Xi Yang, and Yuchun Rao. 2022. "Genetic Engineering Technologies for Improving Crop Yield and Quality" Agronomy 12, no. 4: 759. https://doi.org/10.3390/agronomy12040759
APA StyleYe, R., Yang, X., & Rao, Y. (2022). Genetic Engineering Technologies for Improving Crop Yield and Quality. Agronomy, 12(4), 759. https://doi.org/10.3390/agronomy12040759





