Evaluation and Identification of Stable Chickpea Lines for Yield-Contributing Traits from an Association Mapping Panel
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Statistical Analysis
3. Results
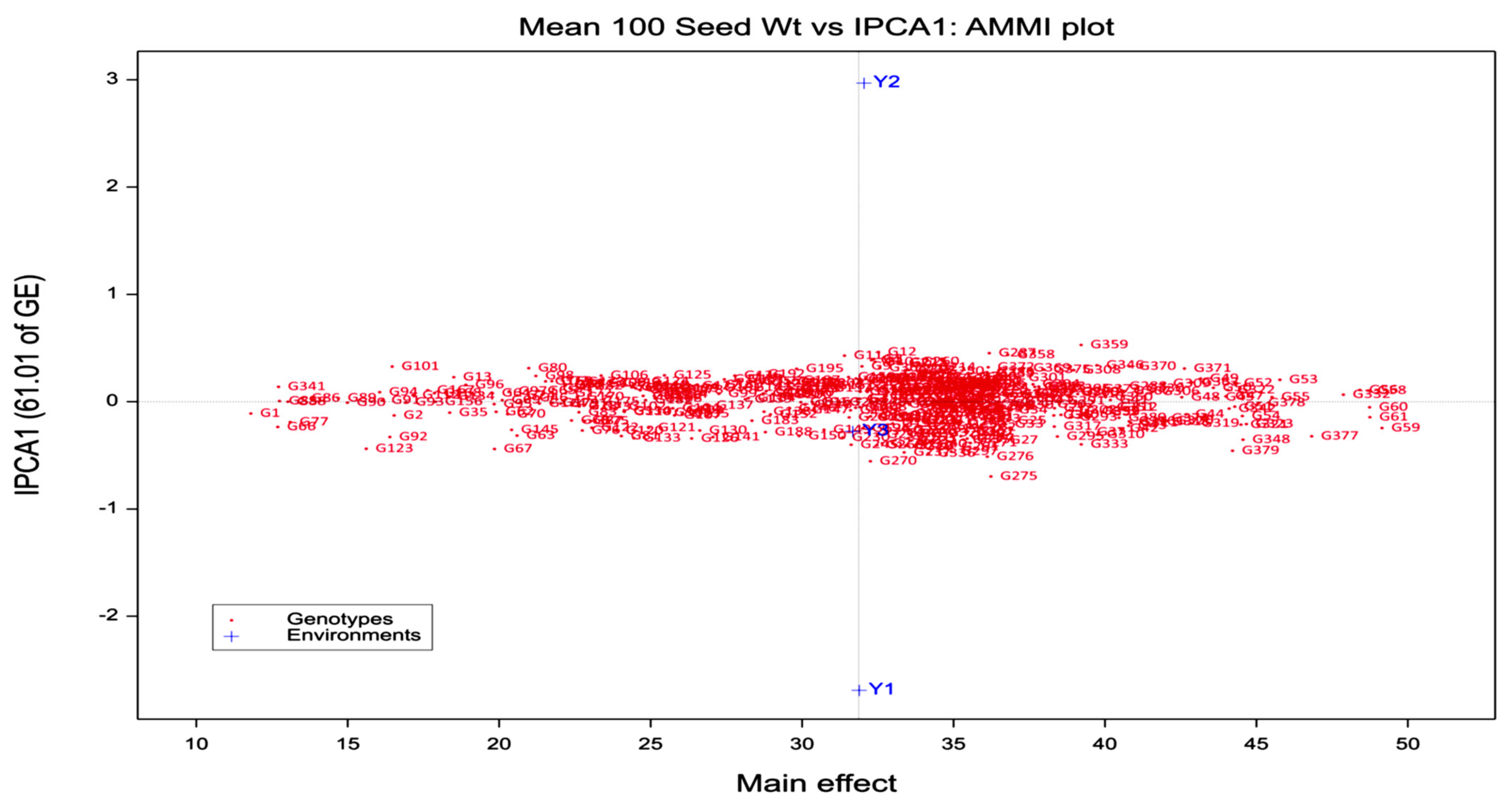
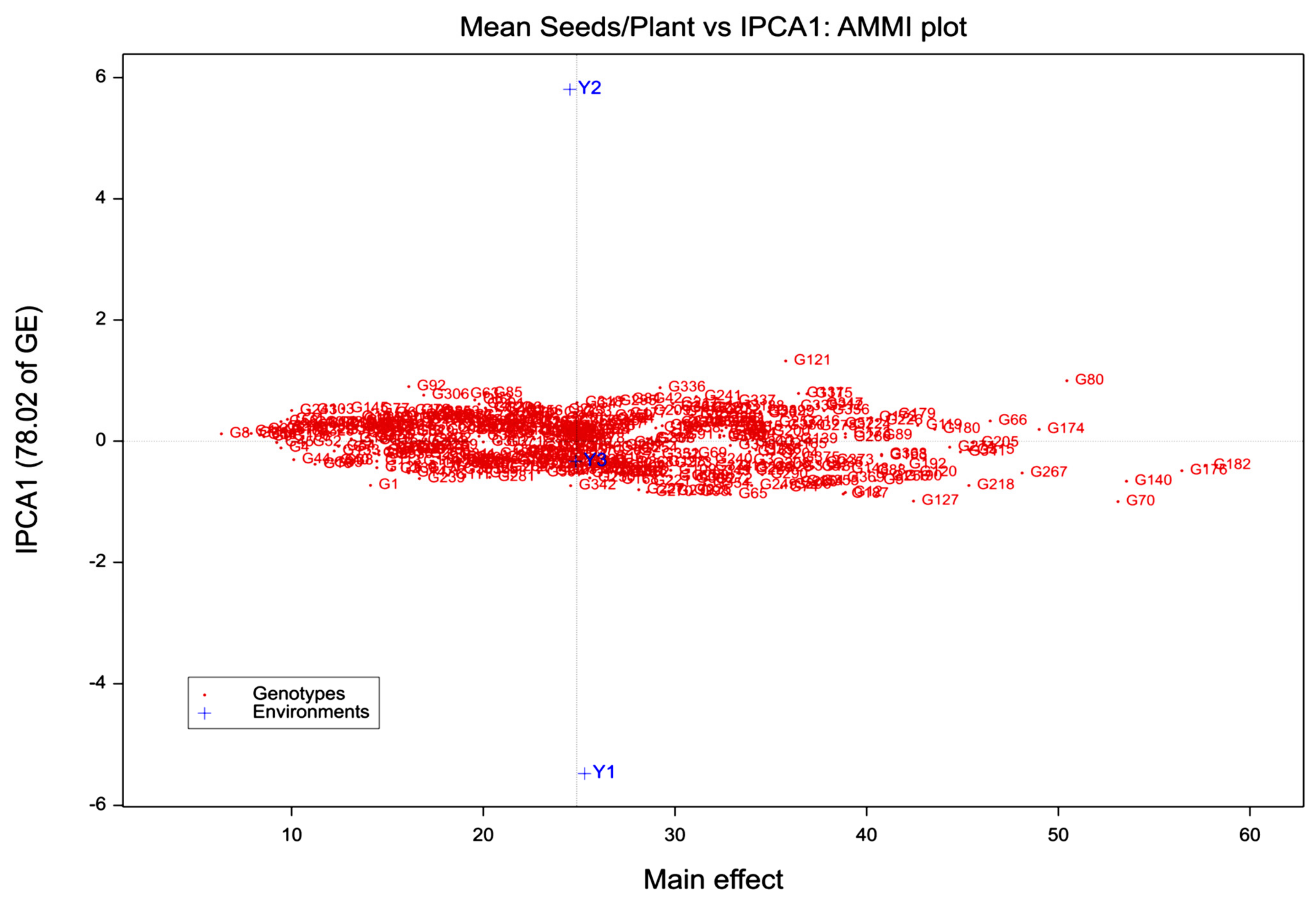
3.1. AMMI Analysis
3.2. Ascertaining High-Yielding and Stable Genotypes
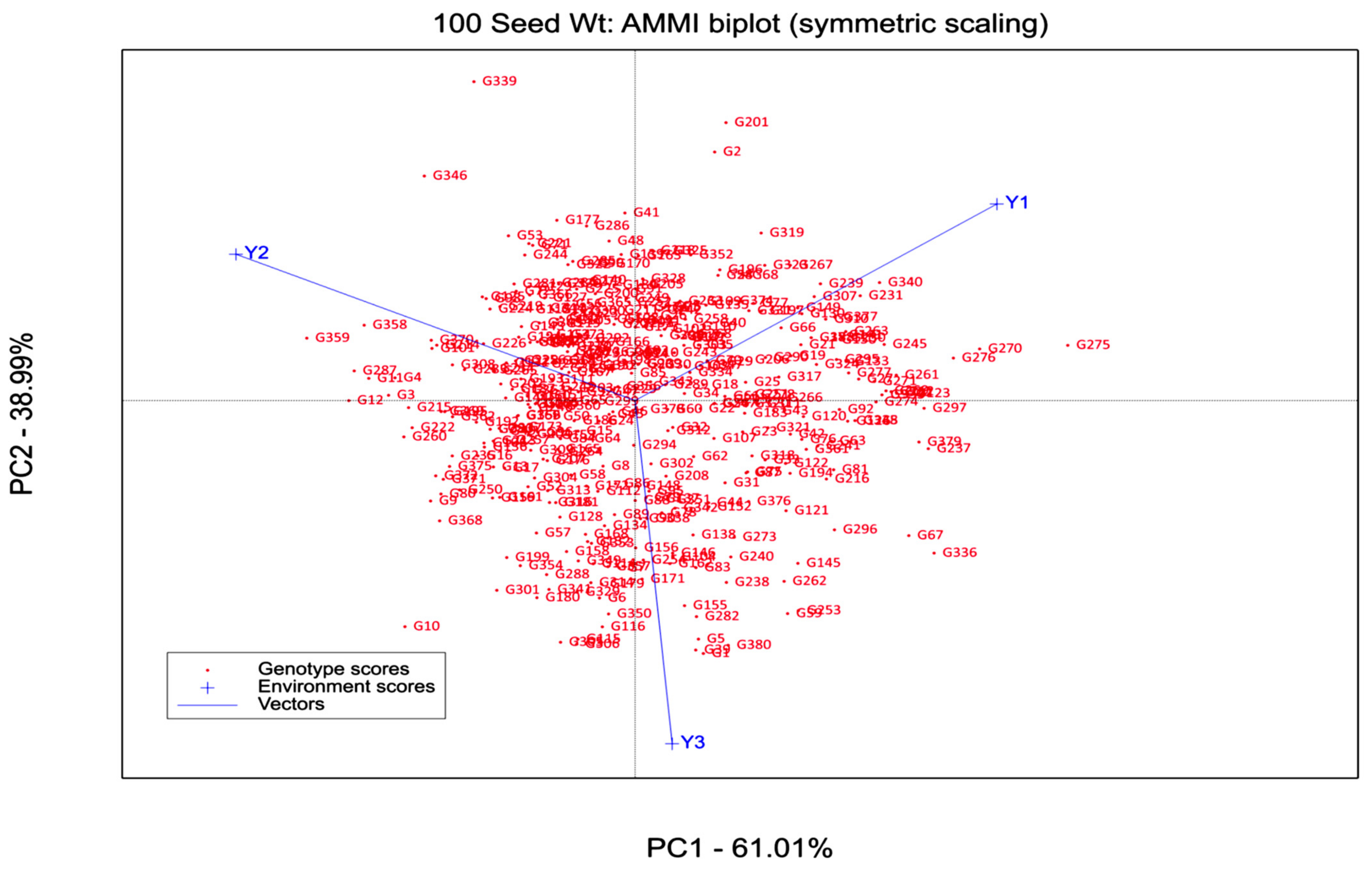
3.3. AMM2 Biplot
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Haider, M.; Haider, S.Q. Assessment of protein-calorie malnutrition. Clin. Chem. 1984, 30, 1286–1299. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, A.; Khalil, I.A.; Ateeq, N.; Sayyar Khan, M. Nutritional quality of important food legumes. Food Chem. 2006, 97, 331–335. [Google Scholar] [CrossRef]
- UNICEF Data. From Promise to Impact Ending Malnutrition By 2030. 2016. Available online: https://data.unicef.org/wp-content/uploads/2016/06/130565-1.pdf. (accessed on 24 December 2018).
- Maiti, R.K.; Wesche-Ebeling, P. Vegetative and reproductive growth and productivity. In Advances in Chickpea Science; Maiti, R., Wesche-Ebeling, P., Eds.; Science Publishers: Enfield, NH, USA, 2001; Volume 67–104. [Google Scholar]
- FAOSTAT. 2020. Available online: http://www.fao.org/faostat/en/#data/QC (accessed on 24 December 2018).
- Gil, J.; Nadal, S.; Luna, D.; Moreno, M.T.; Haro, A.D. Variability of some physico-chemical characters in Desi and Kabuli chickpea types. J. Sci. Food Agril. 1996, 71, 179–184. [Google Scholar] [CrossRef]
- Ibrikci, H.; Knewtson, S.J.; Grusak, M.A. Chickpea leaves as a vegetable green for humans: Evaluation of mineral composition. J. Sci. Food Agric. 2003, 83, 945–950. [Google Scholar] [CrossRef]
- Gauch, H.G., Jr. Model selection and validation for yield trials with interaction. Biometrics 1988, 1, 705–715. [Google Scholar] [CrossRef]
- Vaghela, M.D.; Poshiya, V.K.; Savaliya, J.J.; Davada, B.K.; Mungra, K.D. Studies on character association and path analysis for seed yield and its components in chickpea (Cicer arietinum L.). Legume Res. 2009, 32, 245–249. [Google Scholar]
- Monpara, B.A.; Gaikwad, S.R. Combining high seed number and weight to improve seed yield potential of chickpea in India. Afr. Crop Sci. J. 2014, 22, 1–8. [Google Scholar]
- Tiwari, A.; Babbar, A.; Pal, N. Genetic variability, correlation and path analysis in yield and yield components in chickpea (Cicer arietinum L.) genotypes under late sown condition. Int. J. Agric. Res. 2016, 8, 2884–2886. [Google Scholar]
- Dumbre, A.D.; Deshmukh, R.B.; Navale, P.A. Analysis of genetic variability in chickpea. J. Maharashtra Agril. Univ. 1984, 9, 350. [Google Scholar]
- Saki, A.; Zaman, M.; Tuhina-Khatun, M.; Kamal, M.; Begum, H. Genetic Variability, Correlation and Path Co-Efficient Analysis for Agronomic Traits in Chickpea (Cicer arietinum L.). Agriculturists 2010, 7, 12–21. [Google Scholar] [CrossRef]
- Babbar, A.; Prakash, V.; Tiwari, P.; Iquebal, M.A. Genetic variability for chickpea (Cicer arietinum L.) under late sown season. Legume Res. Int. J. 2012, 35, 1–7. [Google Scholar]
- Salgotra, S.K. Association Studies for Seed Yield and Its Attributing Traits in Desi Chickpea (Cicer arietinum L.) Varieties. Legume Genom. Genet. 2016, 7, 1–6. [Google Scholar] [CrossRef]
- Kumar, N.; Bharadwaj, C.; Satyavathi, C.T.; Pal, M.; Kumar, T.; Singhal, T.; Sachdeva, S.; Jain, P.K.; Patil, B.S.; Soren, K.R. Morpho Physiological Characterization and Grouping (SAHN) of Chickpea Genotypes for Salinity Tolerance. Vegetos 2017, 30, 3. [Google Scholar] [CrossRef]
- Biçer, B.T. The effect of seed size on yield and yield components of chickpea and lentil. Afr. J. Biotechnol. 2009, 8, 1482–1487. [Google Scholar]
- Zobel, R.W.; Wright, M.J.; Gauch, H.G., Jr. Statistical Analysis of a Yield Trial. Agron. J. 1988, 80, 388–393. [Google Scholar] [CrossRef]
- Gauch, H.G., Jr. AMMI analysis on yield trials. In CIMMYT Wheat Special Report; CIMMYT: Veracruz, México, 1992. [Google Scholar]
- Purchase, J.L. Parametric Snalysis to Describe Genotype x Environment Interaction and Yield Stability in Winter Wheat. Ph.D. Thesis, University of the Free State, Bloemfontein, Africa, 1997. [Google Scholar]
- Gauch, H.G. A Simple Protocol for AMMI Analysis of Yield Trials. Crop Sci. 2013, 53, 1860–1869. [Google Scholar] [CrossRef]
- Thangavel, P.; Anandan, A.; Eswaran, R. AMMI analysis to comprehend genotype-by-environment (G × E) interactions in rainfed grown mungbean (Vigna radiata L.). Aust. J. Crop Sci. 2011, 5, 1767–1775. [Google Scholar]
- Farshadfar, E.; Sutka, J. Biplot analysis of genotype-environment interaction in durum wheat using the AMMI model. Acta Agron. Hung. 2006, 54, 459–467. [Google Scholar] [CrossRef]
- Osiru, M.O.; Olanya, O.M.; Adipala, E.; Kapinga, R.; Lemaga, B. Yield stability analysis of Ipomoea batatus L. cultivars in diverse environments. Aust. J. Crop Sci. 2009, 3, 213–220. [Google Scholar]
- Annicchiarico, P. Additive main effects and multiplicative interaction (AMMI) analysis of genotype-location interaction in variety trials repeated over years. Theor. Appl. Genet. 1997, 94, 1072–1077. [Google Scholar] [CrossRef]
- McLaren, C.G.; Chaudhary, R.C. Use of additive main effects and multiplicative Interaction models to analyse multilocation rice varietal trials. Oryza 1998, 34, 306–318. [Google Scholar]
- Ise, K.; Youquan, S.; Jishin, L.; Kudo, S.; Tano, H.; Sunohora, Y. Genotype by environment interaction analysis for rice yield in Yunan, China. Jpn. J. Trop. Agric. 2001, 45, 22–32. [Google Scholar]
- Das, S.; Misra, R.C.; Patnaik, M.C. G x E interaction of mid-late rice genotypes in LR and AMMI model and evaluation of adaptability and yield stability. Env. Ecol. 2009, 27, 529–535. [Google Scholar]
- Nachit, M.M.; Nachit, G.; Ketata, H.; Gauch, H.G.; Zobel, R.W. Use of AMMI and linear regression models to analyze genotype-environment interaction in durum wheat. Theor. Appl. Genet. 1992, 83, 597–601. [Google Scholar] [CrossRef] [PubMed]
- Rad, M.N.; Kadir, M.A.; Rafii, M.Y.; Jaafar, H.Z.; Naghavi, M.R.; Ahmadi, F. Genotype environment interaction by AMMI and GGE biplot analysis in three consecutive generations of wheat (Triticum aestivum) under normal and drought stress conditions. Aust. J. Crop Sci. 2013, 7, 956–961. [Google Scholar]
- Pawar, V.Y.; Patil, H.T.; Patil, H.S. AMMI analysis for grain yield stability of pearl millet (Penniseum glaucum L.) genotypes. Indian J. Genet. 2012, 72, 79–82. [Google Scholar]
- Anuradha, N.; Satyavathi, C.T.; Meena, M.C.; Sankar, S.M.; Bharadwaj, C.; Bhat, J.; Singh, O.; Singh, S.P. Evaluation of pearl millet [Pennisetum glaucum (L.) R. Br.] for grain iron and zinc content in different agro climatic zones of India. Indian J. Genet. Plant Breed. 2017, 77, 65–73. [Google Scholar] [CrossRef]
- Sabaghpour, S.H.; Razavi, F.; Danyali, S.F.; Tobe, D.; Ebadi, A. Additive Main Effect and Multiplicative Interaction Analysis for Grain Yield of Chickpea (Cicer arietinum L.) in Iran. Int. Sch. Res. Not. 2012, 2012, 1–6. [Google Scholar] [CrossRef][Green Version]
- Balapure, M.M.; Mhase, L.B.; Kute, N.S.; Pawar, V.Y. AMMI analysis for stability of chickpea. Legum. Res. -Int. J. 2016, 39, 301–304. [Google Scholar] [CrossRef]
- Kanouni, H.; Farayedi, Y.; Saeid, A.; Sabaghpour, S.H. Stability Analyses for Seed Yield of Chickpea (Cicer arietinum L.) Genotypes in the Western Cold Zone of Iran. J. Agric. Sci. 2015, 7, 219. [Google Scholar] [CrossRef][Green Version]
- Akter, A.; Jamil, H.M.; Umma, K.M.; Islam, M.R.; Hossain, K.; Mamunur, R.M. AMMI Biplot Analysis for Stability of Grain Yield in Hybrid Rice (Oryza sativa L.). J. Rice Res. 2014, 2, 126. [Google Scholar] [CrossRef]
- Freeman, G.H. Modern statistical methods for analyzing genotype-environment interactions. In Genotype × Environment Interaction and Plant Breeding; Kang, M.S., Ed.; Louisiana State University Agricultural Center: Baton Rouge, LA, USA, 1990; pp. 118–125. [Google Scholar]
- Gauch, H.G.; Zobel, R.W. AMMI analysis of yield trials. In Genotype by Environment Interaction; Kang, M.S., Gauch, H.G., Jr, Eds.; CRC Press: Boca Raton, FL, USA, 1996; pp. 85–122. [Google Scholar]

| Cropping Season (October–April) | Rainfall (mm) | Temperature (°C) | |
|---|---|---|---|
| Minimum | Maximum | ||
| 2014–15 | 315.8 | 6.6 | 34.8 |
| 2015–16 | 22.0 | 6.1 | 38.7 |
| 2016–17 | 127.7 | 5.3 | 38.0 |
| Source | d.f. | Seed Weight | Seed Number | ||
|---|---|---|---|---|---|
| MSS | Variance (%) | MSS | Variance (%) | ||
| Genotypes | 379 | 606.3 ** | 93.08 | 606.3 ** | 93.08 |
| Environments | 2 | 457.4 ** | 0.37 | 457.4 ** | 0.37 |
| Rep within Env | 6 | 31.7 ** | 31.7 ** | ||
| G × E | 758 | 13.4 ** | 4.1 | 13.4 ** | 4.1 |
| Error | 2274 | 2.6 | 2.6 | ||
| Total | 3419 | 72.2 | 72.2 | ||
| Source | d.f. | Seed Weight | Seed Number | ||||
|---|---|---|---|---|---|---|---|
| MSS | % GE Explained | % Cumulative | MSS | % GE explained | % Cumulative | ||
| Treatments | 1139 | 211.5 ** | 211.5 ** | ||||
| Genotypes | 379 | 606.3 ** | 606.3 ** | ||||
| Environments | 2 | 457.4 ** | 457.4 ** | ||||
| G X E | 758 | 13.4 ** | 13.4 ** | ||||
| IPCA1 | 380 | 24.4 ** | 61.01 | 61.01 | 24.4 ** | 78.02 | 78.02 |
| IPCA2 | 378 | 2.3 ** | 38.99 | 100 | 2.3 ** | 21.98 | 100 |
| Residual | 0 | 0 | 0 | ||||
| Error | 2274 | 2.6 | 2.6 | ||||
| Total | 3419 | 72.2 | 72.2 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shimray, P.W.; Bharadwaj, C.; Patil, B.S.; Sankar, S.M.; Kumar, N.; Reddy, S.P.P.; Singhal, T.; Hegde, V.; Parida, S.K.; Roorkiwal, M.; et al. Evaluation and Identification of Stable Chickpea Lines for Yield-Contributing Traits from an Association Mapping Panel. Agronomy 2022, 12, 3115. https://doi.org/10.3390/agronomy12123115
Shimray PW, Bharadwaj C, Patil BS, Sankar SM, Kumar N, Reddy SPP, Singhal T, Hegde V, Parida SK, Roorkiwal M, et al. Evaluation and Identification of Stable Chickpea Lines for Yield-Contributing Traits from an Association Mapping Panel. Agronomy. 2022; 12(12):3115. https://doi.org/10.3390/agronomy12123115
Chicago/Turabian StyleShimray, Philanim Wungmarong, C. Bharadwaj, B. S. Patil, S. Mukesh Sankar, Neeraj Kumar, Sneha Priya Pappula Reddy, Tripti Singhal, Venkatraman Hegde, Swarup K. Parida, Manish Roorkiwal, and et al. 2022. "Evaluation and Identification of Stable Chickpea Lines for Yield-Contributing Traits from an Association Mapping Panel" Agronomy 12, no. 12: 3115. https://doi.org/10.3390/agronomy12123115
APA StyleShimray, P. W., Bharadwaj, C., Patil, B. S., Sankar, S. M., Kumar, N., Reddy, S. P. P., Singhal, T., Hegde, V., Parida, S. K., Roorkiwal, M., Varshney, R. K., & Verma, P. (2022). Evaluation and Identification of Stable Chickpea Lines for Yield-Contributing Traits from an Association Mapping Panel. Agronomy, 12(12), 3115. https://doi.org/10.3390/agronomy12123115









