Abstract
Phytophthora root rot, caused by Phytophthora sojae (P. sojae), is one of the most devastating diseases limiting soybean production worldwide. microRNAs (miRNAs) play major roles in regulating plant defense against pathogens. To understand the roles of soybean miRNAs during P. sojae infection, we analyzed four small RNA libraries from two soybean germplasms before and after P. sojae isolate JS08-12 infection. The cultivar Nannong 10-1 was resistant to JS08-12, whereas the 06-070583 line was susceptible to JS08-12. In total, 528 known and 555 putative novel miRNAs in soybean were identified from 97 million reads; 74 known miRNAs and 75 novel miRNAs that might be specifically related to Nannong10-1 responses to P. sojae; and 55 known and 43 novel miRNAs expressed before and after infection in the susceptible line 06-070583. qRT-PCR provided similar miRNA expression patterns to those obtained by the small-RNA sequencing of the four libraries. Then, the potential target genes of these differentially expressed miRNA were predicted, which encoded transcriptional factors, resistance proteins and transporters. Finally, we focused on the targets of the three legume-specific miRNAs (gma-miR1508, gma-miR1509, and gma-miR1510) and charted the miRNA–target interactions and networks based on the published degradome data.
1. Introduction
Soybean (Glycine max) is one of the most cultivated crops worldwide. Its seeds, which are used as a food source, contain approximately 40% protein and 20% oil. However, like any other major crop, the production of soybean is greatly affected by pathogens such as fungi, bacteria, viruses, and oomycetes. Oomycetes are eukaryotic microorganisms that can cause devastating diseases in plants and serious economic losses in some crops []. Phytophthora sojae, an oomycete, causes phytophthora root rot (PRR), which is responsible for overwhelming damage to soybean production throughout the world.
microRNA(miRNA), a class of non-coding endogenous small RNA, have been proven to be the key players in regulating plant growth and development, immunity against pathogens, and responses to environmental stresses at the transcriptional and post-transcriptional level by degrading corresponding mRNA or inhibiting its translation in plants [,,,]. High-throughput technology has predicted and discovered thousands of miRNAs in many different plant species [,]. Most of the predicted miRNAs are involved in regulating the developmental and productive processes of plants []. Over the last decade or so, several studies have confirmed that miRNA also responds to infection by the pathogens of oomycetes. In 2011, Guo et al. [] studied the miRNA involved in soybean—P. sojae interactions using microarrays and identified the differentially expressed miRNAs under P. sojae infection. Cui et al. (2017) [] reported that gma-miR1510a/b suppressed the expression of an NB-LRR domain gene and reduced resistance to P. sojae. The overexpression of Sp-miR396a-5p made tobacco more susceptible to P. nicotianae, indicating the importance of miRNA in regulating plant disease resistance []. Some biotrophic oomycetes, for example, H. arabidopsidis [,] and P. capsici [], are controlled by the overexpression of miRNA; in these two instances, miR393 produces resistance in Arabidopsis. Wong et al. [] also used Illumina sequencing technology to analyze and identify the miRNAs involved in the interaction between soybean and P. sojae, and by knocking down the level of mature miR393 enhanced the susceptibility of soybean to P. sojae and drastically reduced the expression of isoflavonoid biosynthetic genes.
As the original soybean-producing country, China holds numerous soybean germplasms, and a large number of PRR-resistance germplasms have been identified in previous studies [,]. The P. sojae isolate JS08-12 (virulent formula: 1a, 1b, 1c,1d, 1k, 2, 3a, 3b, 3c, 4, 5, 6, and 7) was used to inoculate Nannong 10-1 and 06-070583 by hypocotyl inoculations. The soybean line Nannong 10-1 is a resistant line with 100% living seedlings, and 06-070583 is a susceptible line with 100% dead seedlings []. The above two germplasms are native to China. A single dominant gene, RpsJS, which is located on soybean chromosome 18 (molecular linkage group G), is responsible for the PRR resistance in Nannong 10-1. With the objective of gaining deeper insights into the P. sojae resistance mechanisms in soybean as well as understanding the molecular interactions between soybean and pathogens, we constructed four libraries from the resistant cultivar Nannong 10-1 and the susceptible line 06-070583, uninfected (control) and infected by the P. sojae isolate JS08-12. By the high-throughput sequencing of small-RNA in these libraries and by analyzing the target genes of the miRNA, the results of the present study will help elucidate the regulatory and defense mechanisms of soybean microRNA and the potential targets under P. sojae attack.
2. Materials and Methods
2.1. Plant Materials and P. sojae Isolate
The soybean germplasm Nannnong 10-1 and 06-070583 were used as plant materials and were obtained from the National Center for Soybean Improvement, China. P. sojae isolate JS08-12 was obtained from the Key Laboratory of the Monitoring and Management of Plant Diseases and Insects, Ministry of Agriculture, Nanjing Agricultural University, China.
2.2. Inoculation Method
Thirty seedlings of Nannnong 10-1 and 06-070583 were inoculated with P. sojae via a slant board assay, as described by Burnham et al. (2003) [] with a few modifications. Thirty root-section samples were collected at 12 h after inoculation 5 mm below and above the lesion margin from each seedling with characteristic lesions. For mock inoculated plants, sections were taken from the same position as in the inoculated samples. The root samples were immediately treated with liquid nitrogen and stored at −80 °C in a refrigerator.
2.3. RNA Extraction, Library Construction, and Illumina Sequencing
For miRNA sequencing, the total RNA of 4 samples (S01—mock of Nannong 10-1; S02—JS08-12 treatment of Nannong 10-1; S03—mock of 06-070583; and S04—JS08-12 treatment of 06-070583) was extracted using TRIzol reagent (Invitrogen, Waltham, MA, USA) according to the supplier’s manual. One-percent agrose gel electrophoresis was carried out to confirm the integrity and quality of total RNA. A Nanodrop 2000 spectrophotometer (IMPLEN, Westlake Village, CA, USA) was used to assess RNA purity and concentration. A Qubit 2.0 fluorometer (Thermo Fisher Scientific, Waltham, MA, USA) and Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA) were used to measure the concentration and integrity of the RNA, respectively. The qualified RNA samples were selected for the construction of the following four small-RNA libraries using the Next Ultra Small RNA Sample Library Prep Kit for Illumina (NEB, Ipswich, NJ, USA) (Supplementary Figure S1). The Illumina HiSeq 2500 (Illumina, CA, USA) platform was employed to sequence the four small-RNA samples at Biomarker Technology Co. (Shanghai, China).
2.4. Alignment and Mapping of the Small-RNA Sequence Data
The length of the sequence read was set to 50 nucleotides (nt). Raw reads with low sequencing quality and a length shorter than 18 nt or longer than 30 nt were eliminated to produce clean data. To identify known non-coding RNAs and repetitive sequences, clean reads were aligned to ribosome RNA (rRNA) sequences in Silva (http://www.arb-silva.de, 1 Jaunary 2019) [], transfer RNA (tRNA) sequences in GtRNAdb (http://lowelab.ucsc.edu/GtRNAdb, 1 Jaunary 2019) [], small nuclear RNA (snRNA) and small nucleolar RNA (snoRNA) sequences in Rfam [] (http://rfam.xfam.org, 22 Jaunary 2019), and repetitive sequences in Repbase (http://www.girinst.org/repbase, 30 Jaunary 2019) [] using Bowtie software. The clean reads were also aligned to the soybean reference genome using miRDeep2 [,].
2.5. Identification of Known and Novel miRNAs
The identification of known miRNAs was used to align the clean reads to miRBase release 22 (http://www.mirbase.org, 1 May 2018) []. The identification of novel miRNA in soybean was performed according to the criteria for plant miRNA annotations []. First, the reads obtained by the sequencing were mapped onto the soybean reference genome by miRDeep2, and the secondary structure of miRNA precursors was predicted by the RNAfold program [].
2.6. microRNA Expression Analysis
The expression of each miRNA was calculated and normalized using the TPM algorithm []. DESeq [] was used to analyze the differentially expressed miRNAs. To detect the P. sojae-responsive miRNAs of each genotype, differential expression analysis was performed for the same genotype subjected to JS08-12. The criteria for differential expression were established as |log2 (fold change)| > 1 and FDR < 0.05 (Supplementary Figures S3–S5).
2.7. Validation of Differentially Expressed microRNA by Quantitative Real-Time PCR
The expression of randomly selected miRNAs was validated by stem-loop RT-PCR. Total RNA samples were treated with DNase I (Takara, Dalian, China) to remove residual genomic DNA. RNA was reverse-transcribed to cDNA, and qRT-PCR was carried out using Mir-X miRNA qRT-PCR SYBR® Kits (Takara, Dalian, China), according to the supplier’s instructions. The PCR cycling conditions were: 95 °C for 10 s, 40 cycles of 95 °C for 5 s, and 60 °C for 20 s. All reactions were repeated three times, and snoR1 served as the internal control. The relative expression level of miRNA was calculated using the 2−ΔΔCT method [].
2.8. Prediction and Confirmation of microRNA Target Genes
The target genes of miRNA were predicted by TargetFinder software [] with default parameters. The function and annotation of target genes was carried out using the Blast to Swiss-Prot (http://www.uniprot.org/ accessed on 1 Jaunary 2019), NCBI non-redundant protein sequence (Nr) (ftp://ftp.ncbi.nih.gov/blast/db/ accessed on 1 Jaunary 2019), and Pfam databases (http://pfam.xfam.org/ accessed on 1 Jaunary 2019) []. Then, DPMIND (http://202.195.246.60/DPMIND/ accessed on 1 Jaunary 2019) [], which is a degradome-based plant miRNA-target interaction and network database, provided us with a comprehensive retrieval and analysis platform to study the miRNAs and their verified targets in soybean.
2.9. Functional Annotations of the Predicted Targets of the Differentially Expressed miRNA
Gene Ontology (GO) annotations, the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis, Clusters of Orthologous Groups of proteins (COG), Swiss-Prot, the Non-Redundant (NR) Protein Database, KOG, and the Pfam database were employed to investigate the putative biological functions of the target genes and biological processes possibly regulated by miRNAs in soybean.
3. Results
3.1. Resistance Response of Two Soybean Germplasms to P. sojae
Seven-year-old seedlings of Nannong 10-1 and 06-070583 were inoculated with P. sojae JS08-12 (virulent formula; 1a, 1b, 1c,1d, 1k, 2, 3a, 3b, 3c, 4, 5, 6, and 7). The phenotypes of the two lines showed that Nannong 10-1 was a resistant line with 100% living seedlings, and 06-070583 was a susceptible line with 100% dead seedlings ([]; Figure 1). The first response, according to previous research, can occur as early as 12 h after inoculation with P. sojae []. Hence, we chose to compare the roles of miRNA in the defense against P. sojae between Nannong 10-1 and 06-070583 at 0 h and 12 h post-inoculation.

Figure 1.
Phenotypes of the two soybean germplasms treated with the P. sojae isolate JS08-12 by hypocotyl inoculation. (A) Nannong 10-1; (B) 06-070583.
3.2. High-Throughput Sequencing and Small-RNA Data Analysis
In the present study, four small RNA libraries were constructed from the roots of Nannong 10-1 and 06-070583 seedlings treated with P. sojae JS08-12 for 0 h and 12 h, respectively (S01: Nannong 10-1 mock; S02: Nannong 10-1 with P. sojae treatment; S03: 06-070583 mock; S04: 06-070583 with P. sojae treatment). Moreover, the four sRNA libraries were sequenced by Illumina technology and provided a total of 97,924,844 sRNA raw reads(Table 1). After low-quality and adapter sequences were removed, 69,325,641 reads ranging from 18 to 30 nucleotides (nt) were used for further analyses. Then, the known non-coding RNAs, including rRNA, snRNA, snoRNA, and tRNA, were annotated and removed. The numbers and proportions of the different types of small RNA are shown in Table 2.

Table 1.
Statistics of sRNA (small RNA) sequences from the four libraries.

Table 2.
sRNA classification annotation statistics.
These unannotated reads were mapped onto the G. max database using miRDeep2 computational software []. Figure 2 shows the locations of the mapped reads in different chromosomes of soybean. As reported in Table 3, 21, 22, and 24 nt were most abundant, which was consistent with previous studies involving small-RNA-sequencing analyses in soybean [,,]. The length distribution between the two cultivars and P. sojae infection libraries was fairly similar. Small RNAs in the 21 nt and 22 nt classes showed higher abundance than those of the 24 nt class.
Figure 2.
Mapped reads displayed on different soybean chromosomes. (A) S01—Nannong 10-1 mock; (B) S02—Nannong 10-1 at 12 h post-inoculation (hpi); (C) S03—06-070583 mock; (D) S04—06-070583 at 12 hpi.

Table 3.
The number of differentially expressed miRNAs.
3.3. Identification of Conserved and Novel miRNA
Due to the specificity of dicer enzymes and dicer-like (DCL) enzymes, the final generation of mature miRNA is mainly concentrated in the 20 nt to 24 nt classes. In this study, 555 new miRNAs and 528 known miRNAs were predicted in the four samples. The length distributions of the known and new miRNAs identified are shown in Figure 3, vary from 19 to 25 nt, with the majority in the 20-21 nt. Since the AGO protein of the miRNA pathway preferentially associates 21 nt small RNAs starting with a 5′ uridine, so a strong bias in the identification and cutting of precursor miRNA at the 5′ end of the first base pair of U. The typical miRNA base proportion was obtained by analyzing the base preference of miRNA. For the first base preference of the 5′ end and the base preference of the mature sequences of the total miRNAs, see Figure 3b,c.
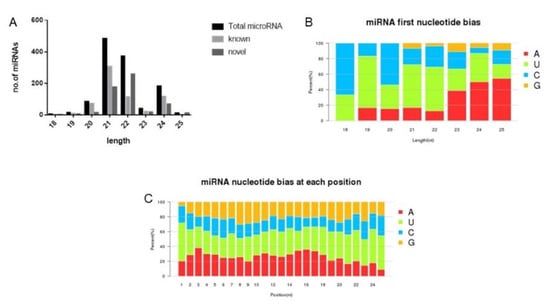
Figure 3.
Characterizations of known and novel soybean miRNAs. (A) Size distribution of mature miRNAs, including known and novel miRNAs. (B) Distribution of the first nucleotide of known and novel miRNAs. (C) Distribution of nucleotide bias at each position of known and novel miRNAs.
3.4. Identification of Differentially Expressed miRNAs upon P. sojae Infection
To detect the effect of P. sojae on G. max miRNA expression, we performed a differential expression analysis between the libraries with and without P. sojae treatment. All miRNAs with more than one normalized read were analyzed by fold changes and FDR. Differential miRNA expression was considered to be indicted by an FDR lower than 0.01 and a fold change higher than two. A total of 74 conserved miRNAs (belonging to 34 families) and 75 novel miRNAs significantly changed in response to P. sojae in Nannong 10-1 (Figure 4 and Table S1). Among them, there were 18 upregulated and 131 downregulated miRNAs. For the upregulated miRNAs, 12 conserved miRNAs (gma-miR1535a, gma-miR166m, gma-miR171c-3p, gma-miR171c-5p, gma-miR171n, gma-miR171o-3p, gma-miR171p, gma-miR171q, gma-miR319f, gma-miR4374a, gma-miR5037c, and gma-miR862a), belonging to seven families, and 6 novel miRNAs were identified. Sixty-two conserved miRNAs (belonging to 30 families) and sixty-nine novel miRNAs were downregulated in P. sojae-treated tissues of Nannong 10-1.
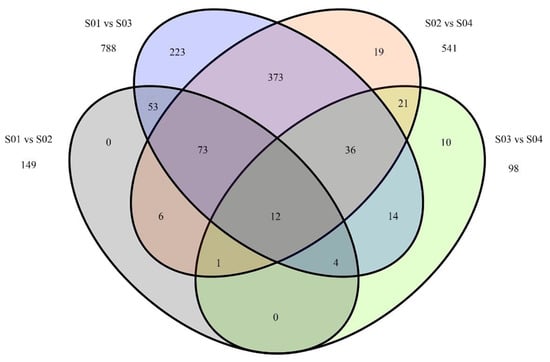
Figure 4.
Venn diagram illustrating specific Phytophthora sojae-responsive miRNAs in Nannong 10-1 (S01—Nannong 10-1 mock; S02—Nannong 10-1 at 12 hpi) and 06-070583 (S03—06-070583 mock; S04—06-070583 at 12 hpi).
We also analyzed the differentially expressed miRNAs between 06-070583 with and without P. sojae treatment. Fifty-five conserved miRNAs and forty-three novel miRNAs were altered between the two samples (Table S2). Nine conserved miRNAs and thirty-nine novel miRNAs were upregulated in P. sojae-treated G. max roots; forty-six conserved miRNAs (belonging to 13 families) and four novel miRNAs were downregulated in P. sojae-treated G. max roots.
3.5. Validation of Sequencing Data by Stem-Loop qRT-PCR
To confirm the expression of the identified miRNAs and detect the dynamic responses to P. sojae infection, 11 significantly expressed miRNA families were selected for stem-loop qRT-PCR to validate their expression patterns in resistant and susceptible plants, and the results coincided with the RNA-seq data, proving their validity. Ten miRNAs were downregulated in the resistant cultivar Nannong10-1, and one microRNA was upregulated in the susceptible line 06-070583 post P. sojae inoculation (Table 4).

Table 4.
Real-time RT-PCR analysis of P. sojae-responsive miRNAs.
3.6. Target-Gene Prediction and Functional Annotations of the Predicted Targets of P. sojae-Responsive miRNAs in Resistant and Susceptible Soybean Germplasms
In total, 1484 target genes were predicted for 585 microRNAs (Table S3). The division of known and novel microRNAs resulted in 356 known miRNAs with 957 targeted genes and 229 novel miRNAs with 587 predicted target genes. Among the 1484 target genes, 1443 were annotated (Table S4). The numbers of annotated miRNA target genes in the different samples are shown in Table S5. The GO annotation showed that these target genes could participate in various cellular processes, such as “defense response”, “the oxidation–reduction process”, “protein phosphorylation”, and “the regulation of transcription of DNA-template” (Figure 5). Based on the results of TOPGO, the two most basic “cellular component” categories were “nucleus” and “integral component of membrane”; in “biological process”, the top two categories were “the regulation of transcription, DNA-template” and “defense response”; and in “molecular function”, the top two categories were “DNA binding” and “ATP binding” in the resistant cultivar. Additionally TOPGO showed that the basic “biological processes” included “metabolic processes”, “cellular processes”, and “biological regulation”, whereas the “molecular processes” included “binding” and “catalytic activity” and the cellular components included “cells”, “cell parts”, and “organelles”(Figure 6). Meanwhile, 21 KEGG pathways were classified in the resistant cultivar Nannong 10-1, and 10 KEGG pathways in the susceptible line 06-070583, and the most common pathways were “zeatin biosynthesis”, “plant hormone signal transduction”, “ribosome”, “cyanoamino acid metabolism”, “nucleotide excision repair”, and “phenylpropanoid biosynthesis”, which are shown below. We also found several pathways related particularly to disease response (Figure 7) [,].
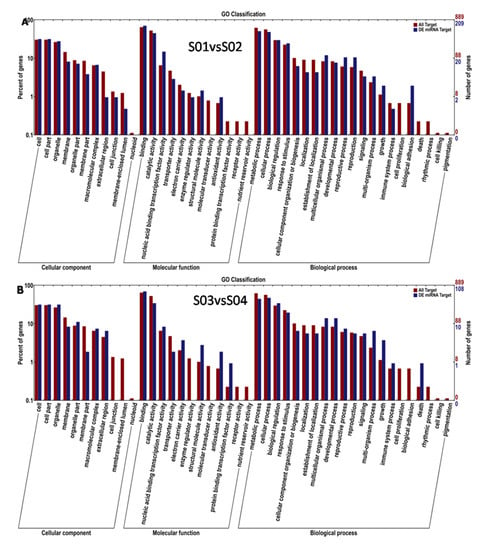
Figure 5.
GO analysis of miRNA target genes identified in S01 vs. S02 (A) and S03 vs. S04 (B). The digits on the left y-axis and right y-axis indicate the percentage and enrichment of miRNA targets, respectively. Only the target genes identified for miRNAs by RNA sequencing were considered.
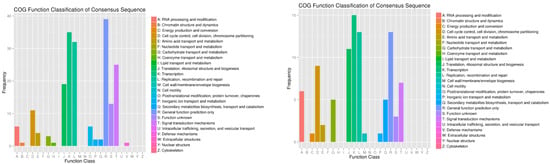
Figure 6.
COG annotation classification of differentially expressed miRNA target genes.
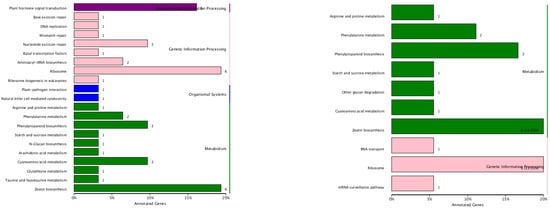
Figure 7.
KEGG classification of differentially expressed targets of miRNAs. The labels on the y-axis provide the names of the metabolic pathways, and the digits on the x-axis are the numbers of genes annotated for this pathway and the proportion of the total number of annotated genes.
3.7. Regulatory Networks of Leguminous-Specific miRNA Targets
Here, we noted that some leguminous-specific miRNAs, including gma-miR1508a/c, gma-miR1509a/b, and gma-miR1510a/b, were downregulated in the resistant cultivar Nannong 10-1 treated with P. sojae strain JS08-12 (see Supplementary Figure S2). Thus, we used DPMIND software to ascertain the network between the above miRNAs and their targets according to published degradome data. Strong evidence showed that Glyma. 15G017000.1 was the target gene of gma-miR1508a, the homologous gene to AT1G52190.1 in Arabidopsis thaliana, which encodes major facilitator superfamily proteins (Figure 8). gma-miR1510b-3p has several target genes (Figure 9): Glyma. 13G194900.1 (homologous to AT5G36930.2 in Arabidopsis thaliana); Glyma. 15G232400.1 and Glyma. 15G232400.2 (homologous to AT5G41540.1); Glyma. 04G219600.1 (homologous to AT5G36930.2); Glyma. 12G163000.1 (homologous to AT1G31540.1); Glyma. 19G055000.1 (homologous to AT5G36930.2); Glyma. 19G055000.2 (homologous to AT5G48770.1); Glyma. 19G055000.3 (homologous to AT5G17680.1); Glyma. 19G055000.4 (homologous to AT5G36930.2); Glyma. 19G055000.5 (homologous to AT5G36930.2); and Glyma. 19G055000.6 (homologous to AT5G36930.2). All the abovementioned genes encode disease-resistance family proteins (TIR-NBS-LRR class). These predicted target genes may play key roles in the soybean—P. sojae interaction. gma-miR1509a/b had three predicted target genes: Glyma. 04G195800.1, Glyma. 14G075600.1, and Glyma. 14G076300.1; there was particularly strong evidence for Glyma. 04G195800.1 (Figure 10). However, it is regrettable that there were no annotations for these genes.
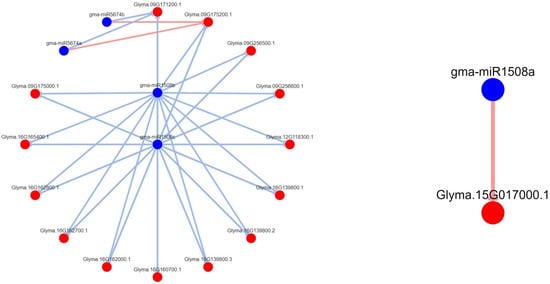
Figure 8.
Degradome-based gma-miR1508a/b/c–target interactions and network. Note: we used DPMIND software (http://cbi.njau.edu.cn/DPMIND/network.php?race_id=3847, 1 January 2022) to obtain the network. Note: blue circles indicate miRNA, red circles indicate targets, red lines indicate strong evidence, blue lines indicate weak evidence.
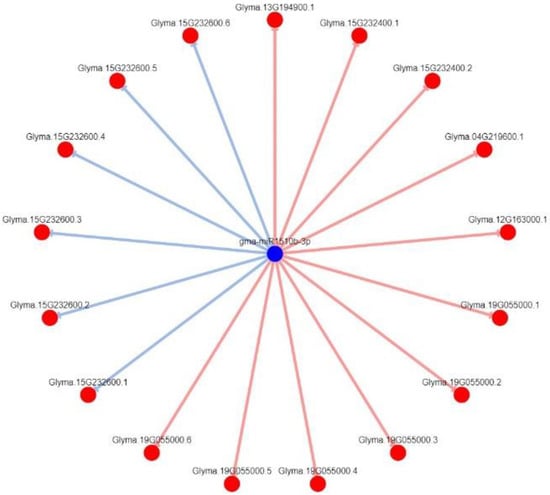
Figure 9.
Degradome-based gma-miR1510b-5p/3p–target interactions and network. Note: we used DPMIND software (http://cbi.njau.edu.cn/DPMIND/network.php?race_id=3847, 1 January 2022) to obtain the network. Note: blue circles indicate miRNA, red circles indicate targets, red lines indicate strong evidence, blue lines indicate weak evidence.
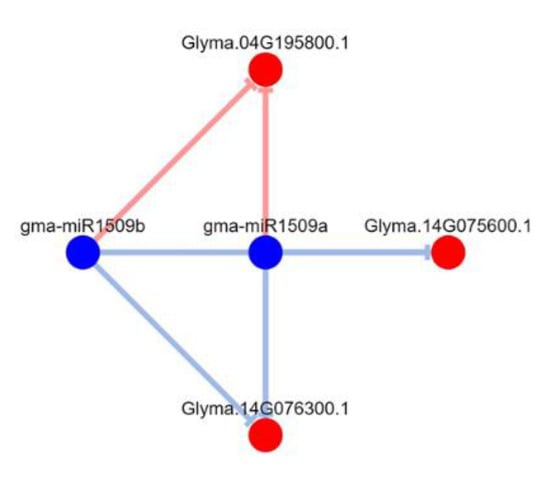
Figure 10.
Degradome-based gma-miR1509–target interactions and network. Note: we used DPMIND software (http://cbi.njau.edu.cn/DPMIND/network.php?race_id=3847, 1 January 2022) to obtain the network. Note: blue circles indicate miRNA, red circles indicate targets, red lines indicate strong evidence, blue lines indicate weak evidence.
4. Discussion
Although many conserved and legume-specific miRNAs have been identified in soybean using high-throughput sequencing and bioinformatic analysis [,,,], fewer studies have been published on microRNAs and their role in the interactions between soybean and oomycetes [,,,]. When we inoculated the resistant line Nannong 10-1 (containing the RpsJS gene) and the susceptible line 06-070583 with P. sojae JS08-12 (virulent formula; 1a, 1b, 1c, 1d, 1k, 2, 3a, 3b, 3c, 4, 5, 6 and 7) and analyzed them, we obtained 1083 known and putatively new microRNAs []. Upon further analysis, we obtained 74 known miRNAs and 75 novel miRNAs that were presumably involved in Nannong 10-1 responses to P. sojae, as well as 55 known and 43 novel miRNAs that were likely involved in the response of the susceptible line 06-070583 to P. sojae before and after infection. Subsequently, we predicted the target genes of these microRNAs. Finally, we focused on the legume-specific miRNAs gma-miR1508, gma-miR1509, and gma-miR1510 and used the published degradome data to analyze the resistance-related genes.
When investigating resistance-related genes, researchers focus on resistance (R) proteins, which lead to effector-triggered immunity (ETI) in plant cells upon the detection of pathogen effectors []. Most R proteins contain nucleotide-binding (NB) and protein-binding leucine-rich repeat (LRR) domains. An examination of these NB-LRR proteins shows the presence of either a TIR (Toll interleukin 1 receptor) or CC (coiled coil) domain, which detect and recognize biotic effectors, i.e., fungi, oomycetes, bacteria, and viruses, to stimulate the plant defense system [,]. Remarkably, other 22 nt miRNAs targeting the NBS-LRR class of R genes have been found to be abundant and diverse in legumes and Solanum species [,]. For example, secondary siRNAs are produced via RDR6 and DCL4 by miR482, miR2109, and miR1507, which perform a silencing effect [,]. Hence, to target and regulate the rapidly evolving R genes, diversifying the secondary siRNAs is, in all probability, the most effective way to maximize their response []. Additional examples of these 22 nt miRNAs include: miR2118 (the passenger strand of soybean miR482), which targets the conserved P-loop motif of TIR-NBS-LRR; miR2109, which targets the TIR-1 motif of TIR-NBS-LRR; and miR1507, which targets the kinase-2 motif of CC-NBS-LRR []. In another experiment, two miRNAs (miR2109 and miR1507) were produced when soybean was infected with active P. sojae, but not when it was infected with heat-inactivated P. sojae, hinting at these miRNAs’ possible role in ETI; the up- and downregulation of these miRNAs proved this hypothesis to be true []. To confirm this phenomenon, when pathogen effectors suppressed RNAi, the upregulation of miRNA targets (which, in this case, were the R genes) provided an abundance of R proteins to fight the pathogen []. Hence, our identification of the targeted genes gma-miR1508a (Figure 8) and gma-miR1510b-3p (Figure 9) through degradome analysis proved the involvement of said microRNAs in the defense against P. sojae. A previous study [] showed that GmTNL16, encoding NBS-LRR-type proteins, was targeted by gma-miR1510; overexpressed GmTNL16 and silenced gma-miR1510 could improve the resistance to P. sojae in soybean hairy roots. Furthermore, JA- and SA-pathway-related genes, including JAZ, COI1, TGA, and PR genes, responded to P. sojae treatment.
microRNA are important regulators of plant hormone signals. ABA is a key plant hormone that plays an important role under multiple stress conditions by mediating the expression of stress-related genes and inducing stomatal closure, acting as the basic responder to environmental changes [,]. Several key genes in the microRNA biogenesis pathway, including HYL1, DCL1, HEN1, SE, and HASTY, were impaired with ABA-hypersensitive mutants during germination [,], suggesting that microRNAs are involved in ABA signaling. Moreover, miRNA159a was proven to be involved in ABA signal transduction []. The downregulation of gma-miR159d in cyanoamino acid metabolism was observed in the differential expression analysis of the resistant cultivar Nannong 10-1 during our data mining study. This suggested that gma-miR159d is suppressed when attacked by P. sojae, which in turn kick starts ABA production.
Many growth-related pathways, such as leaf and root architecture and vascular development, are modulated by a hormone named auxin, which is positively regulated by TIR (transport inhibitor response 1), as it promotes Aux/IAA proteins through ubiquitination. miR393 targets TIR, hence controlling auxin production. An increased level of miR393 would downregulate auxin signals and reduce plant growth. In the resistant cultivar Nannong 10-1, the downregulation of TIR1, which was targeted by gma-miR393 family members, strongly suggested the downregulation of ubiquitin mediated proteolysis, inhibiting cellular development and plant growth (Supplementary Figure S6a). The alternate responsive expression pattern suggested the instigation of a wide range of regulatory mechanisms by the miR393 family (Table S1).
gma-miR172k and gma-miR172f are inhibited in the resistant cultivar Nannong 10-1, and the target genes maybe regulate the ABF signals (Table S1). This suggested a positive correlation with stomatal closure upon P. sojae attack. When we investigated the susceptible line, a novel microRNA was found to be responsible for the downregulation of PP2C, suggesting an innate immunity signal in susceptible soybean (Supplementary Figure S6b).
We know that miR396a overexpression leads to a lowered stomatal density in Arabidopsis, proving to be an important factor in stress responses [,]. miR166 is crucial for cell development, as it regulates the class-3 homeodomain-leucine zipper transcription factors, which partially control lateral root development, auxiliary meristem initiation, and leaf polarity [,,,]. Both of these microRNAs were downregulated in the resistant line upon P. sojae attack, suggesting innate stress response activation (Supplementary Table S1).
Cyanide, a nitrogen-rich compound that is effectively toxic to mammals, is naturally produced in plants, algae, fungi, and bacteria. The processing of cyanide is achieved through cyanogenesis, whereby cyanide is degraded and nitrogen is released, which is then used in growth. Cyanogenesis in a wide range of plants constitutes a chemical defense system against herbivores and pathogens [,]. The cyanoamino acid metabolism pathway, involving six novel microRNAs downregulated in the resistant cultivar and upregulated in the susceptible line, showed a negative correlation with pathogen-responsive genes in the Nannong 10-1 cultivar (Supplementary Figure S7a,b). These microRNAs showed the same pattern in starch and sucrose metabolism along with phenylpropanoid biosynthesis (Supplementary Figure S8a,b), suggesting the wide range of deactivated pathways controlled by these six novel microRNAs.
Furthermore, the downregulation of MKK1/2 by a novel microRNA was observed in the KEGG analysis, suggesting that through the downregulation of MKK1/2 as part of the plant–pathogen interaction, the signal was interrupted to deactivate pathogen-responsive genes and WRKY33, which in turn controls the oomycete defense-related genes (Supplementary Figure S9a,b).
5. Conclusions
Our results revealed the basic structure of microRNAs involved in plant metabolism under P. sojae attack and provided the functional annotation of eight novel microRNAs involved in soybean metabolism. Based on these findings, we could conclude that miRNAs are not just regulators but are key players in defense against P. sojae. This could be a breakthrough for understanding new aspects of plant defense mechanisms, as this draft could provide a framework for function identification of miRNAs and the development of new methods for improving P. sojae resistance in soybean.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/agronomy12122922/s1, Table S1: Differentially expressed miRNAs in Nannong 10-1 in response to P. sojae, Table S2: Differentially expressed miRNAs in 06-070583 in response to P. sojae, Table S3: miRNA target gene information, Table S4: Differential expression of miRNA target gene annotation, Table S5: Samples of different miRNA target gene annotation results, Figure S1: sRNA flowchart of sequencing information analysis, Figure S2: Homologue species of miRNA families found in different plants, Figure S3: RPKM and TPM distribution of four libraries along with their Pearson correlation, Figure S4: Correlation plot between the different samples, Figure S5: Differentially expressed miRNA volcano map, Figure S6a-b: Plant–hormone signaling pathway in S01 vs. S02 along with their DEGs, Figure S7: Cyanoamino acid metabolism showing the downregulated DEGs in S01 vs. S02 and S03 vs. S04, Figure S8: KEGG pathway of phenylpropanoid biosynthesis for differentially expressed miRNA target genes in S01 vs. S02 and S03 vs. S04, Figure S9: Plant–pathogen interaction pathway in S01 vs. S02 and S03 vs. S04.
Author Contributions
Conceptualization, H.X., N.G. and J.Z.; methodology, N.G. and J.X.; validation, N.Z., J.S. and X.C.; data curation, R.S. and S.D.; writing—original draft preparation, A.T.; writing—review and editing, N.G.; supervision, H.X.; funding acquisition, J.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Core Technology Development for Breeding Program of Jiangsu Province (JBGS-2021-059, JBGS-2021-014); the National Key R&D Program of China (2021YFD1201605); the Fundamental Research Funds for the Central Universities (KYZZ2022003); Jiangsu Agriculture Science and Technology Innovation Fund (CX (20) 2015); the National Natural Science Foundation of China (32072082, 31301340); the China Agriculture Research System of MOF and MARA, Program for Changjiang Scholars and Innovative Research Teams in Universities (PCSIRT_17R55); the Jiangsu Collaborative Innovation Center for Modern Crop Production; and the Cyrus Tang Innovation Center for Seed Industry.
Data Availability Statement
The data supporting the reported results will be made available upon request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kamoun, S. Molecular genetics of pathogenic oomycetes. Eukaryot. Cell 2003, 2, 191–199. [Google Scholar] [CrossRef]
- Mattick, J.S. Challenging the dogma: The hidden layer of non-protein-coding RNAs in complex organisms. Bioessays 2003, 25, 930–939. [Google Scholar] [CrossRef] [PubMed]
- Reinhart, B.J.; Weinstein, E.G.; Rhoades, M.W.; Bartel, B.; Bartel, D.P. MicroRNAs in plants. Genes Dev. 2002, 16, 1616–1626. [Google Scholar] [CrossRef] [PubMed]
- Röther, S.; Meister, G. Small RNAs derived from longer non-coding RNAs. Biochimie 2011, 93, 1905–1915. [Google Scholar] [CrossRef] [PubMed]
- Borges, F.; Martienssen, R.A. The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol. 2015, 16, 727–741. [Google Scholar] [CrossRef]
- Nazarov, P.V.; Kreis, S. Integrative approaches for analysis of mRNA and microRNA high-throughput data. Comput. Struct. Biotechnol. J. 2021, 19, 1154–1162. [Google Scholar] [CrossRef]
- Zhu, E.; Zhao, F.; Xu, G.; Hou, H.; Zhou, L.; Li, X.; Sun, Z.; Wu, J. mirTools: microRNA profiling and discovery based on high-throughput sequencing. Nucleic Acids Res. 2010, 38 (Suppl. S2), W392–W397. [Google Scholar] [CrossRef]
- Jones-Rhoades, M.W.; Bartel, D.P.; Bartel, B. MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006, 57, 19–53. [Google Scholar] [CrossRef]
- Guo, N.; Ye, W.-W.; Wu, X.-L.; Shen, D.-Y.; Wang, Y.-C.; Xing, H.; Dou, D.-L. Microarray profiling reveals microRNAs involving soybean resistance to Phytophthora sojae. Genome 2011, 54, 954–958. [Google Scholar] [CrossRef]
- Cui, X.; Yan, Q.; Gan, S.; Xue, D.; Dou, D.; Guo, N.; Xing, H. Overexpression of gma-miR1510a/b suppresses the expression of a NB-LRR domain gene and reduces resistance to Phytophthora sojae. Gene 2017, 621, 32–39. [Google Scholar] [CrossRef]
- Chen, L.; Luan, Y.; Zhai, J. Sp-miR396a-5p acts as a stress-responsive genes regulator by conferring tolerance to abiotic stresses and susceptibility to Phytophthora nicotianae infection in transgenic tobacco. Plant Cell Rep. 2015, 34, 2013–2025. [Google Scholar] [CrossRef] [PubMed]
- Robert-Seilaniantz, A.; MacLean, D.; Jikumaru, Y.; Hill, L.; Yamaguchi, S.; Kamiya, Y.; Jones, J.D.G. The microRNA miR393 re-directs secondary metabolite biosynthesis away from camalexin and towards glucosinolates. Plant J. 2011, 67, 218–231. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, K. Chemical genomic analyses of plant-pathogen interactions. Ph.D. Thesis, University of Toronto, Toronto, ON, Canada, 2012. [Google Scholar]
- Hou, Y.; Zhai, Y.; Feng, L.; Karimi, H.Z.; Rutter, B.D.; Zeng, L.; Choi, D.S.; Zhang, B.; Gu, W.; Chen, X.; et al. A Phytophthora effector suppresses trans-kingdom RNAi to promote disease susceptibility. Cell Host Microbe 2019, 25, 153–165.e5. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.; Gao, L.; Yang, Y.; Zhai, J.; Arikit, S.; Yu, Y.; Duan, S.; Chan, V.; Xiong, Q.; Yan, J.; et al. Roles of small RNA s in soybean defense against Phytophthora sojae infection. Plant J. 2014, 79, 928–940. [Google Scholar] [CrossRef] [PubMed]
- Carter, T.E., Jr.; Nelson, R.L.; Sneller, C.H.; Cui, Z. Genetic diversity in soybean. Soybeans Improv. Prod. Uses 2004, 16, 303–416. [Google Scholar]
- Giachero, M.L.; Declerck, S.; Marquez, N. Phytophthora root rot: Importance of the disease, current and novel methods of control. Agronomy 2022, 12, 610. [Google Scholar] [CrossRef]
- Sun, J.; Li, L.; Zhao, J.; Huang, J.; Yan, Q.; Xing, H.; Guo, N. Genetic analysis and fine mapping of RpsJS, a novel resistance gene to Phytophthora sojae in soybean [Glycine max (L.) Merr.]. Theor. Appl. Genet. 2014, 127, 913–919. [Google Scholar] [CrossRef]
- Burnham, K.D.; Dorrance, A.E.; VanToai, T.T.; Martin, S.K.S. Quantitative trait loci for partial resistance to Phytophthora sojae in soybean. Crop. Sci. 2003, 43, 1610–1617. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Chan, P.P.; Lowe, T.M. GtRNAdb: A database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009, 37 (Suppl. S1), D93–D97. [Google Scholar] [CrossRef]
- Kalvari, I.; Nawrocki, E.P.; Ontiveros-Palacios, N.; Argasinska, J.; Lamkiewicz, K.; Marz, M.; Griffiths-Jones, S.; Toffano-Nioche, C.; Gautheret, D.; Weinberg, Z.; et al. Rfam 14: Expanded coverage of metagenomic, viral and microRNA families. Nucleic Acids Res. 2021, 49, D192–D200. [Google Scholar] [CrossRef] [PubMed]
- Bao, W.; Kojima, K.K.; Kohany, O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 2015, 6, 11. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Liu, G.; Bao, Y.; Wu, Y.; You, Q. Evaluation and application of tools for the identification of known microRNAs in plants. Appl. Plant Sci. 2021, 9, e11414. [Google Scholar] [CrossRef] [PubMed]
- Friedländer, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2018, 47, D155–D162. [Google Scholar] [CrossRef] [PubMed]
- Axtell, M.J.; Meyers, B.C. Revisiting criteria for plant microRNA annotation in the era of big data. Plant Cell 2018, 30, 272–284. [Google Scholar] [CrossRef]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef]
- Zhao, S.; Ye, Z.; Stanton, R. Misuse of RPKM or TPM normalization when comparing across samples and sequencing protocols. Rna 2020, 26, 903–909. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Bo, X.; Wang, S. TargetFinder: A software for antisense oligonucleotide target site selection based on MAST and secondary structures of target mRNA. Bioinformatics 2004, 21, 1401–1402. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2020, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Fei, Y.; Wang, R.; Li, H.; Liu, S.; Zhang, H.; Huang, J. DPMIND: Degradome-based plant miRNA–target interaction and network database. Bioinformatics 2017, 34, 1618–1620. [Google Scholar] [CrossRef] [PubMed]
- Moy, P.; Qutob, D.; Chapman, B.P.; Atkinson, I.; Gijzen, M. Patterns of gene expression upon infection of soybean plants by Phytophthora sojae. Mol. Plant-Microbe Interact. 2004, 17, 1051–1062. [Google Scholar] [CrossRef] [PubMed]
- Tian, B.; Wang, S.; Todd, T.C.; Johnson, C.D.; Tang, G.; Trick, H.N. Genome-wide identification of soybean microRNA responsive to soybean cyst nematodes infection by deep sequencing. BMC Genom. 2017, 18, 572. [Google Scholar] [CrossRef]
- Zhou, Z.; Wang, Z.; Li, W.; Fang, C.; Shen, Y.; Li, C.; Wu, Y.; Tian, Z. Comprehensive analyses of microRNA gene evolution in paleopolyploid soybean genome. Plant J. 2013, 76, 332–344. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Hossain, M.S.; Arikit, S.; Valdés-López, O.; Zhai, J.; Wang, J.; Libault, M.; Ji, T.; Qiu, L.; Meyers, B.C.; et al. Identification of microRNAs and their mRNA targets during soybean nodule development: Functional analysis of the role of miR393j-3p in soybean nodulation. New Phytol. 2015, 207, 748–759. [Google Scholar] [CrossRef]
- Tian, F.; Yang, D.-C.; Meng, Y.-Q.; Jin, J.; Gao, G. PlantRegMap: Charting functional regulatory maps in plants. Nucleic Acids Res. 2020, 48, D1104–D1113. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Zhang, B.; Pan, X.; Stellwag, E.J. Identification of soybean microRNAs and their targets. Planta 2008, 229, 161–182. [Google Scholar] [CrossRef]
- Kulcheski, F.R.; De Oliveira, L.F.; Molina, L.G.; Almerão, M.P.; A Rodrigues, F.; Marcolino, J.; Barbosa, J.F.; Stolf-Moreira, R.; Nepomuceno, A.L.; Marcelino-Guimarães, F.C.; et al. Identification of novel soybean microRNAs involved in abiotic and biotic stresses. BMC Genom. 2011, 12, 307–317. [Google Scholar] [CrossRef]
- Turner, M.; Yu, O.; Subramanian, S. Genome organization and characteristics of soybean microRNAs. BMC Genom. 2012, 13, 169. [Google Scholar] [CrossRef]
- Wang, J.; Liu, C.Y.; Zhang, L.W.; Wang, J.L.; Hu, G.H.; Ding, J.J.; Chen, Q.S. MicroRNAs involved in the pathogenesis of phytophthora root rot of soybean (Glycine max). Agric. Sci. China 2011, 10, 1159–1167. [Google Scholar]
- Zhou, L.; Deng, S.; Xuan, H.; Fan, X.; Sun, R.; Zhao, J.; Wang, H.; Guo, N.; Xing, H. A novel TIR-NBS-LRR gene regulates immune response to Phytophthora root rot in soybean. Crop. J. 2022. [Google Scholar] [CrossRef]
- Dodds, P.; Rathjen, J. Plant immunity: Towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 2010, 11, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Pignatta, D.; Bendix, C.; Brunkard, J.O.; Cohn, M.M.; Tung, J.; Sun, H.; Kumar, P.; Baker, B. MicroRNA regulation of plant innate immune receptors. Proc. Natl. Acad. Sci. USA 2012, 109, 1790–1795. [Google Scholar] [CrossRef]
- Zhai, J.; Jeong, D.H.; De Paoli, E.; Park, S.; Rosen, B.D.; Li, Y.; González, A.J.; Yan, Z.; Kitto, S.L.; Grusak, M.A.; et al. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 2011, 25, 2540–2553. [Google Scholar] [CrossRef] [PubMed]
- Shivaprasad, P.V.; Chen, H.-M.; Patel, K.; Bond, D.; Santos, B.; Baulcombe, D.C. A microRNA superfamily regulates nucleotide binding site–leucine-rich repeats and other mRNAs. Plant Cell 2012, 24, 859–874. [Google Scholar] [CrossRef]
- Lam, H.-M.; Xu, X.; Liu, X.; Chen, W.; Yang, G.; Wong, F.-L.; Li, M.-W.; He, W.; Qin, N.; Wang, B.; et al. Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat. Genet. 2010, 42, 1053–1059. [Google Scholar] [CrossRef]
- Cuperus, J.T.; Carbonell, A.; Fahlgren, N.; Garcia-Ruiz, H.; Burke, R.T.; Takeda, A.; Sullivan, C.M.; Gilbert, S.D.; Montgomery, T.A.; Carrington, J.C. Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat. Struct. Mol. Biol. 2010, 17, 997–1003. [Google Scholar] [CrossRef]
- Wang, L.; Gu, X.; Xu, D.; Wang, W.; Wang, H.; Zeng, M.; Chang, Z.; Huang, H.; Cui, X. miR396-targeted AtGRF transcription factors are required for coordination of cell division and differentiation during leaf development in Arabidopsis. J. Exp. Bot. 2011, 62, 761–773. [Google Scholar] [CrossRef]
- Akdogan, G.; Tufekci, E.D.; Uranbey, S.; Unver, T. miRNA-based drought regulation in wheat. Funct. Integr. Genom. 2016, 16, 221–233. [Google Scholar] [CrossRef] [PubMed]
- Chen, X. MicroRNA biogenesis and function in plants. FEBS Lett. 2005, 579, 5923–5931. [Google Scholar] [CrossRef]
- Chen, X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 2004, 303, 2022–2025. [Google Scholar] [CrossRef] [PubMed]
- Hansen, C.C.; Sørensen, M. Reconfigured cyanogenic glucoside biosynthesis in eucalyptus cladocalyx involves a cytochrome P450 CYP706C55. Plant Physiol. 2018, 178, 1081–1095. [Google Scholar] [CrossRef]
- Liu, D.; Song, Y.; Chen, Z.; Yu, D. Ectopic expression of miR396 suppresses GRF target gene expression and alters leaf growth in Arabidopsis. Physiol. Plant. 2009, 136, 223–236. [Google Scholar] [CrossRef]
- Yu, X.-Z.; Trapp, S.; Zhou, P.-H.; Chen, L. Effect of temperature on the uptake and metabolism of cyanide by weeping willows. Int. J. Phytoremediat. 2007, 9, 243–255. [Google Scholar] [CrossRef] [PubMed][Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).