Intelligent Weed Management Based on Object Detection Neural Networks in Tomato Crops
Abstract
:1. Introduction
2. Materials and Methods
2.1. Image Acquisition
2.2. Image Pre-processing
2.3. RetinaNet Object Detection Neural Network
2.4. Training Model
2.5. Fitness Evaluation
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| NN | Neural Network |
| mAP | Mean Average Precision |
| AP | Average Precision |
| SSWM | Site-Specific Weed Management |
| EU | European Union |
| CNN | Convolutional Neural Networks |
| IoU | Intersection Over Union |
| SVM | Support Vector Machines |
| EPPO | European and Mediterranean Plant Protection Organization |
| SOLNI | Solanum nigrum L. |
| POROL | Portulaca oleracea L. |
| ECHCG | Echinochloa crus galli L. |
| SETIT | Setaria verticillata L. |
| NR | Species plants not recognised by the expert |
| LYPES | Solanum lycopersicum L. |
| TL | Transfer Learning |
| TP | True Positive |
| FP | False Positive |
| FN | False Negative |
Appendix A

References
- Bruinsma, J. World Agriculture: Towards 2015/2030: An FAO Perspective; Routledge: London, UK, 2017. [Google Scholar]
- Qasem, J.R. Weed Seed Dormancy: The Ecophysiology and Survival Strategies. In Seed Dormancy and Germination; IntechOpen: London, UK, 2020. [Google Scholar]
- Machleb, J.; Peteinatos, G.G.; Kollenda, B.L.; Andújar, D.; Gerhards, R. Sensor-based mechanical weed control: Present state and prospects. Comput. Electron. Agric. 2020, 176, 105638. [Google Scholar] [CrossRef]
- Dyrmann, M.; Karstoft, H.; Midtiby, H.S. Plant species classification using deep convolutional neural network. Biosyst. Eng. 2016, 151, 72–80. [Google Scholar] [CrossRef]
- Pantazi, X.-E.; Moshou, D.; Bravo, C. Active learning system for weed species recognition based on hyperspectral sensing. Biosyst. Eng. 2016, 146, 193–202. [Google Scholar] [CrossRef]
- Sabzi, S.; Abbaspour-Gilandeh, Y. Using video processing to classify potato plant and three types of weed using hybrid of artificial neural network and partincle swarm algorithm. Measurement 2018, 126, 22–36. [Google Scholar] [CrossRef]
- Milan, R. Directive 2009/128/EC on the Sustainable Use of Pesticides; European Parliamentary Research Service: Brussels, Belgium, 2018.
- Pérez-Ortiz, M.; Peña, J.M.; Gutiérrez, P.A.; Torres-Sánchez, J.; Hervás-Martínez, C.; López-Granados, F. Selecting patterns and features for between- and within- crop-row weed mapping using UAV-imagery. Expert Syst. Appl. 2016, 47, 85–94. [Google Scholar] [CrossRef] [Green Version]
- Fernández-Quintanilla, C.; Peña, J.M.; Andújar, D.; Dorado, J.; Ribeiro, A.; López-Granados, F. Is the current state of the art of weed monitoring suitable for site-specific weed management in arable crops? Weed Res. 2018, 58, 259–272. [Google Scholar] [CrossRef]
- Tang, J.; Zhang, Z.; Wang, D.; Xin, J.; He, L. Research on weeds identification based on K-means feature learning. Soft Comput 2018, 22, 7649–7658. [Google Scholar] [CrossRef]
- Kamilaris, A.; Prenafeta-Boldú, F.X. Deep learning in agriculture: A survey. Comput. Electron. Agric. 2018, 147, 70–90. [Google Scholar] [CrossRef] [Green Version]
- Olsen, A.; Konovalov, D.A.; Philippa, B.; Ridd, P.; Wood, J.C.; Johns, J.; Banks, W.; Girgenti, B.; Kenny, O.; Whinney, J.; et al. DeepWeeds: A Multiclass Weed Species Image Dataset for Deep Learning. Sci. Rep. 2019, 9, 2058. [Google Scholar] [CrossRef] [Green Version]
- Dyrmann, M. Automatic Detection and Classification of Weed Seedlings under Natural Light Conditions. Ph.D. Thesis, University of Southern Denmark, Odense, Denmark, 2017. [Google Scholar]
- Rezatofighi, H.; Tsoi, N.; Gwak, J.; Sadeghian, A.; Reid, I.; Savarese, S. Generalized Intersection over Union: A Metric and A Loss for Bounding Box Regression. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019. [Google Scholar]
- dos Santos Ferreira, A.; Matte Freitas, D.; Gonçalves da Silva, G.; Pistori, H.; Theophilo Folhes, M. Weed detection in soybean crops using ConvNets. Comput. Electron. Agric. 2017, 143, 314–324. [Google Scholar] [CrossRef]
- Sharpe, S.M.; Schumann, A.W.; Yu, J.; Boyd, N.S. Vegetation detection and discrimination within vegetable plasticulture row-middles using a convolutional neural network. Precis. Agric. 2020, 21, 264–277. [Google Scholar] [CrossRef]
- Osorio, K.; Puerto, A.; Pedraza, C.; Jamaica, D.; Rodríguez, L. A Deep Learning Approach for Weed Detection in Lettuce Crops Using Multispectral Images. AgriEngineering 2020, 2, 471–488. [Google Scholar] [CrossRef]
- Chen, R.; Chu, T.; Landivar, J.A.; Yang, C.; Maeda, M.M. Monitoring cotton (Gossypium hirsutum L.) germination using ultrahigh-resolution UAS images. Precis. Agric. 2018, 19, 161–177. [Google Scholar] [CrossRef]
- Peteinatos, G.G.; Reichel, P.; Karouta, J.; Andújar, D.; Gerhards, R. Weed Identification in Maize, Sunflower, and Potatoes with the Aid of Convolutional Neural Networks. Remote Sens. 2020, 12, 4185. [Google Scholar] [CrossRef]
- Peteinatos, G.G.; Weis, M.; Andújar, D.; Rueda Ayala, V.; Gerhards, R. Potential use of ground-based sensor technologies for weed detection: Ground-based sensor technologies for weed detection. Pest Manag. Sci. 2014, 70, 190–199. [Google Scholar] [CrossRef] [PubMed]
- Subeesh, A.; Bhole, S.; Singh, K.; Chandel, N.S.; Rajwade, Y.A.; Rao, K.V.R.; Kumar, S.P.; Jat, D. Deep convolutional neural network models for weed detection in polyhouse grown bell peppers. Artif. Intell. Agric. 2022, 6, 47–54. [Google Scholar] [CrossRef]
- Zheng, Y.Y.; Kong, J.L.; Jin, X.B.; Su, T.L.; Nie, M.J.; Bai, Y.T. Real-Time Vegetables Recognition System based on Deep Learning Network for Agricultural Robots. In Proceedings of the 2018 Chinese Automation Congress (CAC), Xi’an, China, 30 November–2 December 2018; pp. 2223–2228. [Google Scholar]
- Sattler, T.; Zhou, Q.; Pollefeys, M.; Leal-Taixé, L. Understanding the Limitations of CNN-Based Absolute Camera Pose Regression. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019; pp. 3297–3307. [Google Scholar]
- Lin, T.Y.; Goyal, P.; Girshick, R.; He, K.; Dollár, P. Focal Loss for Dense Object Detection. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 2999–3007. [Google Scholar]
- Li, Z.; Namiki, A.; Suzuki, S.; Wang, Q.; Zhang, T.; Wang, W. Application of Low-Altitude UAV Remote Sensing Image Object Detection Based on Improved YOLOv5. Appl. Sci. 2022, 12, 8314. [Google Scholar] [CrossRef]
- Abdur Rahman, Y.L.; Wang, H. Deep Neural Networks for Weed Detections Towards Precision Weeding. In Proceedings of the 2022 ASABE Annual International Meeting, Houston, TX, USA, 17–20 July 2022; American Society of Agricultural and Biological Engineers: St. Joseph, MI, USA, 2022. [Google Scholar]
- Tannouche, A.; Sbai, K.; Miloud, R.; Agounoune, R.; Abdelhai, R. Real Time Weed Detection using a Boosted Cascade of Simple Features. Int. J. Electr. Comput. Eng. (IJECE) 2016, 6, 2755–2765. [Google Scholar] [CrossRef]
- Sabzi, S.; Abbaspour-Gilandeh, Y.; García-Mateos, G. A fast and accurate expert system for weed identification in potato crops using metaheuristic algorithms. Comput. Ind. 2018, 98, 80–89. [Google Scholar] [CrossRef]
- Yeshe, A.; Gourkhede, P.; Vaidya, P. Blue River Technology: Futuristic Approach of Precision Farming; Just Agriculture: Punjab, India, 2022. [Google Scholar]
- Rakhmatulin, I.; Kamilaris, A.; Andreasen, C. Deep Neural Networks to Detect Weeds from Crops in Agricultural Environments in Real-Time: A Review. Remote Sens. 2021, 13, 4486. [Google Scholar] [CrossRef]
- Correa, J.M.L.; Todeschini, M.; Pérez, D.S.; Karouta, J.; Bromberg, F.; Ribeiro, A.; Andújar, D. 8. Multi species weed detection with Retinanet one-step network in a maize field. In Precision Agriculture ’21; Wageningen Academic: Budapest, Hungary, 2021; pp. 79–86. [Google Scholar]
- Zaragoza, C.; Tei, F.; Montemurro, P.; Baumann, D.T.; Dobrzanski, A.; Giovinazzo, R.; Kleifeld, Y.; Rocha, F.; Alaoui, S.B.; Sanseovic, T.; et al. Weeds and weed management in processing tomato. Acta Hortic. 2003, 613, 111–121. [Google Scholar] [CrossRef]
- LabelImg, T. Git Code LabelImg; Github: San Francisco, CA, USA, 2015. [Google Scholar]
- Zaidi, S.S.A.; Ansari, M.S.; Aslam, A.; Kanwal, N.; Asghar, M.; Lee, B. A survey of modern deep learning based object detection models. Digit. Signal Process. 2022, 126, 103514. [Google Scholar] [CrossRef]
- Gaiser, H.d.V.M.; Lacatusu, V.; Williamson, A.; Liscio, E.; Henon, Y.; Gratie, C. Fizyr Fizyr/Keras-Retinanet 0.5.1, Zenodo: Geneva, Switzerland, 2019.
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Shorten, C.; Khoshgoftaar, T.M. A survey on Image Data Augmentation for Deep Learning. J. Big Data 2019, 6, 60. [Google Scholar] [CrossRef]
- Chollet, F. Keras; Github: San Francisco, CA, USA, 2015. [Google Scholar]
- Yen, S.-J.; Lee, Y.-S. Under-Sampling Approaches for Improving Prediction of the Minority Class in an Imbalanced Dataset. In Intelligent Control and Automation: International Conference on Intelligent Computing, ICIC 2006, Kunming, China, 16–19 August 2006; Huang, D.-S., Li, K., Irwin, G.W., Eds.; Springer: Berlin/Heidelberg, Germany, 2006; pp. 731–740. [Google Scholar]
- Viola, P.A.; Jones, M.J. Rapid object detection using a boosted cascade of simple features. In Proceedings of the 2001 IEEE Computer Society Conference on Computer Vision and Pattern Recognition, CVPR 2001, Kauai, HI, USA, 8–14 December 2001; Volume 1, p. I. [Google Scholar]
- Montesinos López, O.A.; Montesinos López, A.; Crossa, J. Overfitting, Model Tuning, and Evaluation of Prediction Performance. In Multivariate Statistical Machine Learning Methods for Genomic Prediction; Springer International Publishing: Cham, Switzerland, 2022; pp. 109–139. [Google Scholar]
- Garibaldi-Márquez, F.; Flores, G.; Mercado-Ravell, D.A.; Ramírez-Pedraza, A.; Valentín-Coronado, L.M. Weed Classification from Natural Corn Field-Multi-Plant Images Based on Shallow and Deep Learning. Sensors 2022, 22, 3021. [Google Scholar] [CrossRef]
- Jha, K.; Doshi, A.; Patel, P.; Shah, M. A comprehensive review on automation in agriculture using artificial intelligence. Artif. Intell. Agric. 2019, 2, 1–12. [Google Scholar] [CrossRef]
- Lopez-Martinez, N.; Marshall, G.; De Prado, R. Resistance of barnyardgrass (Echinochloa crus-galli) to atrazine and quinclorac. Pestic. Sci. 1997, 51, 171–175. [Google Scholar] [CrossRef]
- Talbert, R.E.; Burgos, N.R. History and Management of Herbicide-resistant Barnyardgrass (Echinochloa crus-galli) in Arkansas Rice. Weed Technol. 2007, 21, 324–331. [Google Scholar] [CrossRef]
- Jasieniuk, M.; Brûlé-Babel, A.L.; Morrison, I.N. The Evolution and Genetics of Herbicide Resistance in Weeds. Weed Sci. 1996, 44, 176–193. [Google Scholar] [CrossRef]
- Gerhards, R.; Andújar Sanchez, D.; Hamouz, P.; Peteinatos, G.G.; Christensen, S.; Fernandez-Quintanilla, C. Advances in site-specific weed management in agriculture—A review. Weed Res. 2022, 62, 123–133. [Google Scholar] [CrossRef]
- Lati, R.N.; Siemens, M.C.; Rachuy, J.S.; Fennimore, S.A. Intrarow Weed Removal in Broccoli and Transplanted Lettuce with an Intelligent Cultivator. Weed Technol. 2016, 30, 655–663. [Google Scholar] [CrossRef]
- Zhang, W.; Miao, Z.; Li, N.; He, C.; Sun, T. Review of Current Robotic Approaches for Precision Weed Management. Curr. Robot. Rep. 2022, 3, 139–151. [Google Scholar] [CrossRef] [PubMed]
- Etienne, A.; Ahmad, A.; Aggarwal, V.; Saraswat, D. Deep Learning-Based Object Detection System for Identifying Weeds Using UAS Imagery. Remote Sens. 2021, 13, 5182. [Google Scholar] [CrossRef]
- Potena, C.; Nardi, D.; Pretto, A. Fast and Accurate Crop and Weed Identification with Summarized Train Sets for Precision Agriculture. In Intelligent Autonomous Systems 14. IAS 2016. Advances in Intelligent Systems and Computing; Springer: Cham, Switzerland, 2016. [Google Scholar]
- Lu, Y.; Young, S. A survey of public datasets for computer vision tasks in precision agriculture. Comput. Electron. Agric. 2020, 178, 105760. [Google Scholar] [CrossRef]
- Maharana, K.; Mondal, S.; Nemade, B. A review: Data pre-processing and data augmentation techniques. Glob. Transit. Proc. 2022, 3, 91–99. [Google Scholar] [CrossRef]
- Zhang, R.; Zhou, B.; Lu, C.; Ma, M. The Performance Research of the Data Augmentation Method for Image Classification. Math. Probl. Eng. 2022, 2022, 2964829. [Google Scholar] [CrossRef]
- Partel, V.; Charan Kakarla, S.; Ampatzidis, Y. Development and evaluation of a low-cost and smart technology for precision weed management utilizing artificial intelligence. Comput. Electron. Agric. 2019, 157, 339–350. [Google Scholar] [CrossRef]
- Lee, S.H.; Chan, C.S.; Mayo, S.J.; Remagnino, P. How deep learning extracts and learns leaf features for plant classification. Pattern Recognit. 2017, 71, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Rawat, W.; Wang, Z. Deep Convolutional Neural Networks for Image Classification: A Comprehensive Review. Neural Comput. 2017, 29, 2352–2449. [Google Scholar] [CrossRef] [PubMed]
- Hall, D.; Dayoub, F.; Perez, T.; McCool, C. A Rapidly Deployable Classification System Using Visual Data for the Application of Precision Weed Management. arXiv 2018, arXiv:abs/1801.08613. [Google Scholar] [CrossRef] [Green Version]
- Sapkota, B.B.; Hu, C.; Bagavathiannan, M.V. Evaluating Cross-Applicability of Weed Detection Models across Different Crops in Similar Production Environments. Front. Plant Sci. 2022, 13, 837726. [Google Scholar] [CrossRef] [PubMed]
- Wiles, L.J. Beyond patch spraying: Site-specific weed management with several herbicides. Precis. Agric. 2009, 10, 277–290. [Google Scholar] [CrossRef]
- Allmendinger, A.; Spaeth, M.; Saile, M.; Peteinatos, G.G.; Gerhards, R. Precision Chemical Weed Management Strategies: A Review and a Design of a New CNN-Based Modular Spot Sprayer. Agronomy 2022, 12, 1620. [Google Scholar] [CrossRef]
- Gerhards, R.; Oebel, H. Practical experiences with a system for site-specific weed control in arable crops using real-time image analysis and GPS-controlled patch spraying. Weed Res. 2006, 46, 185–193. [Google Scholar] [CrossRef]
- Bürger, J.; Küzmič, F.; Šilc, U.; Jansen, F.; Bergmeier, E.; Chytrý, M.; Cirujeda, A.; Fogliatto, S.; Fried, G.; Dostatny, D.F.; et al. Two sides of one medal: Arable weed vegetation of Europe in phytosociological data compared to agronomical weed surveys. Appl. Veg. Sci. 2022, 25, e12460. [Google Scholar] [CrossRef]
- Timmermann, C.; Gerhards, R.; Kühbauch, W. The Economic Impact of Site-Specific Weed Control. Precis. Agric. 2003, 4, 249–260. [Google Scholar] [CrossRef]
- Tataridas, A.; Kanatas, P.; Chatzigeorgiou, A.; Zannopoulos, S.; Travlos, I. Sustainable Crop and Weed Management in the Era of the EU Green Deal: A Survival Guide. Agronomy 2022, 12, 589. [Google Scholar] [CrossRef]
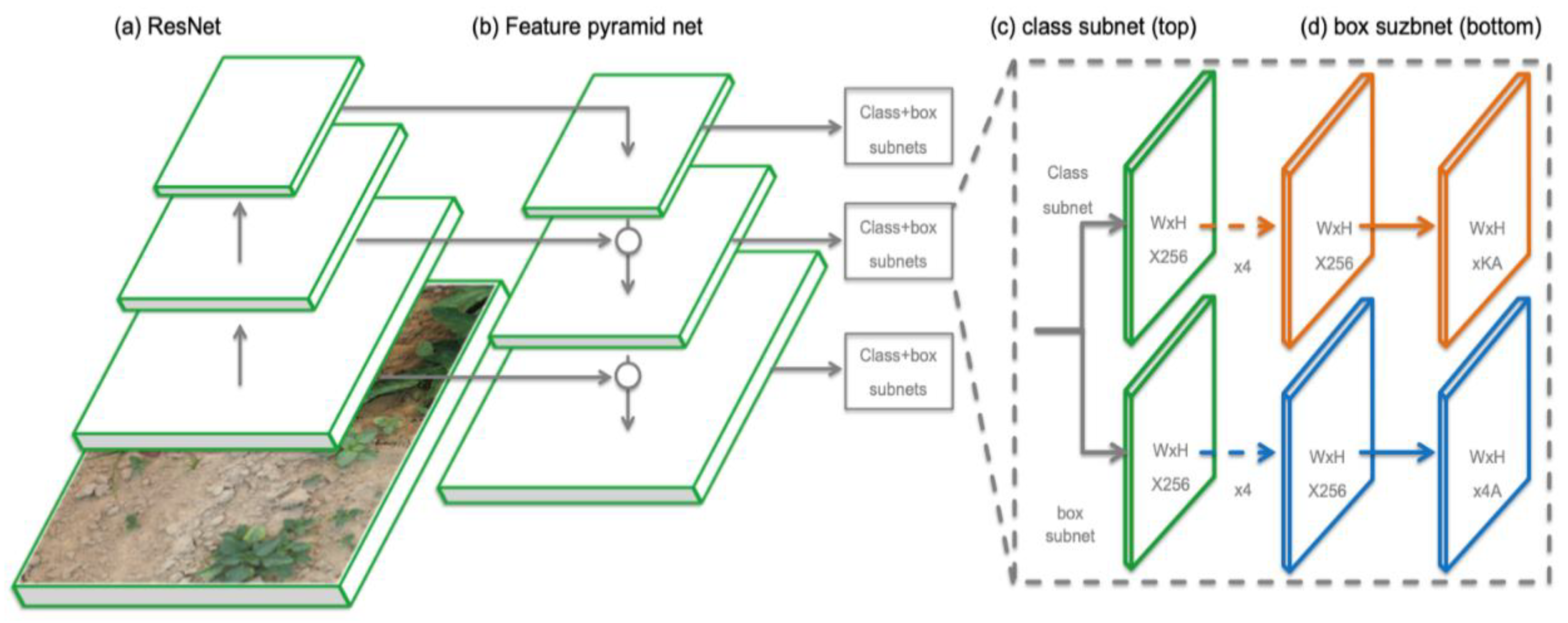
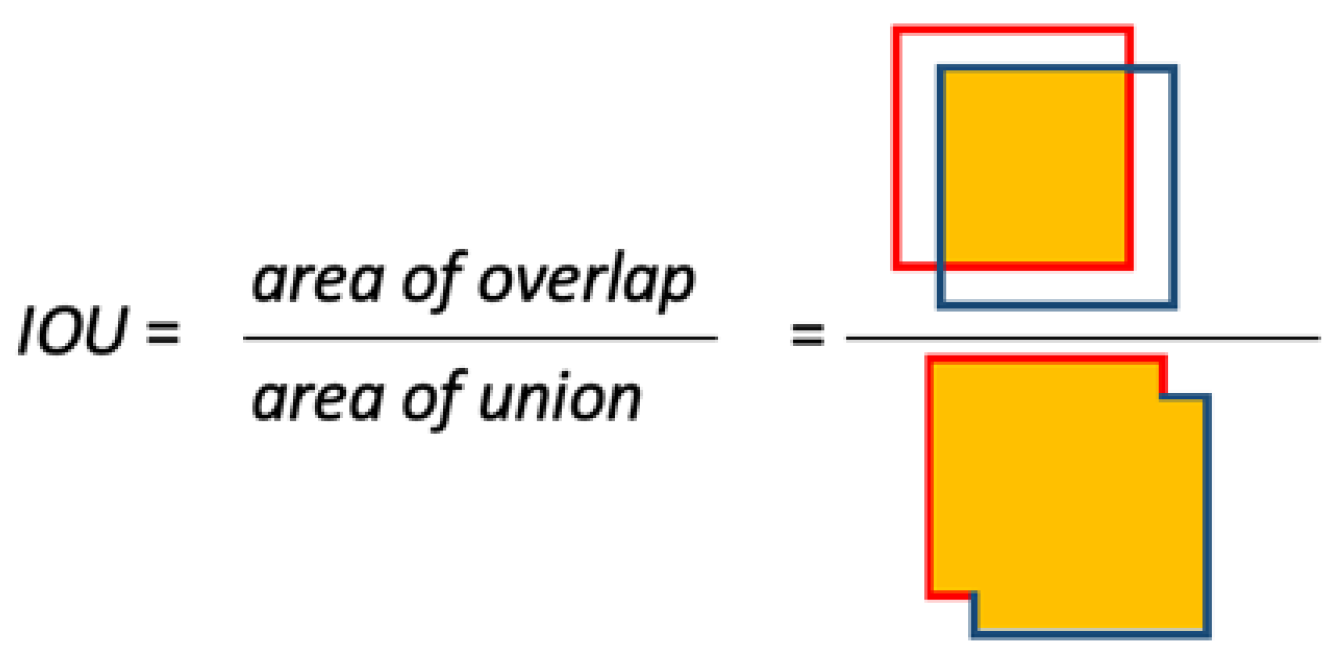



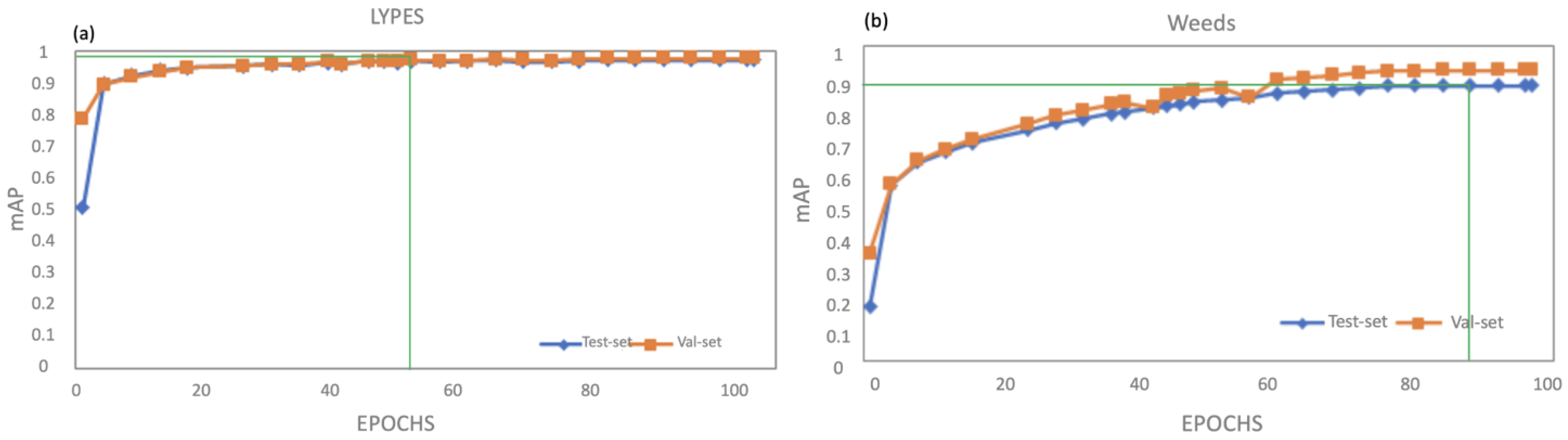
| Species | Label | Training Set | Test Set | Validation Set |
|---|---|---|---|---|
| Solanum nigrum L. | SOLNI | 1917 | 383 | 821 |
| Cyperus rotundus L. | CYPRO | 1691 | 338 | 725 |
| Echinochloa crus galli L. | ECHCG | 895 | 179 | 384 |
| Setaria verticillata L. | SETIT | 157 | 31 | 67 |
| Portulaca oleracea L. | POROL | 506 | 506 | 101 |
| Solanum lycopersicum L. | LYPES | 799 | 160 | 342 |
| Not recognised | NR | 372 | 74 | 159 |
| Species | Label | AP |
|---|---|---|
| Solanum nigrum L. | SOLNI | 0.9209 |
| Cyperus rotundus L. | CYPRO | 0.9322 |
| Echinochloa crus galli L. | ECHCG | 0.9502 |
| Setaria verticillata L. | SETIT | 0.9044 |
| Portulaca oleracea L. | POROL | 0.9776 |
| Solanum lycopersicum L. | LYPES | 0.9842 |
| Not recognised | NR | 0.8234 |
| mAP | ---- | 0.92755 |
| Weed Group | Species Label | mAP |
|---|---|---|
| Dicotyledonous | SOLNI and POROL | 0.9492 |
| Monocotyledonous | CYPRO, SETIT and ECHCG | 0.9533 |
| Label | RetinaNet | YOLOv7 | Faster-RCNN |
|---|---|---|---|
| SOLNI | 0.9209 | 0.8100 | 0.86755 |
| CYPRO | 0.9322 | 0.5533 | 0.90785 |
| ECHCG | 0.9502 | 0.9650 | 0.91056 |
| SETIT | 0.9044 | 0.6349 | 0.89502 |
| POROL | 0.9776 | 0.9323 | 0.92346 |
| LYPES | 0.9842 | 0.9530 | 0.96763 |
| NR | 0.8234 | 0.96735 | 0.97735 |
| Neural Network | mAP | Speed Prediction Sec/Frame | Number of Trained Parameters |
|---|---|---|---|
| RetinaNet | 0.90354 | 0.2354 | 39,336,702 |
| RetinaNet and Augmentation | 0.92755 | ||
| YOLOv7 | 0.65346 | 0.0869 | 36,519,530 |
| YOLOv7 and Augmentation | 0.83085 | ||
| Faster-RCNN | 0.89135 | 1.2863 | 55,338,998 |
| Faster-RCNN and Augmentation | 0.92135 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
López-Correa, J.M.; Moreno, H.; Ribeiro, A.; Andújar, D. Intelligent Weed Management Based on Object Detection Neural Networks in Tomato Crops. Agronomy 2022, 12, 2953. https://doi.org/10.3390/agronomy12122953
López-Correa JM, Moreno H, Ribeiro A, Andújar D. Intelligent Weed Management Based on Object Detection Neural Networks in Tomato Crops. Agronomy. 2022; 12(12):2953. https://doi.org/10.3390/agronomy12122953
Chicago/Turabian StyleLópez-Correa, Juan Manuel, Hugo Moreno, Angela Ribeiro, and Dionisio Andújar. 2022. "Intelligent Weed Management Based on Object Detection Neural Networks in Tomato Crops" Agronomy 12, no. 12: 2953. https://doi.org/10.3390/agronomy12122953
APA StyleLópez-Correa, J. M., Moreno, H., Ribeiro, A., & Andújar, D. (2022). Intelligent Weed Management Based on Object Detection Neural Networks in Tomato Crops. Agronomy, 12(12), 2953. https://doi.org/10.3390/agronomy12122953






