Genetic Diversity Assessment of Sweetpotato Germplasm in China Using InDel Markers
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. DNA Extraction and InDel Genotyping
2.3. Allele Scoring and Data Analysis
3. Results
3.1. InDel Marker Polymorphism and Analysis of Molecular Variance (AMOVA)
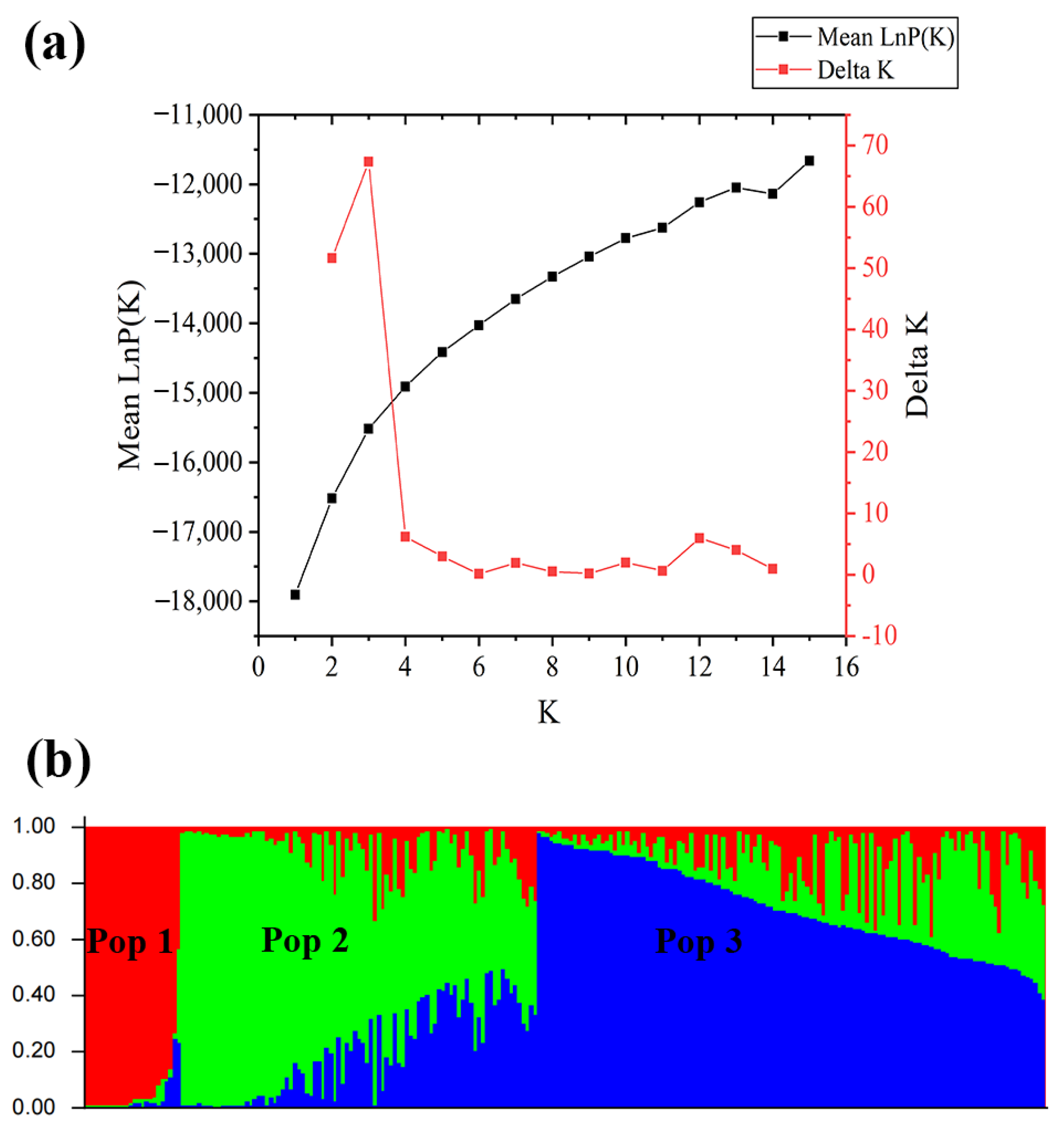
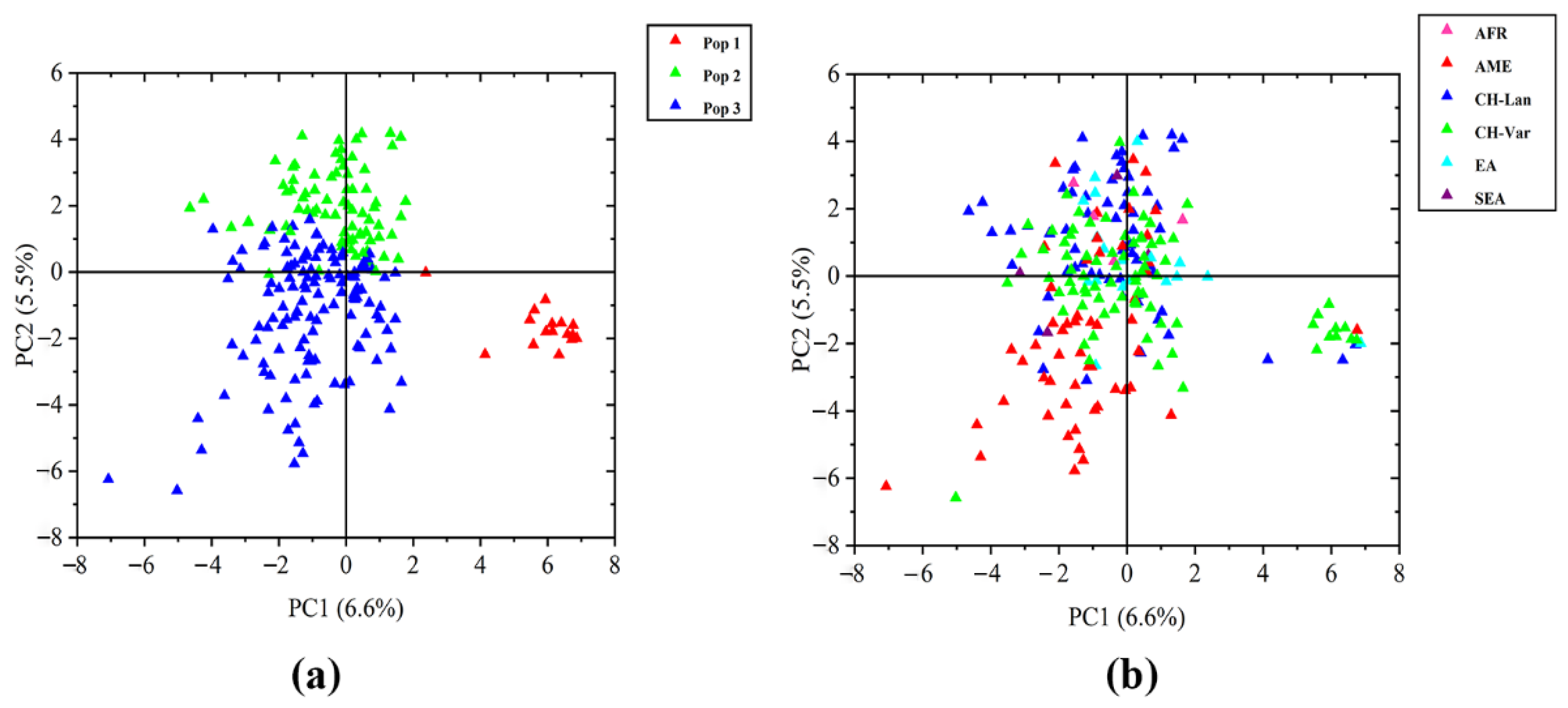
3.2. Population Structure and Principal Component Analysis
3.3. Genetic Distance and Phylogenetic Relationship Analysis
4. Discussion
4.1. Characteristics of InDel Markers and Analysis of Molecular Variance (AMOVA)
4.2. Population Structure and Principal Component Analysis
4.3. Number of Alleles and Genetic Distance among Different Regions
4.4. Phylogenetic Relationship Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Schafleitner, R.; Tincopa, L.R.; Palomino, O.; Rossel, G.; Robles, R.F.; Alagon, R.; Rivera, C.; Quispe, C.; Rojas, L.; Pacheco, J.A.; et al. A sweetpotato gene index established by de novo assembly of pyrosequencing and Sanger sequences and mining for gene-based microsatellite markers. BMC Genom. 2010, 11, 604. [Google Scholar] [CrossRef] [PubMed]
- Bovell-Benjamin, A.C. Sweet Potato: A Review of its Past, Present, and Future Role in Human Nutrition. In Advances in Food and Nutrition Research; Academic Press: Cambridge, MA, USA, 2007; Volume 52, pp. 1–59. [Google Scholar]
- Wadl, P.A.; Olukolu, B.A.; Branham, S.E.; Jarret, R.L.; Yencho, G.C.; Jackson, D.M. Genetic Diversity and Population Structure of the USDA Sweetpotato (Ipomoea batatas) Germplasm Collections Using GBSpoly. Front. Plant Sci. 2018, 9, 1166. [Google Scholar] [CrossRef] [PubMed]
- Food and Agriculture Organization of the United Nations. FAOSTAT Statistical Database. FAO. 2020. Available online: https://www.fao.org/faostat/en/#data (accessed on 27 August 2022).
- Gichuki, S.T.; Berenyi, M.; Zhang, D.; Hermann, M.; Schmidt, J.; Glössl, J.; Burg, K. Genetic diversity in sweetpotato [Ipomoea batatas (L.) Lam.] in relationship to geographic sources as assessed with RAPD markers. Genet. Resour. Crop Evol. 2003, 50, 429–437. [Google Scholar] [CrossRef]
- O’Brien, P.J. The Sweet Potato: Its Origin and Dispersal. Am. Anthropol. 1972, 74, 342–365. [Google Scholar] [CrossRef]
- Feng, J.Y.; Li, M.; Zhao, S.; Zhang, C.; Yang, S.T.; Qiao, S.; Tan, W.F.; Qu, H.J.; Wang, D.Y.; Pu, Z.G. Analysis of evolution and genetic diversity of sweetpotato and its related different polyploidy wild species I. trifida using RAD-seq. BMC Plant Biol. 2018, 18, 181. [Google Scholar] [CrossRef] [PubMed]
- Okada, Y.; Monden, Y.; Nokihara, K.; Shirasawa, K.; Isobe, S.; Tahara, M. Genome-Wide Association Studies (GWAS) for Yield and Weevil Resistance in Sweet potato (Ipomoea batatas (L.) Lam). Plant Cell Rep. 2019, 38, 1383–1392. [Google Scholar] [CrossRef]
- Zhang, L.; Yu, Y.; Shi, T.; Kou, M.; Sun, J.; Xu, T.; Li, Q.; Wu, S.; Cao, Q.; Hou, W.; et al. Genome-wide analysis of expression quantitative trait loci (eQTLs) reveals the regulatory architecture of gene expression variation in the storage roots of sweet potato. Hortic. Res. 2020, 7, 90. [Google Scholar] [CrossRef] [PubMed]
- Yada, B.; Brown-Guedira, G.; Alajo, A.; Ssemakula, G.; Mwanga, R.; Yencho, C. Simple Sequence Repeat Marker Analysis of Genetic Diversity among Progeny of a Biparental Mapping Population of Sweetpotato. HortScience 2015, 50, 1143–1147. [Google Scholar] [CrossRef]
- Zawedde, B.M.; Ghislain, M.; Magembe, E.; Amaro, G.B.; Grumet, R.; Hancock, J. Characterization of the genetic diversity of Uganda’s sweet potato (Ipomoea batatas) germplasm using microsatellites markers. Genet. Resour. Crop Evol. 2015, 62, 501–513. [Google Scholar] [CrossRef]
- Pereira-Dias, L.; Vilanova, S.; Fita, A.; Prohens, J.; Rodríguez-Burruezo, A. Genetic diversity, population structure, and relationships in a collection of pepper (Capsicum spp.) landraces from the Spanish centre of diversity revealed by genotyping-by-sequencing (GBS). Hortic. Res. 2019, 6, 54. [Google Scholar] [CrossRef]
- Anglin, N.L.; Robles, R.; Rossel, G.; Alagon, R.; Panta, A.; Jarret, R.L.; Manrique, N.; Ellis, D. Genetic Identity, Diversity, and Population Structure of CIP’s Sweetpotato (I. batatas) Germplasm Collection. Front. Plant Sci. 2021, 12, 1860. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Kong, L.; Yu, K.; Zhang, F.; Shi, X.; Wang, Y.; Nan, H.; Zhao, X.; Lu, S.; Cao, D.; et al. Development and validation of InDel markers for identification of QTL underlying flowering time in soybean. Crop J. 2018, 6, 126–135. [Google Scholar] [CrossRef]
- Liu, L.; Dang, P.M.; Chen, C.Y. Development and Utilization of InDel Markers to Identify Peanut (Arachis hypogaea) Disease Resistance. Front. Plant Sci. 2015, 6, 988. [Google Scholar] [CrossRef] [PubMed]
- Allen, G.C.; Flores-Vergara, M.A.; Krasynanski, S.; Kumar, S.; Thompson, W.F. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat. Protoc. 2006, 1, 2320–2325. [Google Scholar] [CrossRef]
- Xiao, S.; Dai, X.; Zhao, L.; Zhou, Z.; Zhao, L.; Xu, P.; Gao, B.; Zhang, A.; Zhao, D.; Yuan, R.; et al. Resequencing of sweetpotato germplasm resources reveals key loci associated with multiple agronomic traits. Hortic. Res. 2022. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research--an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Yeh, F.; Boyle, T. Population genetic analysis of co-dominant and dominant markers and quantitative traits. Belg. J. Bot. 1996, 129, 157. [Google Scholar]
- Porras-Hurtado, L.; Ruiz, Y.; Santos, C.; Phillips, C.; Carracedo, A.; Lareu, M.V. An overview of STRUCTURE: Applications, parameter settings, and supporting software. Front. Genet. 2013, 4, 98. [Google Scholar] [CrossRef]
- Tang, Q.-Y.; Zhang, C.-X. Data Processing System (DPS) software with experimental design, statistical analysis and data mining developed for use in entomological research. Insect Sci. 2013, 20, 254–260. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Tomiuk, J.; Graur, D. Nei’s Modified Genetic Identity and Distance Measures and their Sampling Variances. Syst. Biol. 1988, 37, 156–162. [Google Scholar] [CrossRef]
- Moberly, J.G.; Bernards, M.T.; Waynant, K.V. Key features and updates for Origin 2018. J. Cheminf. 2018, 10, 5. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Wu, K.; Yang, M.; Liu, H.; Tao, Y.; Mei, J.; Zhao, Y. Genetic analysis and molecular characterization of Chinese sesame (Sesamum indicum L.) cultivars using Insertion-Deletion (InDel) and Simple Sequence Repeat (SSR) markers. BMC Genet. 2014, 15, 35. [Google Scholar] [CrossRef]
- Monteros-Altamirano, A.; Paredes, D.; Buitrón-Bustamante, J.; Tapia, C.; Peña, G. Genetic diversity of sweet potatoes [Ipomoea batatas (L) Lam.] in Ecuador. Genet. Resour. Crop Evol. 2021, 68, 307–320. [Google Scholar] [CrossRef]
- Ngailo, S.; Shimelis, H.; Sibiya, J.; Amelework, B.; Mtunda, K. Genetic diversity assessment of Tanzanian sweetpotato genotypes using simple sequence repeat markers. S. Afr. J. Bot. 2016, 102, 40–45. [Google Scholar] [CrossRef]
- Liu, B.; Wang, Y.; Zhai, W.; Deng, J.; Wang, H.; Cui, Y.; Cheng, F.; Wang, X.; Wu, J. Development of InDel markers for Brassica rapa based on whole-genome re-sequencing. Theor. Appl. Genet. 2013, 126, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.-s.; Su, W.-j.; Wang, L.-j.; Lei, J.; Chai, S.-s.; Liu, Q.-c. Molecular diversity and genetic structure of 380 sweetpotato accessions as revealed by SSR markers. J. Integr. Agr. 2015, 14, 633–641. [Google Scholar] [CrossRef]
- Su, W.; Wang, L.; Lei, J.; Chai, S.; Liu, Y.; Yang, Y.; Yang, X.; Jiao, C. Genome-wide assessment of population structure and genetic diversity and development of a core germplasm set for sweet potato based on specific length amplified fragment (SLAF) sequencing. PLoS ONE 2017, 12, e0172066. [Google Scholar] [CrossRef]
- Lebot, V. Sweet Potato. In Root and Tuber Crops; Bradshaw, J.E., Ed.; Springer: New York, NY, USA, 2010; pp. 97–125. [Google Scholar]
- Jia, G.; Liu, X.; Schnable, J.C.; Niu, Z.; Wang, C.; Li, Y.; Wang, S.; Wang, S.; Liu, J.; Guo, E.; et al. Microsatellite Variations of Elite Setaria Varieties Released during Last Six Decades in China. PLoS ONE 2015, 10, e0125688. [Google Scholar] [CrossRef]
- Huang, X.; Kurata, N.; Wei, X.; Wang, Z.-X.; Wang, A.; Zhao, Q.; Zhao, Y.; Liu, K.; Lu, H.; Li, W.; et al. A map of rice genome variation reveals the origin of cultivated rice. Nature 2012, 490, 497–501. [Google Scholar] [CrossRef] [PubMed]
- Tang, R.; Cui, D.; Zhou, J.; Li, W.; Ma, X.; Han, B.; Guo, X.; Zhao, Z.; Han, L. Comparative analysis of genetic diversity of rice (Oryza sativa L.) varieties cultivated in different periods in China. Genet. Resour. Crop Evol. 2021, 68, 1439–1451. [Google Scholar] [CrossRef]
- Liu, M.; Xu, Y.; He, J.; Zhang, S.; Wang, Y.; Lu, P. Genetic Diversity and Population Structure of Broomcorn Millet (Panicum miliaceum L.) Cultivars and Landraces in China Based on Microsatellite Markers. Int. J. Mol. Sci. 2016, 17, 370. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Liu, Q.; Wang, Y.; Zhai, H. Analysis of genetic diversity of sweetpotato landraces in China. Sci. Agric. Sin. 2005, 38, 250–257. [Google Scholar]
- Zhang, D.; Cervantes, J.; Huamán, Z.; Carey, E.; Ghislain, M. Assessing genetic diversity of sweet potato (Ipomoea batatas (L.) Lam.) cultivars from tropical America using AFLP. Genet. Resour. Crop Evol. 2000, 47, 659–665. [Google Scholar] [CrossRef]
- Karuri, H.; Ateka, E.; Amata, R.U.; Nyende, A.B.; Muigai, A.; Mwasame, E.; Gichuki, S. Evaluating Diversity among Kenyan Sweet Potato Genotypes Using Morphological and SSR Markers. Int. J. Agric. Biol. 2010, 12, 33–38. [Google Scholar]
- Meng, Y.-S.; Zhao, N.; Li, H.; Zhai, H.; He, S.-Z.; Liu, Q.-C. SSR fingerprinting of 203 sweetpotato (Ipomoea batatas (L.) Lam.) varieties. J. Integr. Agric. 2018, 17, 86–93. [Google Scholar] [CrossRef]
- Gordon, E.; Kaviani, M.; Kagale, S.; Payne, T.; Navabi, A. Genetic diversity and population structure of synthetic hexaploid-derived wheat (Triticum aestivum L.) accessions. Genet. Resour. Crop Evol. 2019, 66, 335–348. [Google Scholar] [CrossRef]

| Name | Primer Sequences | Size (bp) | Bands | PIC 1 | Na | Ne | Ave.het |
|---|---|---|---|---|---|---|---|
| Ib-1-6 | F: TGAGATCCACGTTGAAGGTAGG | 188–221 | 3 | 0.565 | 3 | 1.643 | 0.392 |
| R: TGTTAAAGAAATGAGCTACCCACTG | |||||||
| Ib-1-7 | F: GCTAAAAAGTTCCAAAAACCTCCC | 190–220 | 3 | 0.523 | 3 | 1.622 | 0.384 |
| R: TCTTTGTCTCTGAAGTCTGGGC | |||||||
| Ib-2-6 | F: TCAACGGCTATTTCGTGTTCTC | 160–300 | 5 | 0.602 | 4 | 1.998 | 0.500 |
| R: TTTAGGGCAATCTCATGGGTCC | |||||||
| Ib-2-7 | F: TCAACTCGTACAAATCCGAATCCT | 160–170 | 2 | 0.363 | 2 | 1.786 | 0.440 |
| R: CCGGAGAAAACGCCTTTGTTAA | |||||||
| Ib-3-1 | F: TACGGTCGTGGGATTCCAAAAA | 173–181 | 2 | 0.322 | 2 | 1.481 | 0.325 |
| R: ACATTTGTTTCCTACTTGTGGTTGT | |||||||
| Ib-3-2 | F: GTGTGGGACTGTGTAATCACCA | 104–135 | 5 | 0.757 | 4 | 2.057 | 0.514 |
| R: ACAGTAAAGCCAGAAGTGCCAT | |||||||
| Ib-4-5 | F: TCCCTTCGTAGTACTTTTAGAGCT | 166–176 | 2 | 0.313 | 2 | 1.419 | 0.295 |
| R: ACAACCAGAAAGAAGTCATGCA | |||||||
| Ib-4-6 | F: TGTCAGATGAATAGATGGCGTCA | 162–183 | 3 | 0.443 | 3 | 1.648 | 0.393 |
| R: CCAGTTGCCATGGTAAAAGCAA | |||||||
| Ib-5-4 | F: TGGAAAGAGCTAGTAGATTAGCCT | 180–188 | 2 | 0.244 | 2 | 1.232 | 0.188 |
| R: AGGCCCTGTCTATACGAGATCA | |||||||
| Ib-5-5 | F: CCAACCTCAACTATTGCATGCC | 168–261 | 3 | 0.526 | 3 | 1.862 | 0.463 |
| R: TGTTTTCGCTACGTACTGCCTA | |||||||
| Ib-6-1 | F: CTTGACCACAGGGACTAGCATT | 184–259 | 4 | 0.589 | 4 | 2.102 | 0.524 |
| R: ATCAAGACAAATGGCACCTTGC | |||||||
| Ib-6-2 | F: CAGTGGAGGCTAGGTGAGAAAG | 139–205 | 8 | 0.821 | 4 | 1.258 | 0.205 |
| R: ACAAAGTTGTTCCCCTGTAGCT | |||||||
| Ib-7-7 | F: TACTCGAGAGGGGAATTGAAGC | 168–223 | 3 | 0.442 | 3 | 1.731 | 0.422 |
| R: TGCTGAGCAATAATAAGTGTGGT | |||||||
| Ib-7-8 | F: ATCAATCGATCCTTGGAAGGGT | 180–243 | 4 | 0.627 | 4 | 1.564 | 0.361 |
| R: AGCCTCATTCTTCGTGACAACT | |||||||
| Ib-8-4 | F: TGGGTTGCCTAGCTAAACTGAC | 138–164 | 2 | 0.375 | 2 | 1.998 | 0.499 |
| R: GCAAGAAAAGGTGCAAAGTCCT | |||||||
| Ib-8-5 | F: CCTCTCACCGGATCTAGTGGT | 196–240 | 4 | 0.632 | 3 | 2.405 | 0.584 |
| R: GACGTCACTGACTCAATCCTGA | |||||||
| Ib-9-1 | F: CCCGTCTAATGAATTTTGCTGCT | 197–219 | 3 | 0.531 | 3 | 1.924 | 0.480 |
| R: TTGTGACTGTGTTGCCAATGTC | |||||||
| Ib-9-2 | F: TCCATTTCTATGCACGCCTTTG | 113–157 | 2 | 0.373 | 2 | 1.937 | 0.484 |
| R: ATCTCGACATCTTCCCGACATC | |||||||
| Ib-10-2 | F: GGGAAACTGGATCGTGAAAACC | 154–195 | 3 | 0.586 | 2 | 1.016 | 0.016 |
| R: GAAACTTCGAACAATGCCCACA | |||||||
| Ib-10-4 | F: GCCACATGATTGTCATCAACCC | 174–214 | 2 | 0.362 | 2 | 1.766 | 0.434 |
| R: GATGGGTTTCTTTCACTGGTGC | |||||||
| Ib-11-1 | F: TGACTATGTTGACCTGACGTGG | 178–240 | 4 | 0.546 | 4 | 2.318 | 0.569 |
| R: AGAGTTTCACGCCTATACCGAT | |||||||
| Ib-11-5 | F: TTAACAGGACCAGAGGCAACAA | 172–209 | 3 | 0.589 | 3 | 2.246 | 0.555 |
| R: ACTGCTCCCCGAATGGTATTTT | |||||||
| Ib-12-2 | F: GCTTGTTATCGGGGTCCTTACT | 153–180 | 2 | 0.143 | 2 | 1.097 | 0.089 |
| R: ACCAATGATGCGCCAAAAGAAA | |||||||
| Ib-12-8 | F: GGCCATGGTTGCTAAAGTCTTG | 193–239 | 3 | 0.459 | 3 | 1.547 | 0.354 |
| R: ATGGATGCTCTGACTCGAGTTC | |||||||
| Ib-13-2 | F: TGTGTTCTTGTTCCTGGAGTGA | 190–221 | 3 | 0.556 | 3 | 1.688 | 0.408 |
| R: ATCTCTGCCGACTCCATTTCTG | |||||||
| Ib-13-13 | F: GGGTAATCTTTCCTTTTGTGGAGT | 144–155 | 2 | 0.358 | 2 | 1.717 | 0.418 |
| R: AGCTCTAAGCAGCAGAACCATT | |||||||
| Ib-14-2 | F: TAAGTAATCTGGGATTGCGCTG | 120–160 | 3 | 0.418 | 3 | 1.547 | 0.354 |
| R: CTCGTGCAAAGACCAACACATT | |||||||
| Ib-14-7 | F: GTCATTGGGGCCTGTTTTTCTC | 113–120 | 2 | 0.332 | 2 | 1.508 | 0.337 |
| R: TCAGCAGTTACCAAACCCATGA | |||||||
| Ib-15-1 | F: AGATCAGAGTCTCATGGGTTCA | 156–163 | 2 | 0.142 | 2 | 1.096 | 0.088 |
| R: TGCGTCACCATGCACTACTAAT | |||||||
| Ib-15-2 | F: TCAATTCAGGAATCCCTAGCGC | 195–250 | 5 | 0.757 | 4 | 2.115 | 0.527 |
| R: ATTTCGGGTGACAATCGATTGC |
| Source | df 1 | SS | MS | Estimated Variation | Percentage Variation% |
|---|---|---|---|---|---|
| Among groups | 5 | 132.293 | 26.459 | 0.422 | 3% |
| Within groups | 234 | 2770.024 | 11.838 | 11.838 | 97% |
| Total | 239 | 2902.317 | 12.260 | 100% |
| Groups | Average 1 | CH-Lan | SEA | AME | EA | AFR | CH-Var |
|---|---|---|---|---|---|---|---|
| CH-Lan | 0.187 | 0.000 | |||||
| SEA | 0.250 | 0.212 | 0.000 | ||||
| AME | 0.216 | 0.219 | 0.232 | 0.000 | |||
| EA | 0.183 | 0.195 | 0.221 | 0.214 | 0.000 | ||
| AFR | 0.169 | 0.176 | 0.197 | 0.207 | 0.185 | 0.000 | |
| CH-Var | 0.206 | 0.202 | 0.225 | 0.224 | 0.202 | 0.192 | 0.000 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, L.; Qi, Z.; Xiao, S.; Tang, F.; Liu, Y.; Deng, Y.; Dai, X.; Zhou, Z.; Ou, W.; Cao, Q. Genetic Diversity Assessment of Sweetpotato Germplasm in China Using InDel Markers. Agronomy 2022, 12, 3074. https://doi.org/10.3390/agronomy12123074
Zhao L, Qi Z, Xiao S, Tang F, Liu Y, Deng Y, Dai X, Zhou Z, Ou W, Cao Q. Genetic Diversity Assessment of Sweetpotato Germplasm in China Using InDel Markers. Agronomy. 2022; 12(12):3074. https://doi.org/10.3390/agronomy12123074
Chicago/Turabian StyleZhao, Lukuan, Zhanghua Qi, Shizhuo Xiao, Fen Tang, Yang Liu, Yitong Deng, Xibin Dai, Zhilin Zhou, Wenjun Ou, and Qinghe Cao. 2022. "Genetic Diversity Assessment of Sweetpotato Germplasm in China Using InDel Markers" Agronomy 12, no. 12: 3074. https://doi.org/10.3390/agronomy12123074
APA StyleZhao, L., Qi, Z., Xiao, S., Tang, F., Liu, Y., Deng, Y., Dai, X., Zhou, Z., Ou, W., & Cao, Q. (2022). Genetic Diversity Assessment of Sweetpotato Germplasm in China Using InDel Markers. Agronomy, 12(12), 3074. https://doi.org/10.3390/agronomy12123074








