Paradigm and Framework of WUS-CLV Feedback Loop in Stem Cell Niche for SAM Maintenance and Cell Identity Transition
Abstract
1. Introduction
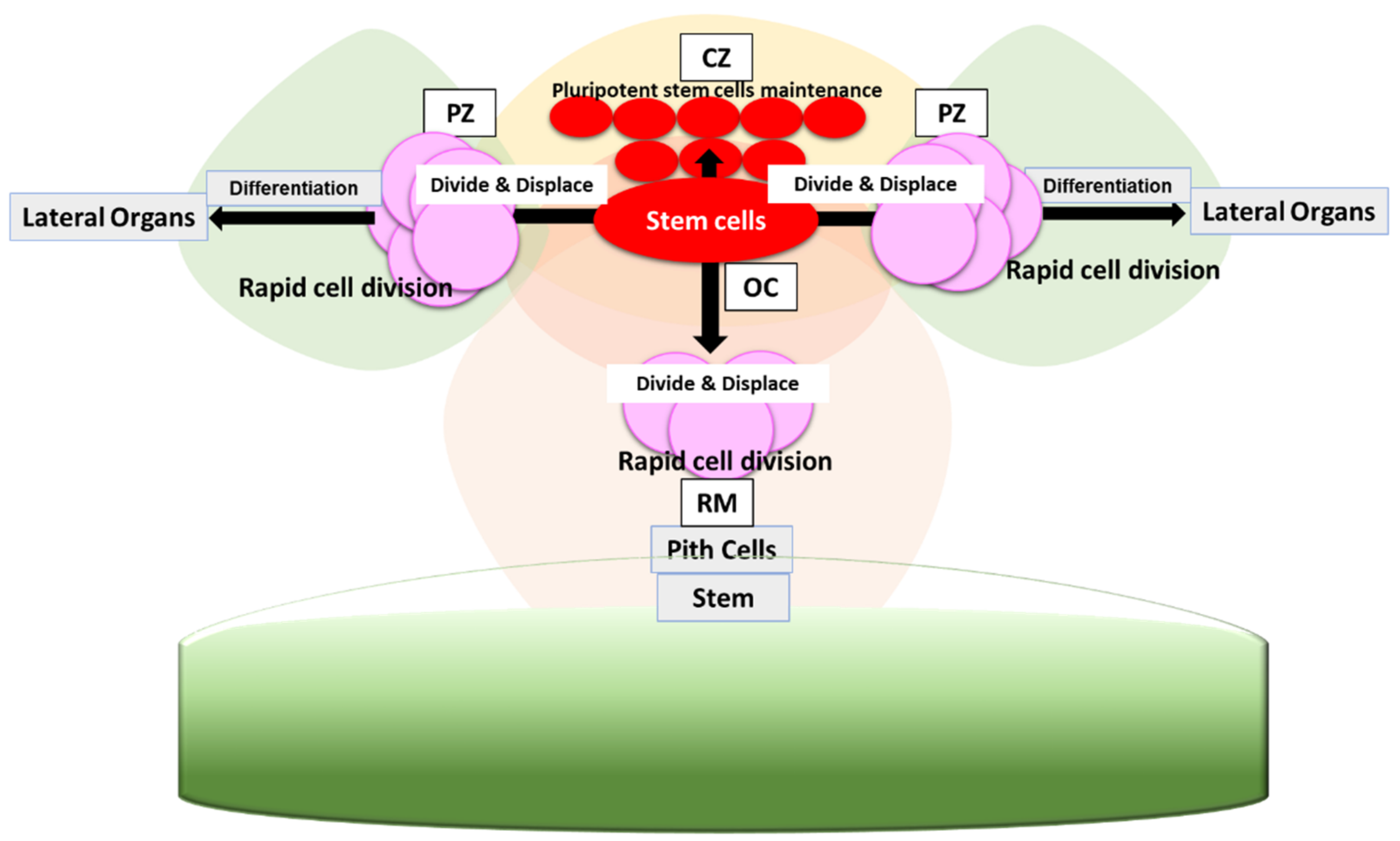
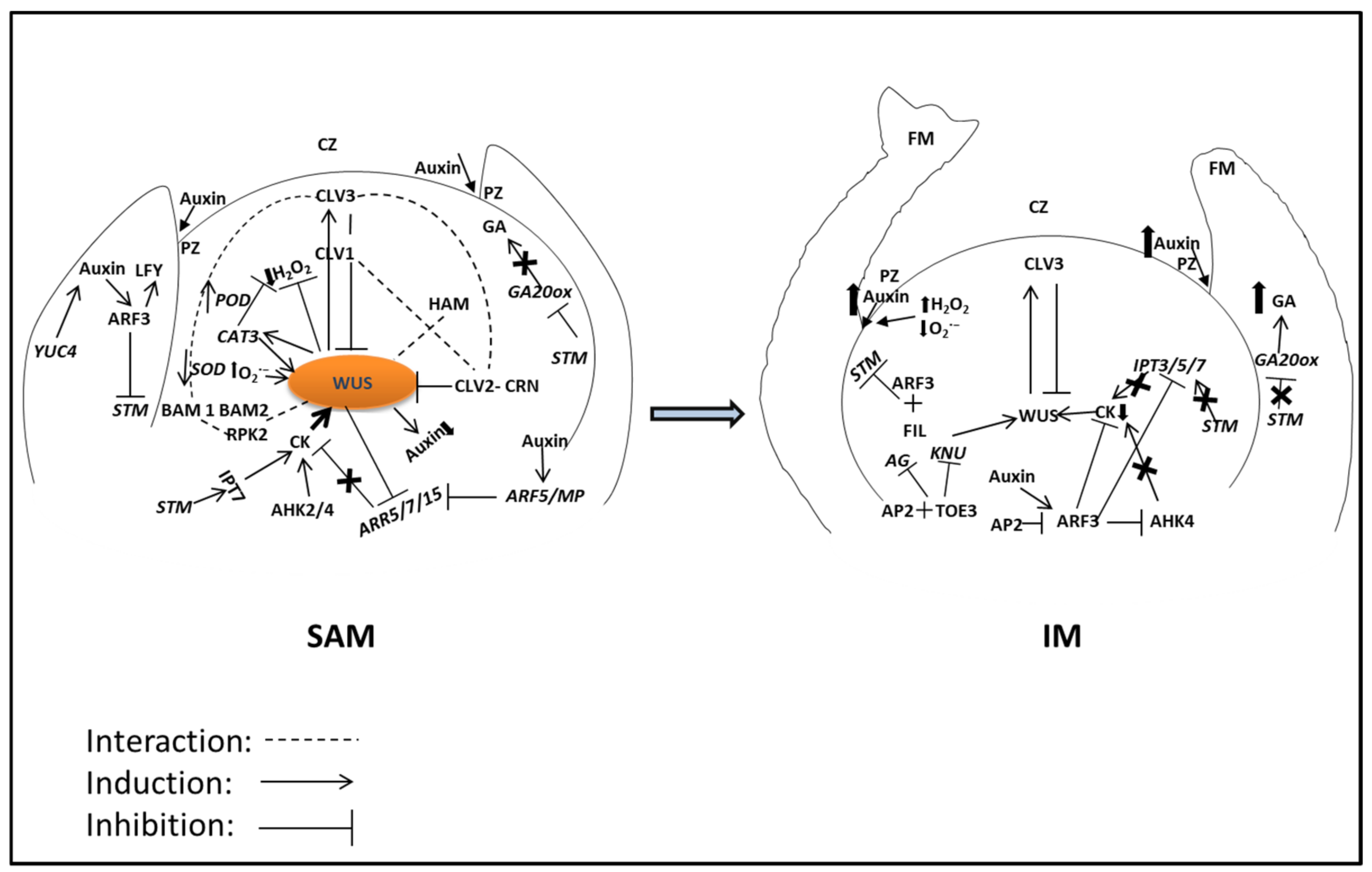
2. WUS-CLV Feedback Loop in SAM Maintenance
3. WUS-CLV in Identity Transition
4. WUS-CLV in Flower Development
5. Phytohormones Crosstalk with WUS-CLV Regulatory Pathway in SAM
6. ROS in SAM Regulation
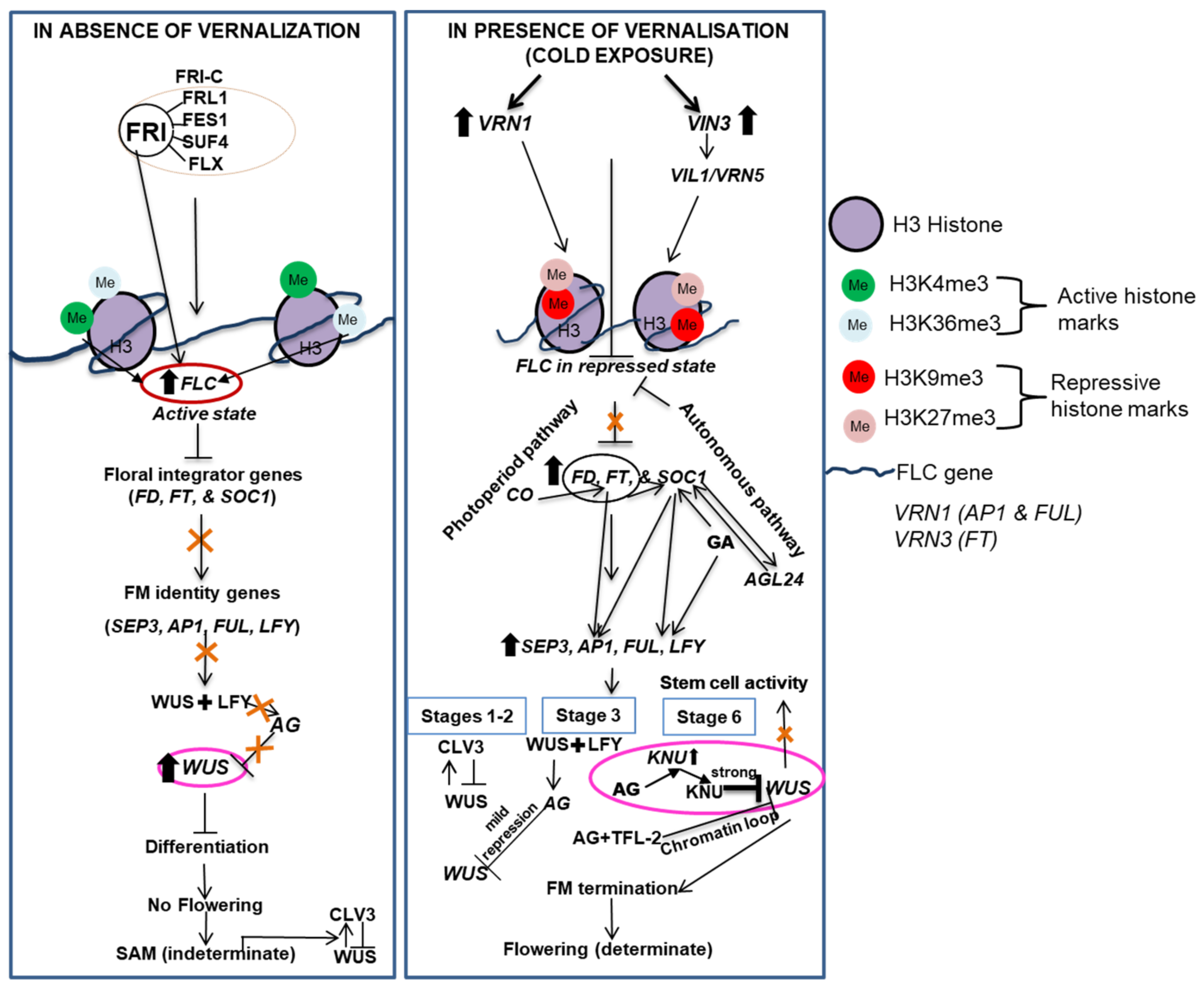
7. Vernalization
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gaillochet, C.; Lohmann, J.U. The never-ending story: From pluripotency to plant developmental plasticity. Development 2015, 142, 2237–2249. [Google Scholar] [CrossRef] [PubMed]
- Scheres, B. Stem cells: A Plant biology perspective. Cell 2005, 122, 499–504. [Google Scholar] [CrossRef] [PubMed]
- Weigel, D.; Jürgens, G. Stem cells that make stems. Nature 2002, 415, 751–754. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.; Guo, Y.; Zhang, H.; Liu, X.; Guo, L. Same actor in different stages: Genes in shoot apical meristem maintenance and floral meristem determinacy in Arabidopsis. Front. Ecol. Evol. 2020, 8, 89. [Google Scholar] [CrossRef]
- Fletcher, J.C. The CLV-WUS Stem cell signaling pathway: A roadmap to crop yield optimization. Plants 2018, 7, 87. [Google Scholar] [CrossRef]
- Whitewoods, C.D.; Cammarata, J.; Venza, Z.N.; Sang, S.; Crook, A.D.; Aoyama, T.; Wang, X.Y.; Waller, M.; Kamisugi, Y.; Cuming, A.C.; et al. CLAVATA was a genetic novelty for the morphological innovation of 3D growth in land plants. Curr. Biol. 2018, 28, 2365–2376.e5. [Google Scholar] [CrossRef]
- Laux, T.; Mayer, K.F.; Berger, J.; Jurgens, G. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 1996, 122, 87–96. [Google Scholar] [CrossRef]
- Schoof, H.; Lenhard, M.; Haecker, A.; Mayer KF, X.; Jürgens, G.; Laux, T. The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 2000, 100, 635–644. [Google Scholar] [CrossRef]
- Wang, X.; Wang, X.; Ren, J.; Ma, Y.; Yin, J. Characterization of tomato transcription factor WUSCHEL and functional study in Arabidopsis. J. Integr. Agric. 2012, 11, 1257–1265. [Google Scholar] [CrossRef]
- Xu, C.; Liberatore, K.L.; MacAlister, C.A.; Huang, Z.; Chu, Y.H.; Jiang, K.; Brooks, C.; Ogawa-Ohnishi, M.; Xiong, G.; Pauly, M.; et al. A cascade of arabinosyltransferases controls shoot meristem size in tomato. Nat. Genetic. 2015, 47, 784–792. [Google Scholar] [CrossRef]
- Nardmann, J.; Werr, W. The shoot stem cell niche in angiosperms: Expression patterns of WUS orthologues in rice and maize imply major modifications in the course of mono-and dicot evolution. Mol. Biol. Evol. 2006, 23, 2492–2504. [Google Scholar] [CrossRef]
- Bommert, P.; Lunde, C.; Nardmann, J.; Vollbrecht, E.; Running, M.; Jackson, D.; Hake, S.; Werr, W. Thick tassel dwarf1 encodes a putative maize ortholog of the Arabidopsis CLAVATA1 leucine-rich repeat receptor-like kinase. Development 2005, 132, 1235–1245. [Google Scholar] [CrossRef]
- Taguchi-Shiobara, F.; Yuan, Z.; Hake, S.; Jackson, D. The fasciated ear2 gene encodes a leucine-rich repeat receptor-like protein that regulates shoot meristem proliferation in maize. Genes Dev. 2001, 15, 2755–2766. [Google Scholar] [CrossRef]
- Je, B.; Gruel, J.; Lee, Y.K.; Bommert, P.; Arevalo, E.D.; Eveland, A.L.; Wu, Q.; Goldshmidt, A.; Meeley, R.; Bartlett, M.; et al. Signaling from maize organ primordia via FASCIATED EAR3 regulates stem cell proliferation and yield traits. Nat. Genet. 2016, 48, 785–791. [Google Scholar] [CrossRef]
- Tanaka, W.; Ohmori, Y.; Ushijima, T.; Matsusaka, H.; Matsushita, T.; Kumamaru, T.; Kawano, S.; Hirano, H.Y. Axillary meristem formation in rice requires the WUSCHEL ortholog TILLERS ABSENT1. Plant Cell 2015, 27, 1173–1184. [Google Scholar] [CrossRef]
- Suzaki, T.; Sato, M.; Ashikari, M.; Miyoshi, M.; Nagato, Y.; Hirano, H.Y. The gene FLORAL ORGAN NUMBER1 regulates floral meristem size in rice and encodes a leucine-rich repeat receptor kinase orthologous to Arabidopsis CLAVATA1. Development 2004, 131, 5649–5657. [Google Scholar] [CrossRef]
- Chu, H.; Qian, Q.; Liang, W.; Yin, C.; Tan, H.; Yao, X.; Yuan, Z.; Yang, J.; Huang, H.; Luo, D.; et al. The FLORAL ORGAN NUMBER4 Gene encoding a putative ortholog of Arabidopsis CLAVATA3 regulates apical meristem size in rice. Plant Physiol. 2006, 142, 1039–1052. [Google Scholar] [CrossRef]
- Liu, X.; Kim, Y.J.; Müller, R.; Yumul, R.E.; Liu, C.; Pan, Y.; Cao, X.; Goodrich, J.; Chen, X. AGAMOUS terminates floral stem cell maintenance in Arabidopsis by directly repressing WUSCHEL through recruitment of Polycomb Group proteins. Plant Cell 2011, 23, 3654–3670. [Google Scholar] [CrossRef]
- Itkin, M.; Seybold, H.; Breitel, D.; Rogachev, I.; Meir, S.; Aharoni, A. TOMATO AGAMOUS-LIKE 1 is a component of the fruit ripening regulatory network. Plant J. 2009, 60, 1081–1095. [Google Scholar] [CrossRef]
- Schmidtiai’, R.J.; Veit, B.; Mandel, M.A.; Mena, M.; Hakeyb, S.; Yanofsky’, M.F. Identification and molecular characterization of ZAG1, the maize homolog of the Arabidopsis floral homeotic gene AGAMOUS. Plant Cell 1993, 5, 729–737. [Google Scholar] [CrossRef]
- Suzaki, T.; Toriba, T.; Fujimoto, M.; Tsutsumi, N.; Kitano, H.; Hirano, H.Y. Conservation and diversification of meristem maintenance mechanism in Oryza sativa: Function of the FLORAL ORGAN NUMBER2 gene. Plant Cell Physiol. 2006, 47, 1591–1602. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, T.; Lee, D.Y.; Miyao, A.; Hirochika, H.; An, G.; Hirano, H.Y. Functional diversification of the two C-class MADS box genes OSMADS3 and OSMADS58 in Oryza sativa. Plant Cell 2006, 18, 15–28. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, K.; Perales, M.; Snipes, S.; Yadav, R.K.; Diaz-Mendoza, M.; Reddy, G.V. DNA-dependent homodimerization, sub-cellular partitioning, and protein destabilization control WUSCHEL levels and spatial patterning. Proc. Natl. Acad. Sci. USA 2016, 113, E6307–E6315. [Google Scholar] [CrossRef] [PubMed]
- Yadav, R.K.; Perales, M.; Gruel, J.; Girke, T.; Jönsson, H.; Reddy, G.V. WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes Dev. 2011, 25, 2025–2030. [Google Scholar] [CrossRef] [PubMed]
- Zuo, J.; Niu, Q.W.; Frugis, G.; Chua, N.H. The WUSCHEL gene promotes vegetative-to-embryonic transition in Arabidopsis. Plant J. 2002, 30, 349–359. [Google Scholar] [CrossRef] [PubMed]
- Gallois, J.L.; Nora, F.R.; Mizukami, Y.; Sablowski, R. WUSCHEL induces shoot stem cell activity and developmental plasticity in the root meristem. Genes Dev. 2004, 18, 375–380. [Google Scholar] [CrossRef]
- Xu, Y.Y.; Wang, X.M.; Li, J.; Li, J.H.; Wu, J.S.; Walker, J.C.; Xu, Z.H.; Chong, K. Activation of the WUS gene induces ectopic initiation of floral meristems on mature stem surface in Arabidopsis thaliana. Plant Mol. Bio. 2005, 57, 773–784. [Google Scholar] [CrossRef]
- Zhou, Y.; Yan, A.; Han, H.; Li, T.; Geng, Y.; Liu, X.; Meyerowitz, E.M. HAIRY MERISTEM with WUSCHEL confines CLAVATA3 expression to the outer apical meristem layers. Science 2018, 361, 502–506. [Google Scholar] [CrossRef]
- Kim, H.-J.; Wu, C.-Y.; Yu, H.-M.; Sheen, J.; Lee, H. Dual CLAVATA3 peptides in Arabidopsis shoot stem cell signaling. J. Plant Biol. 2017, 60, 506–512. [Google Scholar] [CrossRef]
- Mitchum, M.G.; Wang, X.; Davis, E.L. Diverse and conserved roles of CLE peptides. Curr. Op. Plant Biol. 2008, 11, 75–81. [Google Scholar] [CrossRef]
- Perales, M.; Rodriguez, K.; Snipes, S.; Yadav, R.K.; Diaz-Mendoza, M.; Reddy, G.V. Threshold-dependent transcriptional discrimination underlies stem cell homeostasis. Proc. Natl. Acad. Sci. USA 2016, 113, E6298–E6306. [Google Scholar] [CrossRef]
- Daum, G.; Medzihradszky, A.; Suzaki, T.; Lohmann, J.U. A mechanistic framework for noncell autonomous stem cell induction in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 14619–14624. [Google Scholar] [CrossRef]
- Brand, U.; Fletcher, J.C.; Hobe, M.; Meyerowitz, E.M.; Simon, R. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 2000, 289, 617–619. [Google Scholar] [CrossRef]
- Reddy, G.V.; Meyerowitz, E.M. Stem-cell homeostasis and growth dynamics can be uncoupled in the Arabidopsis shoot apex. Science 2005, 310, 663–667. [Google Scholar] [CrossRef]
- Plong, A.; Rodriguez, K.; Alber, M.; Chen, W.; Reddy, G.V. CLAVATA3 mediated simultaneous control of transcriptional and post-translational processes provides robustness to the WUSCHEL gradient. Nat. Commun. 2021, 12, 6361. [Google Scholar] [CrossRef]
- Chongloi, G.L.; Prakash, S.; Vijayaraghavan, U. Regulation of meristem maintenance and organ identity during rice reproductive development. J. Exp. Bot. 2019, 70, 1719–1736. [Google Scholar] [CrossRef]
- DeYoung, B.J.; Bickle, K.L.; Schrage, K.J.; Muskett, P.; Patel, K.; Clark, S.E. The CLAVATA1-related BAM1, BAM2 and BAM3 receptor kinase-like proteins are required for meristem function in Arabidopsis. Plant J. 2006, 45, 1–16. [Google Scholar] [CrossRef]
- Nimchuk, Z.L.; Zhou, Y.; Tarr, P.T.; Peterson, B.A.; Meyerowitz, E.M. Plant stem cell maintenance by transcriptional cross-regulation of related receptor kinases. Development 2015, 142, 1043–1049. [Google Scholar] [CrossRef]
- Durbak, A.R.; Tax, F.E. CLAVATA signaling pathway receptors of Arabidopsis regulate cell proliferation in fruit organ formation as well as in meristems. Genetics 2011, 189, 177–194. [Google Scholar] [CrossRef]
- Cao, X.; He, Z.; Guo, L.; Liu, X. Epigenetic mechanisms are critical for the regulation of WUSCHEL expression in floral meristems. Plant Physiol. 2015, 168, 1189–1196. [Google Scholar] [CrossRef]
- Sun, B.; Zhou, Y.; Cai, J.; Shang, E.; Yamaguchi, N.; Xiao, J.; Looi, L.S.; Wee, W.Y.; Gao, X.; Wagner, D.; et al. Integration of transcriptional repression and polycomb-mediated silencing of WUSCHEL in floral meristems. Plant Cell 2019, 31, 1488–1505. [Google Scholar] [CrossRef] [PubMed]
- Deyhle, F.; Sarkar, A.K.; Tucker, E.J.; Laux, T. WUSCHEL regulates cell differentiation during anther development. Dev. Biol. 2007, 302, 154–159. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Sun, M.; Yuan, C.; Han, Y.; Zheng, T.; Cheng, T.; Wang, J.; Zhang, Q. Interactions between WUSCHEL-and CYC2-like transcription factors in regulating the development of reproductive organs in Chrysanthemum morifolium. Int. J. Mol. Sci. 2019, 20, 1276. [Google Scholar] [CrossRef] [PubMed]
- Barton, M.K. Twenty years on: The inner workings of the shoot apical meristem, a developmental dynamo. Dev. Biol. 2010, 341, 95–113. [Google Scholar] [CrossRef] [PubMed]
- Prunet, N.; Morel, P.; Negrutiu, I.; Trehin, C. Time to stop: Flower meristem termination. Plant Physiol. 2009, 150, 1764–1772. [Google Scholar] [CrossRef]
- Endrizzi, K.; Moussian, B.; Haecker, A.; Levin, J.Z.; Laux, T. The SHOOT MERISTEMLESS gene is required for maintenance of undifferentiated cells in Arabidopsis shoot and floral meristems and acts at a different regulatory level than the meristem genes WUSCHEL and ZWILLE. Plant J. 1996, 10, 967–979. [Google Scholar] [CrossRef]
- Long, J.; Barton, M.K. Initiation of axillary and floral meristems in Arabidopsis. Dev. Biol. 2000, 218, 341–353. [Google Scholar] [CrossRef]
- Krizek, B.A.; Blakley, I.C.; Ho, Y.Y.; Freese, N.; Loraine, A.E. The Arabidopsis transcription factor AINTEGUMENTA orchestrates patterning genes and auxin signaling in the establishment of floral growth and form. Plant J. 2020, 103, 752–768. [Google Scholar] [CrossRef]
- Rojo, E.; Sharma, V.K.; Kovaleva, V.; Raikhel, N.V.; Fletcher, J.C. CLV3 is localized to the extracellular space, where it activates the Arabidopsis CLAVATA stem cell signaling pathway. Plant Cell 2002, 14, 969–977. [Google Scholar] [CrossRef]
- Lohmann, J.U.; Hong, R.L.; Hobe, M.; Busch, M.A.; Parcy, F.; Simon, R.; Weigel, D. A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 2001, 105, 793–803. [Google Scholar] [CrossRef]
- Sun, B.; Ito, T. Regulation of floral stem cell termination in Arabidopsis. Front. Plant Sci. 2015, 6, 17. [Google Scholar] [CrossRef]
- Lenhard, M.; Bohnert, A.; Jürgens, G.; Laux, T. Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell 2001, 105, 805–814. [Google Scholar] [CrossRef]
- Payne, T.; Johnson, S.D.; Koltunow, A.M. KNUCKLES (KNU) encodes a C2H2 zinc-finger protein that regulates development of basal pattern elements of the Arabidopsis gynoecium. Development 2004, 131, 3737–3749. [Google Scholar] [CrossRef]
- Sun, B.; Xu, Y.; Ng, K.H.; Ito, T. A timing mechanism for stem cell maintenance and differentiation in the Arabidopsis floral meristem. Genes Dev. 2009, 23, 1791–1804. [Google Scholar] [CrossRef]
- Guo, L.; Cao, X.; Liu, Y.; Li, J.; Li, Y.; Li, D.; Zhang, K.; Gao, C.; Dong, A.; Liu, X. A chromatin loop represses WUSCHEL expression in Arabidopsis. Plant J. 2018, 94, 1083–1097. [Google Scholar] [CrossRef]
- Gross-Hardt, R.; Lenhard, M.; Laux, T. WUSCHEL signaling functions in interregional communication Arabidopsis ovule development. Genes Dev. 2002, 16, 1129–1138. [Google Scholar] [CrossRef]
- Heisler, M.G.; Ohno, C.; Das, P.; Sieber, P.; Reddy, G.V.; Long, J.A.; Meyerowitz, E.M. Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 2005, 15, 1899–1911. [Google Scholar] [CrossRef]
- Landrein, B.; Vernoux, T. Auxin, Chief Architect of the Shoot Apex. In Auxin and Its Role in Plant Development; Zažímalová, E., Petrášek, J., Benková, E., Eds.; Springer-Verlag: Wien, Austria, 2014; pp. 191–212. [Google Scholar] [CrossRef]
- Schaller, G.E.; Bishopp, A.; Kieber, J.J. The yin-yang of hormones: Cytokinin and auxin interactions in plant development. Plant Cell 2015, 27, 44–63. [Google Scholar] [CrossRef]
- Snipes, S.A.; Rodriguez, K.; DeVries, A.E.; Miyawaki, K.N.; Perales, M.; Xie, M.; Reddy, G.V. Cytokinin stabilizes WUSCHEL by acting on the protein domains required for nuclear enrichment and transcription. PLoS Genet. 2018, 14, e1007351. [Google Scholar] [CrossRef]
- Luo, L.; Zeng, J.; Wu, H.; Tian, Z.; Zhao, Z. A molecular framework for auxin-controlled homeostasis of shoot stem cells in Arabidopsis. Mol. Plant 2018, 11, 899–913. [Google Scholar] [CrossRef]
- Ma, Y.; Miotk, A.; Šutiković, Z.; Ermakova, O.; Wenzl, C.; Medzihradszky, A.; Gaillochet, C.; Forner, J.; Utan, G.; Brackmann, K.; et al. WUSCHEL acts as an auxin response rheostat to maintain apical stem cells in Arabidopsis. Nat. Commun. 2019, 10, 5093. [Google Scholar] [CrossRef] [PubMed]
- Reinhardt, D.; Pesce, E.R.; Stieger, P.; Mandel, T.; Baltensperger, K.; Bennett, M.; Kuhlemeier, C. Regulation of phyllotaxis by polar auxin transport. Nature 2003, 426, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Leibfried, A.; To, J.P.C.; Busch, W.; Stehling, S.; Kehle, A.; Demar, M.; Kieber, J.J.; Lohmann, J.U. WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 2005, 438, 1171–1175. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Miwa, K.; Ishikawa, K.; Yamada, H.; Aiba, H.; Mizuno, T. The arabidopsis sensor His-kinase, AHK4, can respond to cytokinins. Plant Cell Physiol. 2001, 42, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Wang, R.; Zi, H.; Li, Y.; Cao, X.; Li, D.; Lin, D.; Tong, J.; Pan, Y.; Jiao, Y.; et al. AUXIN RESPONSE FACTOR3 regulates floral meristem determinacy by repressing cytokinin biosynthesis and signaling. Plant Cell 2018, 30, 324–346. [Google Scholar] [CrossRef]
- Zhao, Z.; Andersen, S.U.; Ljung, K.; Dolezal, K.; Miotk, A.; Schultheiss, S.J.; Lohmann, J.U. Hormonal control of the shoot stem-cell niche. Nature 2010, 465, 1089–1092. [Google Scholar] [CrossRef]
- Bao, S.; Hua, C.; Shen, L.; Yu, H. New insights into gibberellin signaling in regulating flowering in Arabidopsis. J. Integr. Plant Biol. 2020, 62, 118–131. [Google Scholar] [CrossRef]
- Bolduc, N.; Hake, S. The maize transcription factor KNOTTED1 directly regulates the gibberellin catabolism gene ga20x1. Plant Cell 2009, 21, 1647–1658. [Google Scholar] [CrossRef]
- Yanai, O.; Shani, E.; Dolezal, K.; Tarkowski, P.; Sablowski, R.; Sandberg, G.; Samach, A.; Ori, N. Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr. Biol. 2005, 15, 1566–1571. [Google Scholar] [CrossRef]
- Chung, Y.; Zhu, Y.; Wu, M.F.; Simonini, S.; Kuhn, A.; Armenta-Medina, A.; Jin, R.; Østergaard, L.; Gillmor, C.S.; Wagner, D. Auxin Response Factors promote organogenesis by chromatin-mediated repression of the pluripotency gene SHOOTMERISTEMLESS. Nat. Commu. 2019, 10, 886. [Google Scholar] [CrossRef]
- Jung, J.H.; Lee, S.; Yun, J.; Lee, M.; Park, C.M. The miR172 target TOE3 represses AGAMOUS expression during Arabidopsis floral patterning. Plant Sci. 2014, 215, 29–38. [Google Scholar] [CrossRef]
- Kinoshita, A.; Vayssières, A.; Richter, R.; Sang, Q.; Roggen, A.; van Driel, A.D.; Richard, S.S.; Coupland, G. Regulation of shoot meristem shape by photoperiodic signaling and phytohormones during floral induction of Arabidopsis. Elife 2020, 9, e60661. [Google Scholar] [CrossRef]
- Wang, H.; Caruso, L.V.; Downie, A.B.; Perry, S.E. The embryo MADS domain protein AGAMOUS-Like 15 directly regulated expression of a gene encoding an enzymes involved in gibberellin metabolism. Plant Cell 2004, 166, 1206–1219. [Google Scholar] [CrossRef]
- Li, Q.F.; Wang, C.; Jiang, L.; Li, S.; Sun, S.S.M.; He, J.X. An interaction between BZR1 and DELLAs mediates direct signaling crosstalk between brassinosteroids and gibberellins in Arabidopsis. Sci. Signal. 2012, 5, ra72. [Google Scholar] [CrossRef]
- Browse, J. Jasmonate passes puster: A receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 2009, 60, 183–205. [Google Scholar] [CrossRef]
- Fernández-Calvo, P.; Chini, A.; Fernández-Barbero, G.; Chico, J.M.; Gimenez-Ibanez, S.; Geerinck, J.; Eeckhout, D.; Schweizerd, F.; Godoy, M.; Franco-Zorrillae, J.M.; et al. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 2011, 23, 701–715. [Google Scholar] [CrossRef]
- Wang, H.; Li, Y.; Pan, J.; Lou, D.; Hu, Y.; Yu, D. The bHLH transcription factors MYC2, MYC3, and MYC4 are required for jasmonate-mediated inhibition of flowering in Arabidopsis. Mol. Plant 2017, 10, 1461–1464. [Google Scholar] [CrossRef]
- Wu, K.; Zhang, L.; Zhou, C.; Yu, C.W.; Chaikam, V. HDA6 is required for jasmonate response, senescence and flowering in Arabidopsis. J Exp. Bot. 2008, 59, 225–234. [Google Scholar] [CrossRef]
- Zhou, C.; Zhang, L.; Duan, J.; Miki, B.; Wu, K. HISTONE DEACETYLASE19 is involved in jasmonic acid and ethylene signaling of pathogen response in Arabidopsis. Plant Cell 2005, 17, 1196–1204. [Google Scholar] [CrossRef]
- Finkelstein, R.; Reeves, W.; Ariizumi, T.; Steber, C. Molecular aspects of seed dormancy. Annu. Rev. Plant. Biol. 2008, 59, 387–415. [Google Scholar] [CrossRef]
- Wang, Y.; Li, L.; Ye, T.; Lu, Y.; Chen, X.; Wu, Y. The inhibitory effect of ABA on floral transition is mediated by ABI5 in Arabidopsis. J. Exp. Bot. 2013, 64, 675–684. [Google Scholar] [CrossRef] [PubMed]
- Riboni, M.; Robustelli Test, A.; Galbiati, M.; Tonelli, C.; Conti, L. ABA-dependent control of GIGANTEA signaling enables drought escape via up-regulation of FLOWERING LOCUS T in Arabidopsis thaliana. J. Exp. Bot. 2016, 67, 6309–6632. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, R.; Schippers, J.H.M. ROS-mediated redox signaling during cell differentiation in plants. Biochim. Biophys. Acta-Gen. Subj. 2015, 1850, 1497–1508. [Google Scholar] [CrossRef] [PubMed]
- He, L.; He, T.; Farrar, S.; Ji, L.; Liu, T.; Ma, X. Antioxidants maintain cellular redox homeostasis by elimination of reactive oxygen species. Cell. Phy. Biochem. 2017, 44, 532–553. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.; Dong, Z.; Wu, H.; Tian, Z.; Zhao, Z. Redox regulation of plant stem cell fate. EMBO J. 2017, 36, 2844–2855. [Google Scholar] [CrossRef]
- Wang, Y.; Shirakawa, M.; Ito, T. Dynamic changes in reactive oxygen species in the shoot apex contribute to stem cell death in Arabidopsis thaliana. Int. J. Mol. Sci. 2022, 2022, 3864. [Google Scholar] [CrossRef]
- Berrios, L.; Rentsch, J.D. Linking reactive oxygen species (ROS) to abiotic and biotic feedbacks in plant microbiomes: The dose makes the poison. Int. J. Mol. Sci. 2022, 23, 4402. [Google Scholar] [CrossRef]
- Leijten, W.; Koes, R.; Roobeek, I.; Frugis, G. Translating flowering time from Arabidopsis thaliana to Brassicaceae and Asteraceae crop species. Plants 2018, 7, 111. [Google Scholar] [CrossRef]
- Sheldon, C.C.; Rouse, D.T.; Finnegan, E.J.; Peacock, W.J.; Dennis, E.S. The molecular basis of vernalization: The central role of FLOWERING LOCUS C (FLC). Proc. Natl. Acad. Sci. USA 2000, 97, 3753–3758. [Google Scholar] [CrossRef]
- Shindo, C.; Aranzana, M.J.; Lister, C.; Baxter, C.; Nicholls, C.; Nordborg, M.; Dean, C. Role of FRIGIDA and FLOWERING LOCUS C in determining variation in flowering time of Arabidopsis. Plant Physiol. 2005, 138, 1163–1173. [Google Scholar] [CrossRef]
- Choi, K.; Kim, J.; Hwang, H.-J.; Kim, S.; Park, C.; Kim, S.Y.; Lee, I. The FRIGIDA complex activates transcription of FLC, a strong flowering repressor in Arabidopsis by recruiting chromatin modification factors. Plant Cell 2011, 23, 289–303. [Google Scholar] [CrossRef]
- Kim, D.H.; Doyle, M.R.; Sung, S.; Amasino, R.M. Vernalization: Winter and the timing of flowering in plants. Annu. Rev. Cell Dev. Biol. 2009, 25, 277–299. [Google Scholar] [CrossRef]
- Bastow, R.; Mylne, J.; Lister, C.; Lippman, Z.; Martienssen, R.; Dean, C. Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 2004, 427, 164–167. [Google Scholar] [CrossRef]
- Amasino, R.M.; Sung, S. Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 2004, 427, 159–164. [Google Scholar] [CrossRef]
- Yang, H.; Howard, M.; Dean, C. Antagonistic roles for H3K36me3 and H3K27me3 in the cold-induced epigenetic switch at Arabidopsis FLC. Curr. Biol. 2014, 24, 1793–1797. [Google Scholar] [CrossRef]
- Blazquez, M.A.; Green, R.; Nilsson, O.; Sussman, M.R.; Weigel, D. Gibberelllins promote flowering Arabidopsis by activating the LEAFY promoter. Plant Cell 1998, 10, 791–800. [Google Scholar] [CrossRef]
- Liu, C.; Chen, H.; Er, H.L.; Soo, H.M.; Kumar, P.; Han, J.H.; Liou, Y.C.; Yu, H. Direct interaction of AGL24 and SOC1 integrates flowering signals in Arabidopsis. Development 2008, 135, 1481–1491. [Google Scholar] [CrossRef]
- Sheldon, C.C.; Hills, M.J.; Lister, C.; Dean, C.; Dennis, E.S.; Peacock, W.J. Resetting of FLOWERING LOCUS C expression after epigenetic repression by vernalization. Proc. Natl. Acad. Sci. USA 2008, 105, 2214–2219. [Google Scholar] [CrossRef]
- Choi, J.; Hyun, Y.; Kang, M.J.; Yun, H.I.; Yun, J.Y.; Lister, C.; Dean, C.; Amasino, R.M.; Noh, B.; Noh, Y.S.; et al. Resetting and regulation of FLOWERING LOCUS C expression during Arabidopsis reproductive development. Plant J. 2009, 57, 918–931. [Google Scholar] [CrossRef]
- Angel, A.; Song, J.; Dean, C.; Howard, M. A Polycomb-based switch underlying quantitative epigenetic memory. Nature 2011, 476, 105–108. [Google Scholar] [CrossRef]
- Yun, H.; Hyun, Y.; Kang, M.J.; Noh, Y.S.; Noh, B.; Choi, Y. Identification of regulators required for the reactivation of FLOWERING LOCUS C during Arabidopsis reproduction. Planta 2011, 234, 1237–1250. [Google Scholar] [CrossRef] [PubMed]

| Plant | Genes | Orthologs | References |
|---|---|---|---|
| ARABIDOPSIS (dicot) | WUS | Ref. [7] | |
| CLV1 | Ref. [8] | ||
| CLV2 | |||
| CLV3 | |||
| AG | Ref. [18] | ||
| TOMATO (dicot) | WUS | SlWUS | Ref. [9] |
| CLV1 | fasciated and branched(FAB) | Ref. [10] | |
| CLV2 | SlCLV2 | ||
| CLV3 | SlCLV3 | ||
| AG | Tomato Agamous1 (TAG1) | Ref. [19] | |
| MAIZE (monocot) | WUS | ZmWUS1 and ZmWUS2 | Ref. [11] |
| CLV1 | THICK TASSEL DWARF1 (TD1) | Ref. [12] | |
| CLV2 | FASCIATED EAR2 (FEA2) | Ref [13] | |
| CLV3 | ZmCLE7, ZmCLE14, and ZmFON2-LIKE CLE PROTEIN1 (ZmFCP1) | Ref. [14] | |
| AG | Zea mays Agamous 1 (ZAG1) | Ref. [20] | |
| RICE (monocot) | WUS | TILLERS ABSENT1 (TAB1) | Ref. [15] |
| CLV1 | FLORAL ORGAN NUMBER 1 (FON1) | Ref. [16] | |
| CLV2 | - | ||
| CLV3 | FON2, FON4 | Refs. [17,21] | |
| AG | OsMADS3 and OsMADS58 | Ref. [22] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Agarwal, Y.; Shukla, B.; Manivannan, A.; Soundararajan, P. Paradigm and Framework of WUS-CLV Feedback Loop in Stem Cell Niche for SAM Maintenance and Cell Identity Transition. Agronomy 2022, 12, 3132. https://doi.org/10.3390/agronomy12123132
Agarwal Y, Shukla B, Manivannan A, Soundararajan P. Paradigm and Framework of WUS-CLV Feedback Loop in Stem Cell Niche for SAM Maintenance and Cell Identity Transition. Agronomy. 2022; 12(12):3132. https://doi.org/10.3390/agronomy12123132
Chicago/Turabian StyleAgarwal, Yamini, Bhavya Shukla, Abinaya Manivannan, and Prabhakaran Soundararajan. 2022. "Paradigm and Framework of WUS-CLV Feedback Loop in Stem Cell Niche for SAM Maintenance and Cell Identity Transition" Agronomy 12, no. 12: 3132. https://doi.org/10.3390/agronomy12123132
APA StyleAgarwal, Y., Shukla, B., Manivannan, A., & Soundararajan, P. (2022). Paradigm and Framework of WUS-CLV Feedback Loop in Stem Cell Niche for SAM Maintenance and Cell Identity Transition. Agronomy, 12(12), 3132. https://doi.org/10.3390/agronomy12123132








