The Expanded and Diversified Calmodulin-Binding Protein 60 (CBP60) Family in Rice (Oryza sativa L.) Is Conserved in Defense Responses against Pathogens
Abstract
1. Introduction
2. Materials and Methods
2.1. Sequence Analysis
2.2. Plant Materials and Treatments
2.3. qRT-PCR Analysis
2.4. Promoter Mining
2.5. Statistical Analysis
3. Results
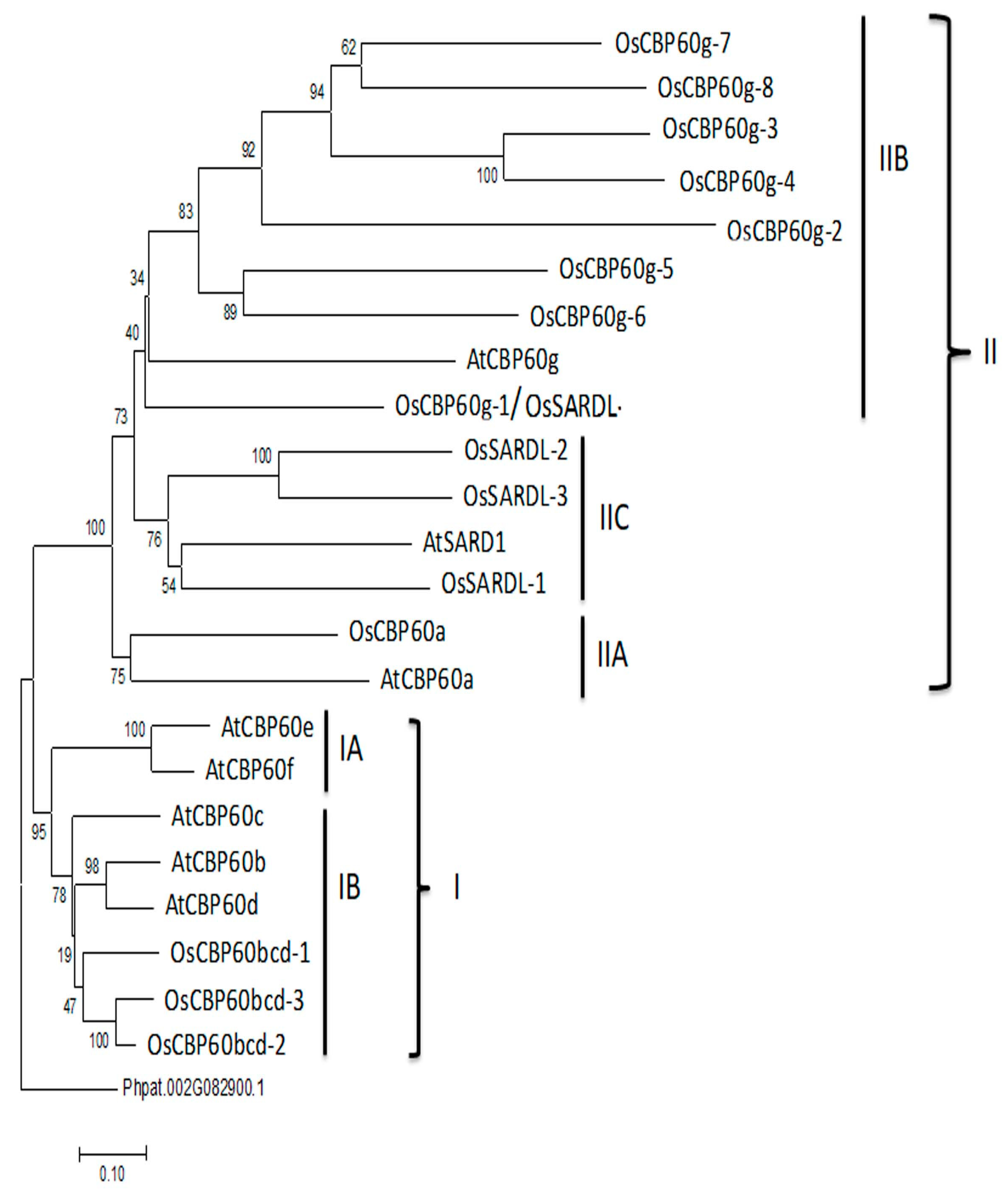
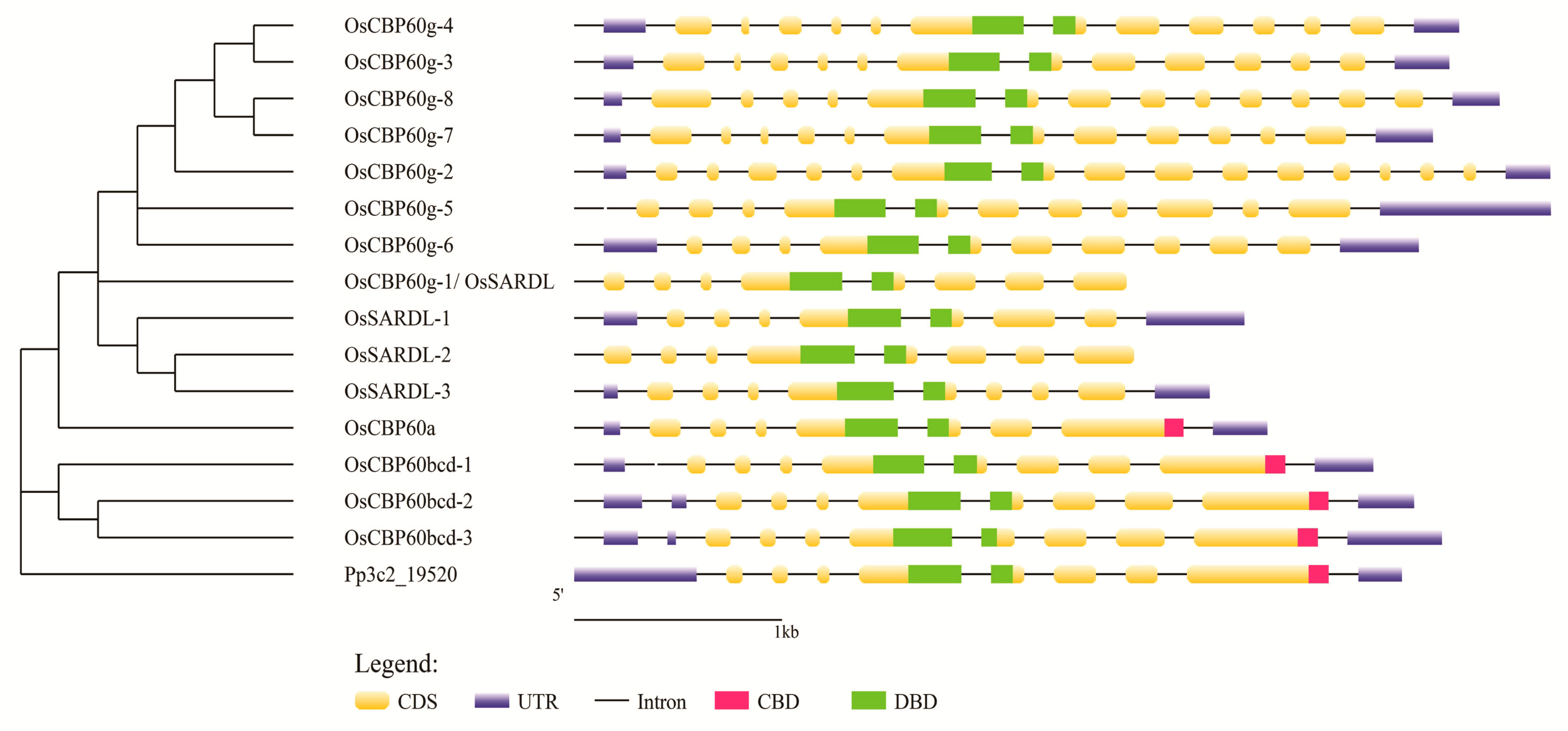
3.1. Phylogenetic Analysis of CBP60 Families in Arabidopsis and Rice
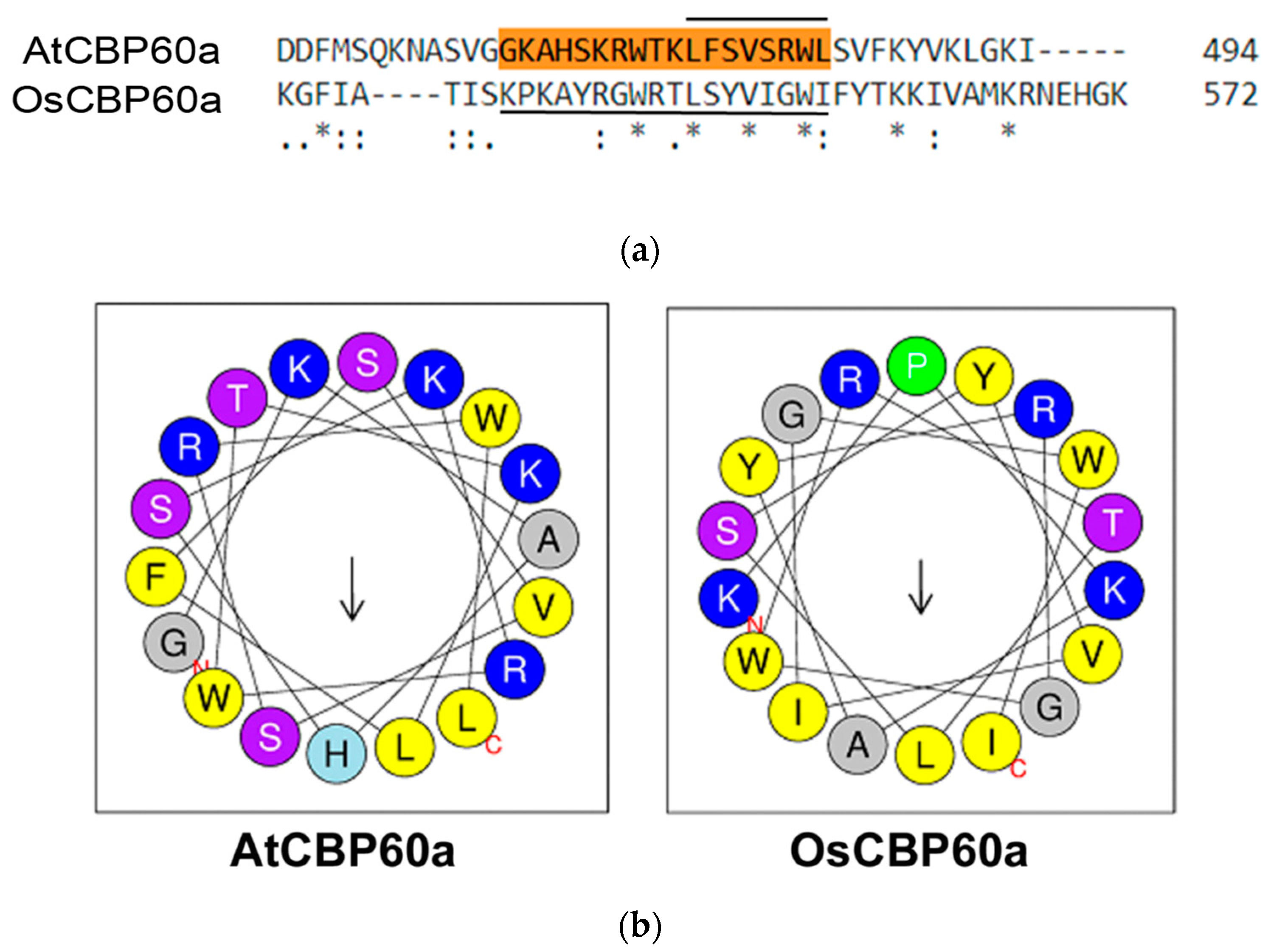
3.2. Predicted CaM- and DNA-Binding Regions in OsCBP60 Proteins
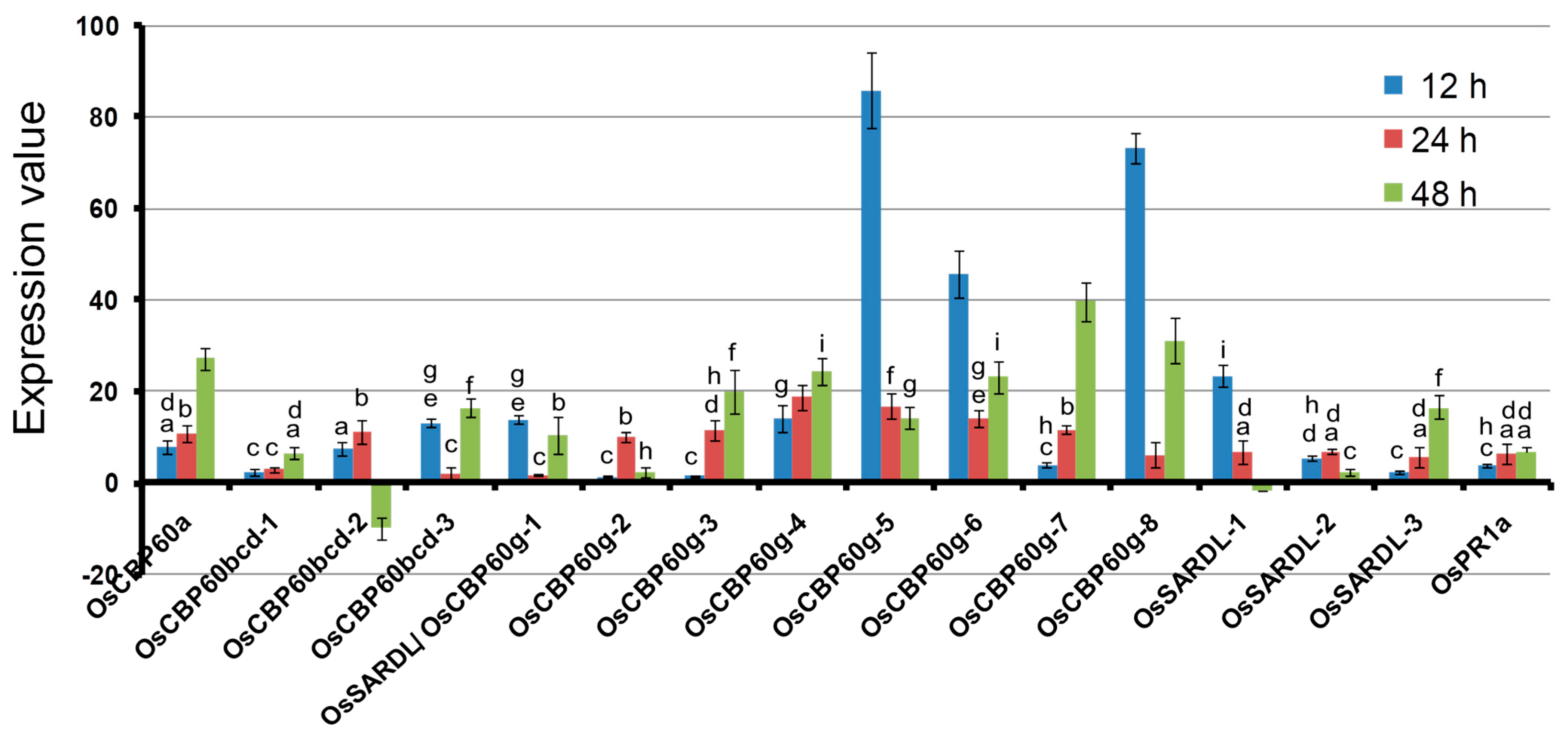
3.3. OsCBP60 Gene Expression Changes in Response to Pathogen Infection
3.4. OsCBP60 Gene Expression Changes in Response to Phytohormones
3.5. Analysis of Transcription Factor Binding Sites in Putative Promoter Regions of CBP60 Genes
3.6. Analysis of BZR1/BES1-Binding Sites in Putative Promoter Regions of OsCBP60 Genes
3.7. Promoter Mining of OsCBP60g-4
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Romero, F.M.; Gatica-Arias, A. CRISPR/Cas9: Development and Application in Rice Breeding. Rice Sci. 2019, 26, 265–281. [Google Scholar] [CrossRef]
- Savary, S.; Willocquet, L.; Pethybridge, S.J.; Esker, P.; McRoberts, N.; Nelson, A. The Global Burden of Pathogens and Pests on Major Food Crops. Nat. Ecol. Evol. 2019, 3, 430–439. [Google Scholar] [CrossRef]
- Fu, C. Genetic Improvement of Resistance to Blast and Bacterial Blight of the Elite Maintainer Line Rongfeng B in Hybrid Rice (Oryza sativa L.) by Using Marker-Assisted Selection. African J. Biotechnol. 2012, 11, 13104–13114. [Google Scholar] [CrossRef]
- Liu, W.; Wang, G.-L. Plant innate immunity in rice: A defense against pathogen infection. Nat. Sci. Rev. 2016, 3, 295–308. [Google Scholar] [CrossRef]
- Liu, T.; Song, T.; Zhang, X.; Yuan, H.; Su, L.; Li, W.; Xu, J.; Liu, S.; Chen, L.; Chen, T.; et al. Unconventionally Secreted Effectors of Two Filamentous Pathogens Target Plant Salicylate Biosynthesis. Nat. Commun. 2014, 5, 4686. [Google Scholar] [CrossRef] [PubMed]
- Bari, R.; Jones, J.D.G. Role of Plant Hormones in Plant Defence Responses. Plant Mol. Biol. 2009, 69, 473–488. [Google Scholar] [CrossRef]
- Mohanta, T.K.; Bashir, T.; Hashem, A.; Abd_Allah, E.F.; Khan, A.L.; Al-Harrasi, A.S. Early Events in Plant Abiotic Stress Signaling: Interplay Between Calcium, Reactive Oxygen Species and Phytohormones. J. Plant Growth Regul. 2018, 37, 1033–1049. [Google Scholar] [CrossRef]
- Nguyen, H.; Thi Mai To, H.; Lebrun, M.; Bellafiore, S.; Champion, A. Jasmonates—the Master Regulator of Rice Development, Adaptation and Defense. Plants 2019, 8, 339. [Google Scholar] [CrossRef]
- Lozano-Durán, R.; Zipfel, C. Trade-off between Growth and Immunity: Role of Brassinosteroids. Trends Plant Sci. 2015, 20, 12–19. [Google Scholar] [CrossRef]
- Sahni, S.; Prasad, B.D.; Liu, Q.; Grbic, V.; Sharpe, A.; Singh, S.P.; Krishna, P. Overexpression of the Brassinosteroid Biosynthetic Gene DWF4 in Brassica Napus Simultaneously Increases Seed Yield and Stress Tolerance. Sci. Rep. 2016, 6, 28298. [Google Scholar] [CrossRef]
- Wang, J.; Shi, H.; Zhou, L.; Peng, C.; Liu, D.; Zhou, X.; Wu, W.; Yin, J.; Qin, H.; Ma, W.; et al. OsBSK1-2, an Orthologous of AtBSK1, Is Involved in Rice Immunity. Front. Plant Sci. 2017, 8, 908. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhu, K.; Wang, Z.; Zhang, H.; Gu, J.; Liu, L.; Yang, J.; Zhang, J. Brassinosteroids Function in Spikelet Differentiation and Degeneration in Rice. J. Integr. Plant Biol. 2019, 61, 943–963. [Google Scholar] [CrossRef] [PubMed]
- Yuan, D.P.; Zhang, C.; Wang, Z.Y.; Zhu, X.F.; Xuan, Y.H. RAVL1 Activates Brassinosteroids and Ethylene Signaling to Modulate Response to Sheath Blight Disease in Rice. Phytopathology 2018, 108, 1104–1113. [Google Scholar] [CrossRef]
- Yang, T.; Poovaiah, B. Calcium/Calmodulin-Mediated Signal Network in Plants. Trends Plant Sci. 2003, 8, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Hetherington, A.M.; Brownlee, C. THE GENERATION OF Ca2+ SIGNALS IN PLANTS. Annu. Rev. Plant Biol. 2004, 55, 401–427. [Google Scholar] [CrossRef]
- Whalley, H.J.; Knight, M.R. Calcium Signatures Are Decoded by Plants to Give Specific Gene Responses. New Phytol. 2013, 197, 690–693. [Google Scholar] [CrossRef]
- DeFalco, T.A.; Bender, K.W.; Snedden, W.A. Breaking the Code: Ca2+ Sensors in Plant Signalling. Biochem. J. 2010, 425, 27–40. [Google Scholar] [CrossRef]
- Bouché, N.; Yellin, A.; Snedden, W.A.; Fromm, H. Plant-Specific Calmodulin-Binding Proteins. Annu. Rev. Plant Biol. 2005, 56, 435–466. [Google Scholar] [CrossRef]
- Virdi, A.S.; Singh, S.; Singh, P. Abiotic Stress Responses in Plants: Roles of Calmodulin-Regulated Proteins. Front. Plant Sci. 2015, 6, 809. [Google Scholar] [CrossRef]
- Tidow, H.; Nissen, P. Structural Diversity of Calmodulin Binding to Its Target Sites. FEBS J. 2013, 280, 5551–5565. [Google Scholar] [CrossRef]
- Andrews, C.; Xu, Y.; Kirberger, M.; Yang, J.J. Structural Aspects and Prediction of Calmodulin-Binding Proteins. Int. J. Mol. Sci. 2020, 22, 308. [Google Scholar] [CrossRef] [PubMed]
- Zielinski, R.E. Calmodulin and Calmodulin-Binding Proteins in Plants. Annu. Rev. Plant Biol. 1998, 49, 697–725. [Google Scholar] [CrossRef] [PubMed]
- Poovaiah, B.W.; Du, L.; Wang, H.; Yang, T. Recent Advances in Calcium/Calmodulin-Mediated Signaling with an Emphasis on Plant-Microbe Interactions. Plant Physiol. 2013, 163, 531–542. [Google Scholar] [CrossRef] [PubMed]
- Wan, D.; Li, R.; Zou, B.; Zhang, X.; Cong, J.; Wang, R.; Xia, Y.; Li, G. Calmodulin-Binding Protein CBP60g Is a Positive Regulator of Both Disease Resistance and Drought Tolerance in Arabidopsis. Plant Cell Rep. 2012, 31, 1269–1281. [Google Scholar] [CrossRef] [PubMed]
- Truman, W.; Sreekanta, S.; Lu, Y.; Bethke, G.; Tsuda, K.; Katagiri, F.; Glazebrook, J. The CALMODULIN-BINDING PROTEIN60 Family Includes Both Negative and Positive Regulators of Plant Immunity. PLANT Physiol. 2013, 163, 1741–1751. [Google Scholar] [CrossRef]
- Wang, L.; Tsuda, K.; Truman, W.; Sato, M.; Nguyen, L.V.; Katagiri, F.; Glazebrook, J. CBP60g and SARD1 Play Partially Redundant Critical Roles in Salicylic Acid Signaling. Plant J. 2011, 67, 1029–1041. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Zhang, Y.; Li, Y.; Zhang, Q.; Ding, Y.; Zhang, Y. ChIP-Seq Reveals Broad Roles of SARD1 and CBP60g in Regulating Plant Immunity. Nat. Commun. 2015, 6, 10159. [Google Scholar] [CrossRef]
- Wang, L.; Tsuda, K.; Sato, M.; Cohen, J.D.; Katagiri, F.; Glazebrook, J. Arabidopsis CaM Binding Protein CBP60g Contributes to MAMP-Induced SA Accumulation and Is Involved in Disease Resistance against Pseudomonas syringae. PLoS Pathog. 2009, 5, e1000301. [Google Scholar] [CrossRef]
- Li, L.-S.; Ying, J.; Li, E.; Ma, T.; Li, M.; Gong, L.-M.; Wei, G.; Zhang, Y.; Li, S. Arabidopsis CBP60b Is a Central Transcriptional Activator of Immunity. Plant Physiol. 2021, 186, 1645–1659. [Google Scholar] [CrossRef]
- Hu, G.; Hao, M.; Wang, L.; Liu, J.; Zhang, Z.; Tang, Y.; Peng, Q. The Cotton MiR477- CBP60A Module Participates in Plant Defense Against Verticillium dahlia. Mol. Plant-Microbe Interact. 2020, 33, 624–636. [Google Scholar] [CrossRef]
- Zheng, Q.; Majsec, K.; Katagiri, F. Pathogen-driven Coevolution across the CBP60 Plant Immune Regulator Subfamilies Confers Resilience on the Regulator Module. New Phytol. 2022, 233, 479–495. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Park, S.; Gilmour, S.J.; Thomashow, M.F. Roles of CAMTA Transcription Factors and Salicylic Acid in Configuring the Low-Temperature Transcriptome and Freezing Tolerance of Arabidopsis. Plant J. 2013, 75, 364–376. [Google Scholar] [CrossRef]
- Yu, Q.; Liu, Y.-L.; Sun, G.-Z.; Liu, Y.-X.; Chen, J.; Zhou, Y.-B.; Chen, M.; Ma, Y.-Z.; Xu, Z.-S.; Lan, J.-H. Genome-Wide Analysis of the Soybean Calmodulin-Binding Protein 60 Family and Identification of GmCBP60A-1 Responses to Drought and Salt Stresses. Int. J. Mol. Sci. 2021, 22, 13501. [Google Scholar] [CrossRef]
- Sun, Y.; Fan, X.-Y.; Cao, D.-M.; Tang, W.; He, K.; Zhu, J.-Y.; He, J.-X.; Bai, M.-Y.; Zhu, S.; Oh, E.; et al. Integration of Brassinosteroid Signal Transduction with the Transcription Network for Plant Growth Regulation in Arabidopsis. Dev. Cell 2010, 19, 765–777. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Li, L.; Zola, J.; Aluru, M.; Ye, H.; Foudree, A.; Guo, H.; Anderson, S.; Aluru, S.; Liu, P.; et al. A Brassinosteroid Transcriptional Network Revealed by Genome-Wide Identification of BESI Target Genes in Arabidopsis Thaliana. Plant J. 2011, 65, 634–646. [Google Scholar] [CrossRef] [PubMed]
- Bonman, J.M. Durable Resistance to Rice Blast. Oryza 1988, 25, 103–110. [Google Scholar]
- Jha, S.; Tank, H.G.; Prasad, B.D.; Chattoo, B.B. Expression of Dm-AMP1 in Rice Confers Resistance to Magnaporthe oryzae and Rhizoctonia solani. Transgenic Res. 2009, 18, 59–69. [Google Scholar] [CrossRef]
- Prasad, B.D.; Creissen, G.; Lamb, C.; Chattoo, B.B. Overexpression of Rice (Oryza sativa L.) OsCDR1 Leads to Constitutive Activation of Defense Responses in Rice and Arabidopsis. Mol. Plant-Microbe Interact. 2009, 22, 1635–1644. [Google Scholar] [CrossRef]
- Kumari, D.; Parasad, B.D.; Sahni, S.; Ghatak, A. Identification and Functional Characterization of Xanthomonas oryzae Pv. oryzae Isolates. Curr. J. Appl. Sci. Technol. 2020, 78–84. [Google Scholar] [CrossRef][Green Version]
- Tran, T.T.; Pérez-Quintero, A.L.; Wonni, I.; Carpenter, S.C.D.; Yu, Y.; Wang, L.; Leach, J.E.; Verdier, V.; Cunnac, S.; Bogdanove, A.J.; et al. Functional Analysis of African Xanthomonas oryzae Pv. oryzae TALomes Reveals a New Susceptibility Gene in Bacterial Leaf Blight of Rice. PLOS Pathog. 2018, 14, e1007092. [Google Scholar] [CrossRef]
- Kauffman, H.E. An Improved Technique for Evaluating Resistance of Rice Varieties to Xanthomonas oryzae. Plant Dis. Rep. 1973, 57, 537–541. [Google Scholar]
- Yamada, S.; Kano, A.; Tamaoki, D.; Miyamoto, A.; Shishido, H.; Miyoshi, S.; Taniguchi, S.; Akimitsu, K.; Gomi, K. Involvement of OsJAZ8 in Jasmonate-Induced Resistance to Bacterial Blight in Rice. Plant Cell Physiol. 2012, 53, 2060–2072. [Google Scholar] [CrossRef]
- Prasad, B.D.; Sahni, S.; Krishna, P.; Kumari, D.; Mahato, A.K.; Jambhulkar, S.J.; Kumar, P.; Ranjan, T.; Pal, A.K. De Novo Transcriptome Assembly and Identification of Brassinosteroid Biosynthetic Pathway in Safflower. J. Plant Growth Regul. 2022, 41, 1854–1870. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Reddy, V.S.; Ali, G.S.; Reddy, A.S.N. Genes Encoding Calmodulin-binding Proteins in the Arabidopsis Genome* 210. J. Biol. Chem. 2002, 277, 9840–9852. [Google Scholar] [CrossRef] [PubMed]
- O’Neil, K.T.; DeGrado, W.F. How Calmodulin Binds Its Targets: Sequence Independent Recognition of Amphiphilic α-Helices. Trends Biochem. Sci. 1990, 15, 59–64. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, S.; Ding, P.; Wang, D.; Cheng, Y.T.; He, J.; Gao, M.; Xu, F.; Li, Y.; Zhu, Z.; et al. Control of Salicylic Acid Synthesis and Systemic Acquired Resistance by Two Members of a Plant-Specific Family of Transcription Factors. Proc. Natl. Acad. Sci. USA 2010, 107, 18220–18225. [Google Scholar] [CrossRef]
- Yuan, P.; Jauregui, E.; Du, L.; Tanaka, K.; Poovaiah, B. Calcium Signatures and Signaling Events Orchestrate Plant–Microbe Interactions. Curr. Opin. Plant Biol. 2017, 38, 173–183. [Google Scholar] [CrossRef]
- Li, Z.K.; Arif, M.; Zhong, D.B.; Fu, B.Y.; Xu, J.L.; Domingo-Rey, J.; Ali, J.; Vijayakumar, C.H.M.; Yu, S.B.; Khush, G.S. Complex Genetic Networks Underlying the Defensive System of Rice (Oryza sativa L.) to Xanthomonas oryzae Pv. oryzae. Proc. Natl. Acad. Sci. USA 2006, 103, 7994–7999. [Google Scholar] [CrossRef]
- Kong, W.; Ding, L.; Xia, X. Identification and Characterization of Genes Frequently Responsive to Xanthomonas oryzae pv. oryzae and Magnaporthe oryzae Infections in Rice. BMC Genomics 2020, 21, 21. [Google Scholar] [CrossRef]
- Robert-Seilaniantz, A.; Grant, M.; Jones, J.D.G. Hormone Crosstalk in Plant Disease and Defense: More Than Just JASMONATE-SALICYLATE Antagonism. Annu. Rev. Phytopathol. 2011, 49, 317–343. [Google Scholar] [CrossRef]
- Divi, U.K.; Rahman, T.; Krishna, P. Brassinosteroid-Mediated Stress Tolerance in Arabidopsis Shows Interactions with Abscisic Acid, Ethylene and Salicylic Acid Pathways. BMC Plant Biol. 2010, 10, 151. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Busta, L.; Zhang, Q.; Ding, P.; Jetter, R.; Zhang, Y. TGACG-BINDING FACTOR 1 (TGA1) and TGA4 Regulate Salicylic Acid and Pipecolic Acid Biosynthesis by Modulating the Expression of Systemic Acquired Resistance Deficient 1 (SARD1) and CALMODULIN-BINDING PROTEIN 60g (CBP60g). New Phytol. 2018, 217, 344–354. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Lu, Y.; Bethke, G.; Harrison, B.T.; Hatsugai, N.; Katagiri, F.; Glazebrook, J. WRKY70 Prevents Axenic Activation of Plant Immunity by Direct Repression of SARD1. New Phytol. 2018, 217, 700–712. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Huang, J.; Xu, Y.; Verma, V.; Jing, B.; Sun, Y.; Ruiz Orduna, A.; Tian, H.; Huang, X.; Xia, S.; et al. Redundant CAMTA Transcription Factors Negatively Regulate the Biosynthesis of Salicylic Acid and N-Hydroxypipecolic Acid by Modulating the Expression of SARD1 and CBP60g. Mol. Plant 2020, 13, 144–156. [Google Scholar] [CrossRef] [PubMed]
- Xiong, J.; Wan, X.; Ran., M.; Xu, X.; Chen, L.; Yang, F. Brassinosteroids Positively Regulate Plant Immunity via BRI1-EMS-SUPPRESSOR 1-Mediated GLUCAN SYNTHASE-LIKE 8 Transcription. Front. Plant Sci. 2022, 13, 854899. [Google Scholar] [CrossRef]
- Du, L.; Poovaiah, B.W. Ca2+/Calmodulin Is Critical for Brassinosteroid Biosynthesis and Plant Growth. Nature 2005, 437, 741–745. [Google Scholar] [CrossRef]
- Ranty, B.; Aldon, D.; Galaud, J.-P. Plant Calmodulins and Calmodulin-Related Proteins. Plant Signal. Behav. 2006, 1, 96–104. [Google Scholar] [CrossRef]
- Pallegar, P.; Priti Krishna, S. Functional Analysis of Two Brassinosteroid Responsive, Putative Calmodulin-Binding Proteins 60 (CBP60S) in Arabidopsis Thaliana Graduate Program in Biology. Ph.D. Thesis, The University of Western Ontario, London, ON, Canada, 2014. [Google Scholar]
- Singh, A.K.; Singh, P.K.; Arya, M.; Singh, N.K.; Singh, U.S. Molecular Screening of Blast Resistance Genes in Rice Using SSR Markers. Plant Pathol. J. 2015, 31, 12–24. [Google Scholar] [CrossRef]
- Mao, C.; Wang, S.; Jia, Q.; Wu, P. OsEIL1, a Rice Homolog of the Arabidopsis EIN3 Regulates the Ethylene Response as a Positive Component. Plant Mol. Biol. 2006, 61, 141–152. [Google Scholar] [CrossRef]
- Zhou, J.-M.; Zhang, Y. Plant Immunity: Danger Perception and Signaling. Cell 2020, 181, 978–989. [Google Scholar] [CrossRef]
- Hilleary, R.; Paez-Valencia, J.; Vens, C.; Toyota, M.; Palmgren, M.; Gilroy, S. Tonoplast-Localized Ca2+ Pumps Regulate Ca2+ Signals during Pattern-Triggered Immunity in Arabidopsis Thaliana. Proc. Natl. Acad. Sci. USA 2020, 117, 18849–18857. [Google Scholar] [CrossRef] [PubMed]
- Lecourieux, D.; Ranjeva, R.; Pugin, A. Calcium in Plant Defence-signalling Pathways. New Phytol. 2006, 171, 249–269. [Google Scholar] [CrossRef] [PubMed]
- Grant, M.; Brown, I.; Adams, S.; Knight, M.; Ainslie, A.; Mansfield, J. The RPM1 Plant Disease Resistance Gene Facilitates a Rapid and Sustained Increase in Cytosolic Calcium That Is Necessary for the Oxidative Burst and Hypersensitive Cell Death. Plant J. 2000, 23, 441–450. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Ali, G.S.; Simons, K.A.; Hou, J.; Yang, T.; Reddy, A.S.N.; Poovaiah, B.W. Ca2+/Calmodulin Regulates Salicylic-Acid-Mediated Plant Immunity. Nature 2009, 457, 1154–1158. [Google Scholar] [CrossRef] [PubMed]
- Vlot, A.C.; Dempsey, D.A.; Klessig, D.F. Salicylic Acid, a Multifaceted Hormone to Combat Disease. Annu. Rev. Phytopathol. 2009, 47, 177–206. [Google Scholar] [CrossRef]
- Wang, J.; Liu, X.; Zhang, A.; Ren, Y.; Wu, F.; Wang, G.; Xu, Y.; Lei, C.; Zhu, S.; Pan, T.; et al. A Cyclic Nucleotide-Gated Channel Mediates Cytoplasmic Calcium Elevation and Disease Resistance in Rice. Cell Res. 2019, 29, 820–831. [Google Scholar] [CrossRef] [PubMed]
- Boonburapong, B.; Buaboocha, T. Genome-Wide Identification and Analyses of the Rice Calmodulin and Related Potential Calcium Sensor Proteins. BMC Plant Biol. 2007, 7, 2229. [Google Scholar] [CrossRef][Green Version]
- Chinpongpanich, A.; Phean-O-Pas, S.; Thongchuang, M.; Qu, L.-J.; Buaboocha, T. C-Terminal Extension of Calmodulin-like 3 Protein from Oryza sativa L.: Interaction with a High Mobility Group Target Protein. Acta Biochim. Biophys. Sin. 2015, 47, 880–889. [Google Scholar] [CrossRef]
- Choi, M.S.; Kim, M.C.; Yoo, J.H.; Moon, B.C.; Koo, S.C.; Park, B.O.; Lee, J.H.; Koo, Y.D.; Han, H.J.; Lee, S.Y.; et al. Isolation of a Calmodulin-Binding Transcription Factor from Rice (Oryza sativa L.). J. Biol. Chem. 2005, 280, 40820–40831. [Google Scholar] [CrossRef]
- Chung, J.-S.; Koo, S.C.; Jin, B.J.; Baek, D.; Yeom, S.-I.; Chun, H.J.; Choi, M.S.; Cho, H.M.; Lee, S.H.; Jung, W.-H.; et al. Rice CaM-Binding Transcription Factor (OsCBT) Mediates Defense Signaling via Transcriptional Reprogramming. Plant Biotechnol. Rep. 2020, 14, 309–321. [Google Scholar] [CrossRef]
- Gain, H.; Nandi, D.; Kumari, D.; Das, A.; Dasgupta, S.B.; Banerjee, J. Genome-wide Identification of CAMTA Gene Family Members in Rice (Oryza sativa L.) and in Silico Study on Their Versatility in Respect to Gene Expression and Promoter Structure. Funct. Integr. Genomics 2022, 22, 193–214. [Google Scholar] [CrossRef]
- Wang, Y.; Shen, C.; Jiang, Q.; Wan, Z.; Gao, C.; Wang, W. Seed priming with calcium chloride enhances stress tolerance in rice seedlings. Plant Sci. 2022, 323, 111381. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Wu, Z.; Tian, H.; Li, X.; Zhang, Y. Arabidopsis CALMODULIN-BINDING PROTEIN 60b Plays Dual Roles in Plant Immunity. Plant Commun. 2021, 2, 100213. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, G.K.; Rakwal, R.; Jwa, N.-S. Differential Induction of Three Pathogenesis-Related Genes, PR10, PR1b and PR5 by the Ethylene Generator Ethephon under Light and Dark in Rice (Oryza sativa L.) Seedlings. J. Plant Physiol. 2001, 158, 133–137. [Google Scholar] [CrossRef]
- Mitsuhara, I.; Iwai, T.; Seo, S.; Yanagawa, Y.; Kawahigasi, H.; Hirose, S.; Ohkawa, Y.; Ohashi, Y. Characteristic Expression of Twelve Rice PR1 Family Genes in Response to Pathogen Infection, Wounding, and Defense-Related Signal Compounds (121/180). Mol. Genet. Genomics 2008, 279, 415–427. [Google Scholar] [CrossRef] [PubMed]
- Matić, S.; Bagnaresi, P.; Biselli, C.; Orru’, L.; Amaral Carneiro, G.; Siciliano, I.; Valé, G.; Gullino, M.L.; Spadaro, D. Comparative Transcriptome Profiling of Resistant and Susceptible Rice Genotypes in Response to the Seedborne Pathogen Fusarium Fujikuroi. BMC Genomics 2016, 17, 608. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, M.W.; Roux, M.; Petersen, M.; Mundy, J. MAP Kinase Cascades in Arabidopsis Innate Immunity. Front. Plant Sci. 2012, 3, 169. [Google Scholar] [CrossRef]
- Ali, S.; Mir, Z.A.; Bhat, J.A.; Tyagi, A.; Chandrashekar, N.; Yadav, P.; Rawat, S.; Sultana, M.; Grover, A. Isolation and Characterization of Systemic Acquired Resistance Marker Gene PR1 and Its Promoter from Brassica Juncea. 3 Biotech 2018, 8, 10. [Google Scholar] [CrossRef]
- Yang, D.-L.; Yang, Y.; He, Z. Roles of Plant Hormones and Their Interplay in Rice Immunity. Mol. Plant 2013, 6, 675–685. [Google Scholar] [CrossRef]
- Silverman, P.; Seskar, M.; Kanter, D.; Schweizer, P.; Metraux, J.P.; Raskin, I. Salicylic Acid in Rice (Biosynthesis, Conjugation, and Possible Role). Plant Physiol. 1995, 108, 633–639. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yu, Q.; Gao, S.; Yu, N.; Zhao, L.; Wang, J.; Zhao, J.; Huang, P.; Yao, L.; Wang, M.; et al. Disruption of the Primary Salicylic Acid Hydroxylases in Rice Enhances Broad-spectrum Resistance against Pathogens. Plant. Cell Environ. 2022, 45, 2211–2225. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Morea, F.A.; He, P.; Shan, L.; Russinova, E. It Takes Two to Tango—Molecular Links between Plant Immunity and Brassinosteroid Signalling. J. Cell Sci. 2020, 133. [Google Scholar] [CrossRef] [PubMed]
- Qi, G.; Chen, H.; Wang, D.; Zheng, H.; Tang, X.; Guo, Z.; Cheng, J.; Chen, J.; Wang, Y.; Bai, M.; et al. The BZR1-EDS1 ModuleRegulates Plant Growth-Defense Coordination. Mol. Plant 2021, 14, 2072–2087. [Google Scholar] [CrossRef]
- Ahmad, F.; Singh, A.; Kamal, A. Crosstalk of Brassinosteroids with Other Phytohormones under Various Abiotic Stresses. J. Appl. Biol. Biotechnol. 2018, 6. [Google Scholar] [CrossRef]
- Divi, U.K.; Rahman, T.; Krishna, P. Gene Expression and Functional Analyses in Brassinosteroid-Mediated Stress Tolerance. Plant Biotechnol. J. 2016, 14, 419–432. [Google Scholar] [CrossRef]
- Peres, A.; Soares, J.; Tavares, R.; Righetto, G.; Zullo, M.; Mandava, N.; Menossi, M. Brassinosteroids, the Sixth Class of Phytohormones: A Molecular View from the Discovery to Hormonal Interactions in Plant Development and Stress Adaptation. Int. J. Mol. Sci. 2019, 20, 331. [Google Scholar] [CrossRef]
- Pan, G.; Liu, Y.; Ji, L.; Zhang, X.; He, J.; Huang, J.; Qiu, Z.; Liu, D.; Sun, Z.; Xu, T.; et al. Brassinosteroids Mediate Susceptibility to Brown Planthopper by Integrating with the Salicylic Acid and Jasmonic Acid Pathways in Rice. J. Exp. Bot. 2018, 69, 4433–4442. [Google Scholar] [CrossRef]
- Hu, P.; Zhou, W.; Cheng, Z.; Fan, M.; Wang, L.; Xie, D. JAV1 Controls Jasmonate-Regulated Plant Defense. Mol. Cell 2013, 50, 504–515. [Google Scholar] [CrossRef]
- Gupta, A.; Bhardwaj, M.; Tran, L.-S.P. Jasmonic Acid at the Crossroads of Plant Immunity and Pseudomonas Syringae Virulence. Int. J. Mol. Sci. 2020, 21, 7482. [Google Scholar] [CrossRef]



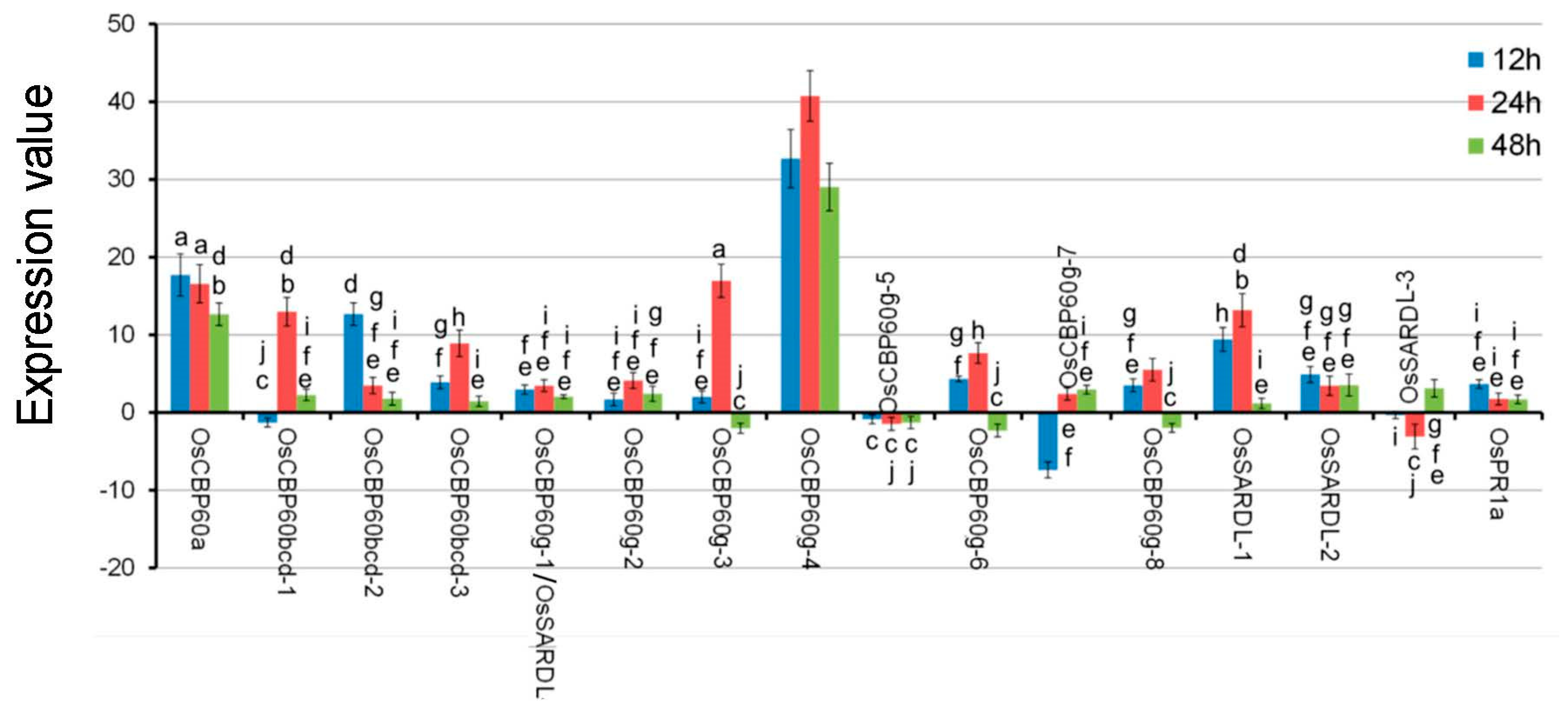

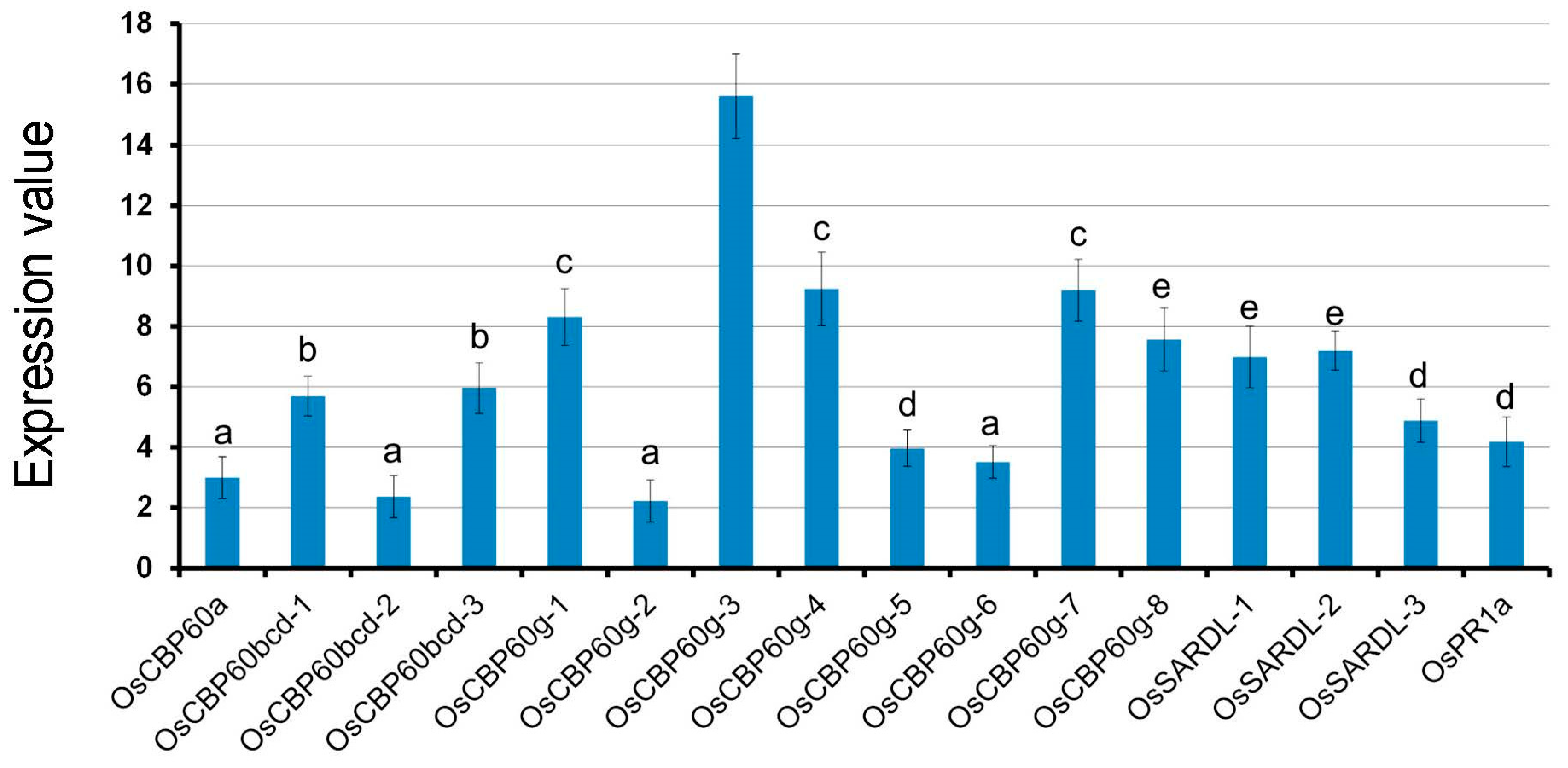

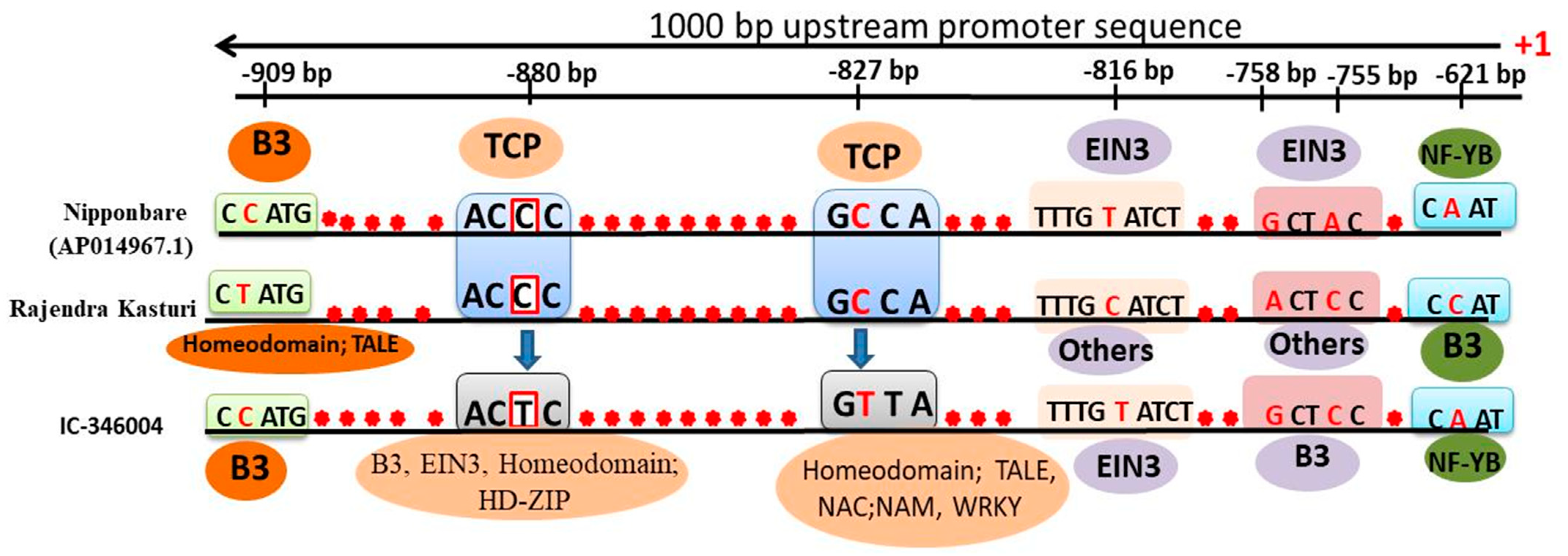
| S.No. | Gene ID | Proposed Name | Predicted Localisation a | Predicted CBD (Consensus *) |
|---|---|---|---|---|
| 1 | LOC_Os02g08120 | OsCBP60bcd-1 | Nucleus | C-terminus |
| 2 | LOC_Os02g35470 | OsCBP60bcd-2 | Nucleus | C-terminus |
| 3 | LOC_Os04g36660 | OsCBP60bcd-3 | Nucleus | C-terminus |
| 4 | LOC_Os03g32160 | OsCBP60a | Nucleus | C-terminus |
| 5 | LOC_Os01g04280 | OsSARDL-1 | Nucleus | none |
| 6 | LOC_Os08g27170 | OsSARDL-2 | Nucleus | none |
| 7 | LOC_Os09g13890 | OsSARDL-3 | Nucleus | none |
| 8 | LOC_Os03g18960 | OsCBP60g-1/OsSARDL | Nucleus | none |
| 9 | LOC_Os03g56660 | OsCBP60g-2 | Nucleus | none |
| 10 | LOC_Os11g44600 | OsCBP60g-3 | Nucleus | none |
| 11 | LOC_Os11g44680 | OsCBP60g-4 | Chloroplast | none |
| 12 | LOC_Os12g36110 | OsCBP60g-5 | Nucleus | none |
| 13 | LOC_Os12g36910 | OsCBP60g-6 | Nucleus | none |
| 14 | LOC_Os12g36920 | OsCBP60g-7 | Nucleus | none |
| 15 | LOC_Os12g36940 | OsCBP60g-8 | Nucleus | none |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumari, D.; Prasad, B.D.; Sahni, S.; Nonhebel, H.M.; Krishna, P. The Expanded and Diversified Calmodulin-Binding Protein 60 (CBP60) Family in Rice (Oryza sativa L.) Is Conserved in Defense Responses against Pathogens. Agronomy 2022, 12, 3060. https://doi.org/10.3390/agronomy12123060
Kumari D, Prasad BD, Sahni S, Nonhebel HM, Krishna P. The Expanded and Diversified Calmodulin-Binding Protein 60 (CBP60) Family in Rice (Oryza sativa L.) Is Conserved in Defense Responses against Pathogens. Agronomy. 2022; 12(12):3060. https://doi.org/10.3390/agronomy12123060
Chicago/Turabian StyleKumari, Diksha, Bishun Deo Prasad, Sangita Sahni, Heather M. Nonhebel, and Priti Krishna. 2022. "The Expanded and Diversified Calmodulin-Binding Protein 60 (CBP60) Family in Rice (Oryza sativa L.) Is Conserved in Defense Responses against Pathogens" Agronomy 12, no. 12: 3060. https://doi.org/10.3390/agronomy12123060
APA StyleKumari, D., Prasad, B. D., Sahni, S., Nonhebel, H. M., & Krishna, P. (2022). The Expanded and Diversified Calmodulin-Binding Protein 60 (CBP60) Family in Rice (Oryza sativa L.) Is Conserved in Defense Responses against Pathogens. Agronomy, 12(12), 3060. https://doi.org/10.3390/agronomy12123060






