Epigenetic and Genetic Contribution for Expression Bias of Homologous Alleles in Polyploid Sugarcane
Abstract
1. Introduction
2. Materials and Methods
2.1. Library Construction and Sequencing
2.2. DNA Methylation Sequencing Data Analysis
2.3. Transcriptome Sequencing Data Analysis
2.4. Ka/Ks Analysis
2.5. GO Enrichment Analysis
2.6. Genetic Substitution Rate Calculation
3. Results
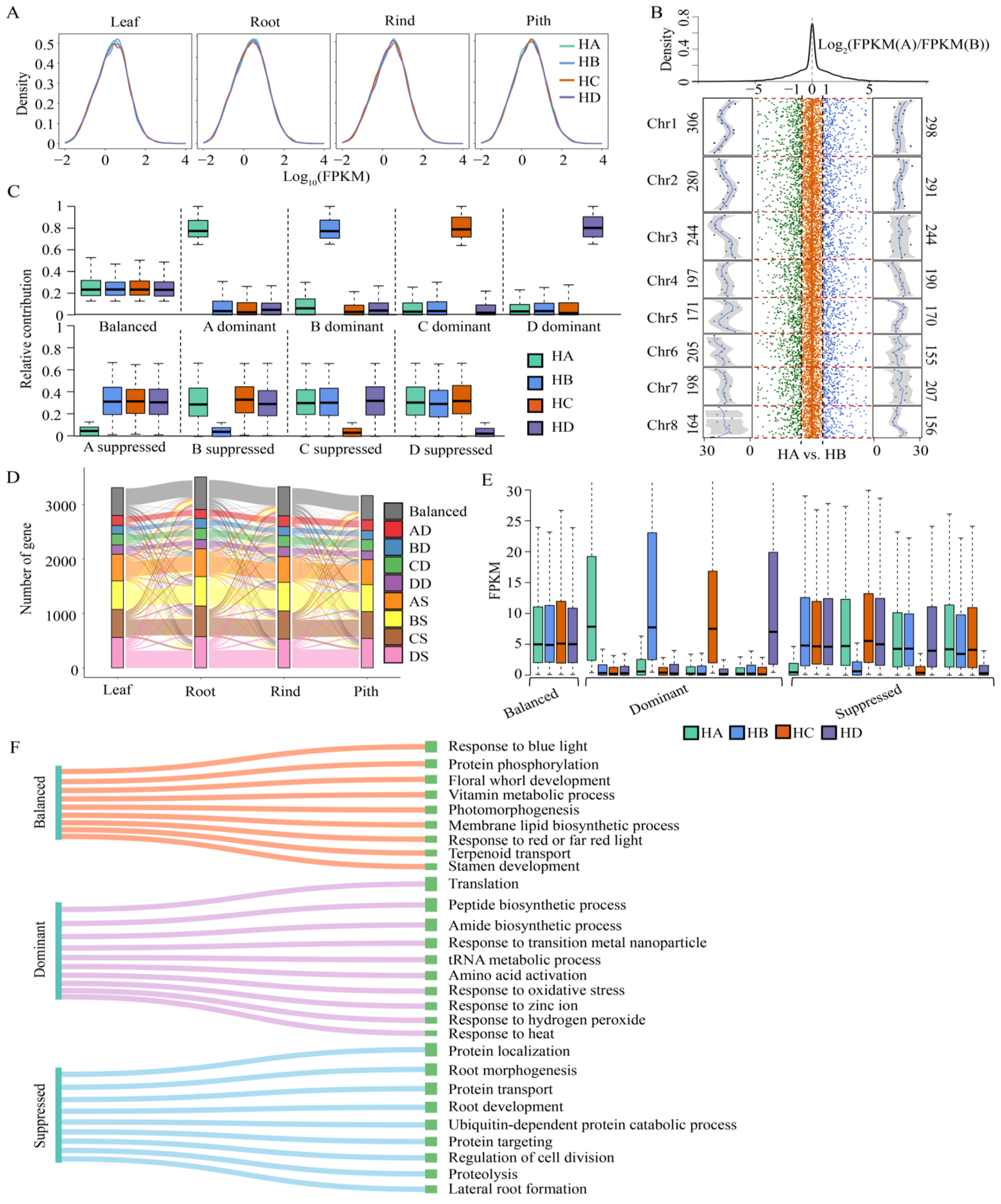
3.1. Expression Patterns of Homologous Alleles in Autopolyploid Sugarcane
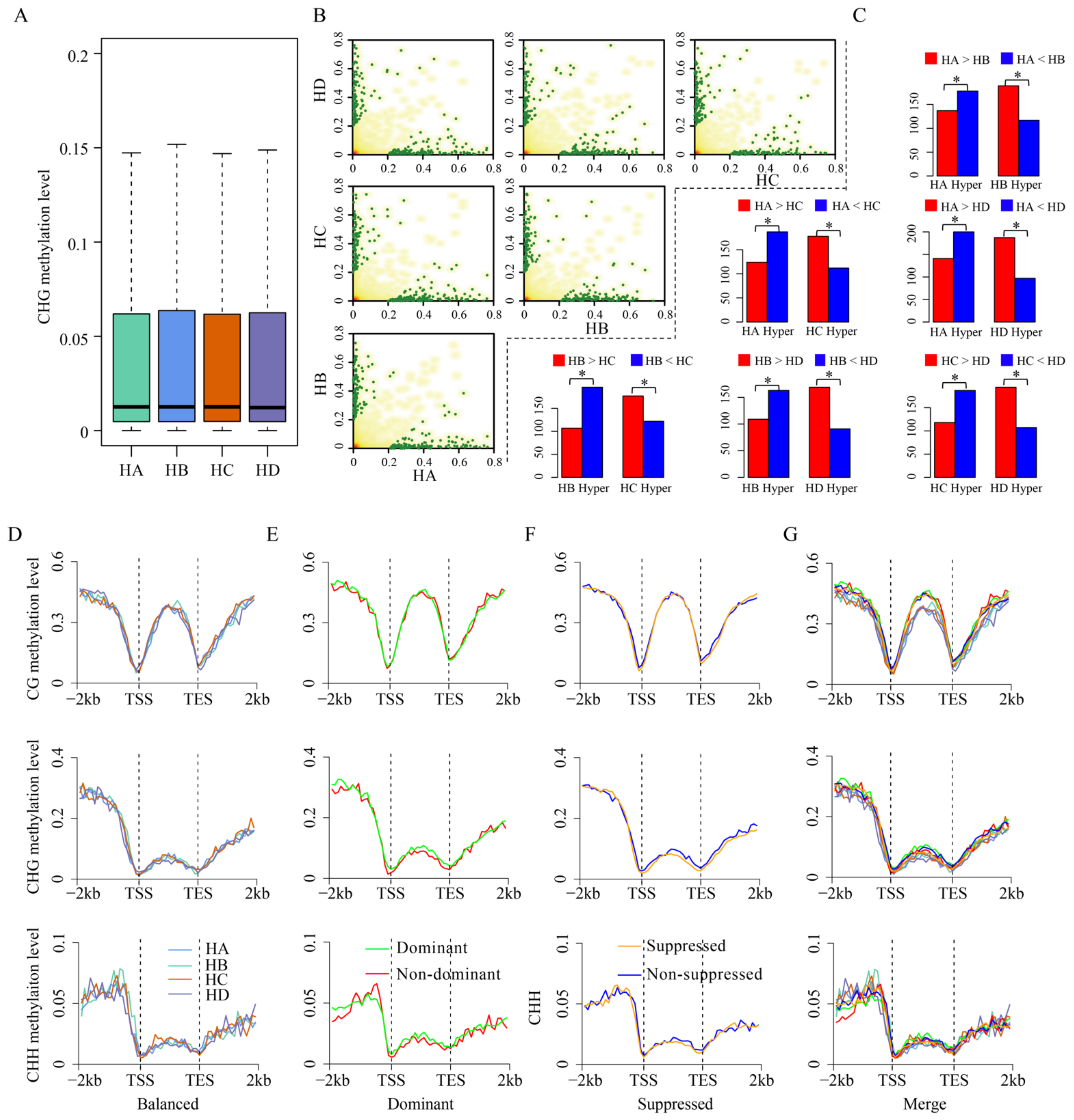
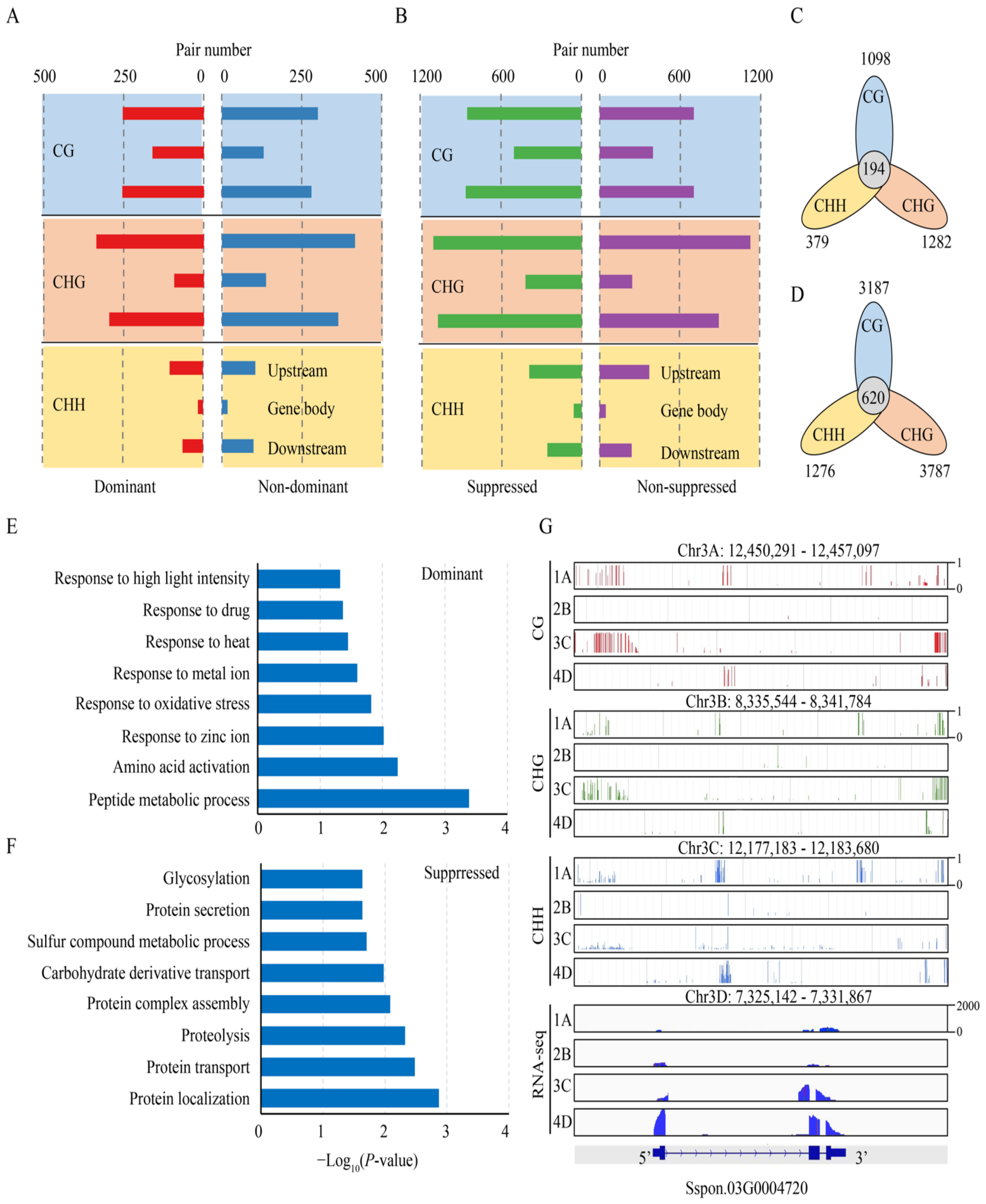
3.2. Role of DNA Methylation in Homologous Alleles Expression Bias
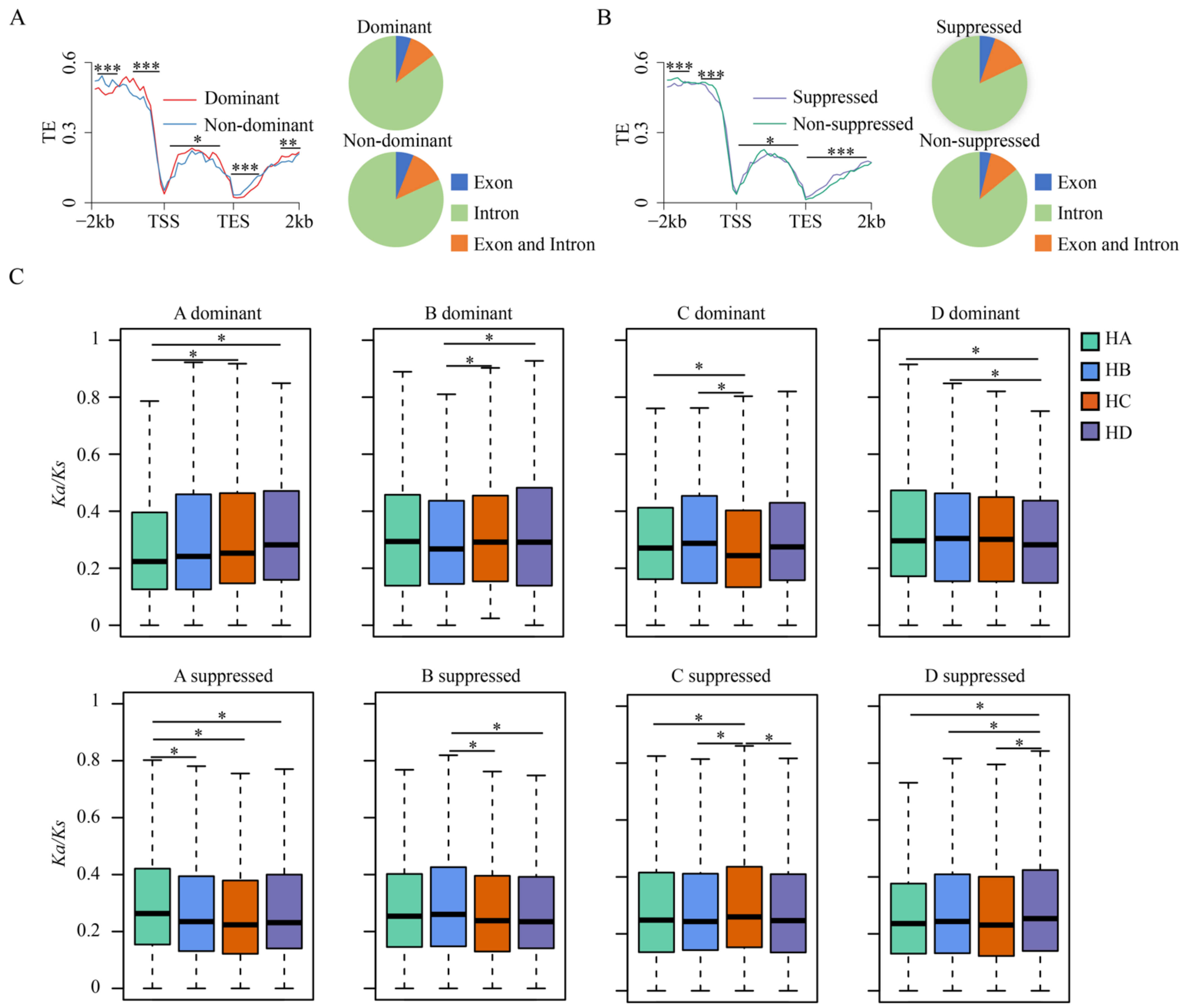
3.3. Variation of TE Insertions and Selection Pressure of Tetrads
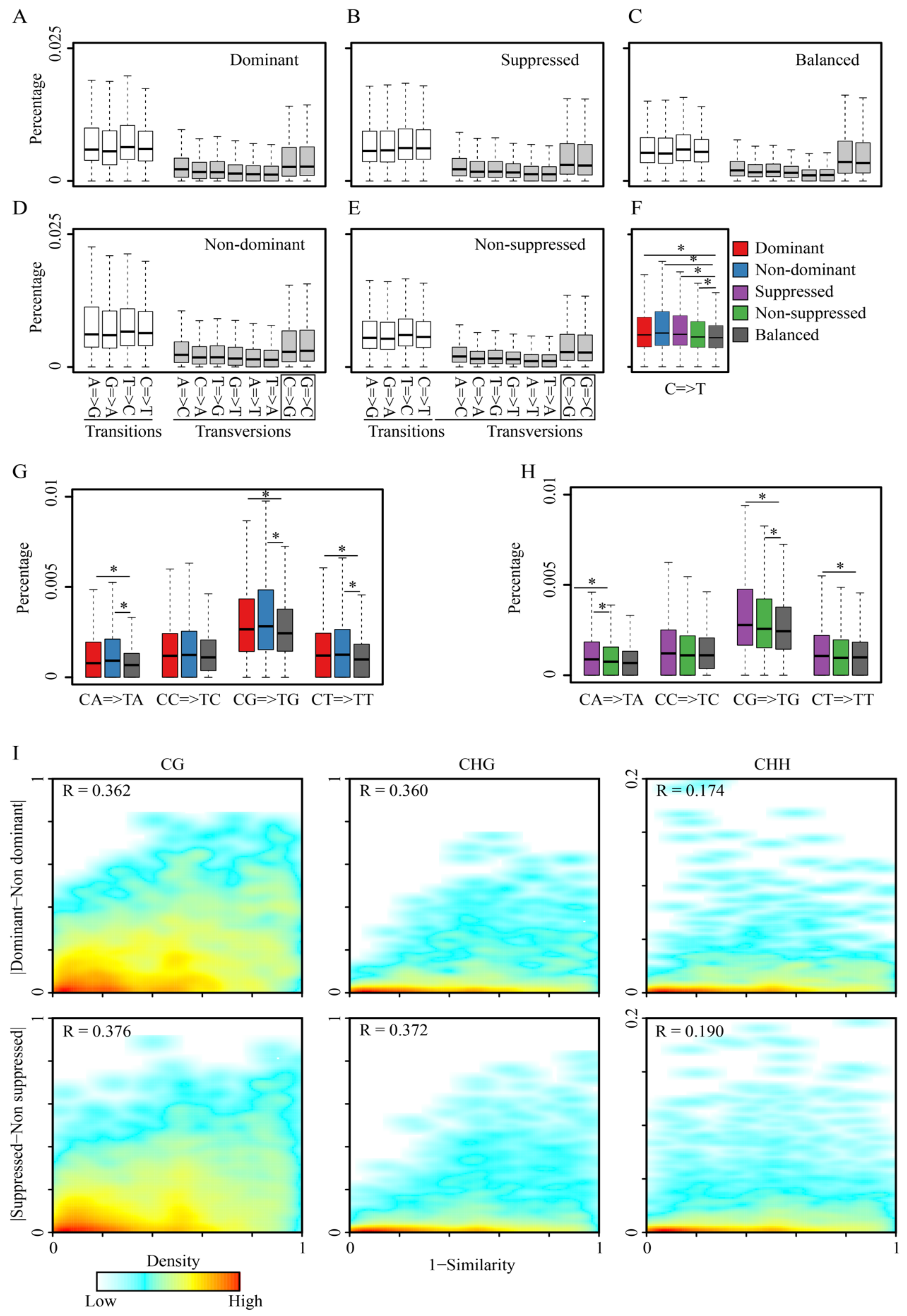
3.4. Variation of CG to TG Substitution Rates among Tetrad Alleles
3.5. DNA Methylation Contributes to Sugarcane Agronomic Traits
4. Discussions and Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Meena, M.R.; Appunu, C.; Arun Kumar, R.; Manimekalai, R.; Vasantha, S.; Krishnappa, G.; Kumar, R.; Pandey, S.K.; Hemaprabha, G. Recent Advances in Sugarcane Genomics, Physiology, and Phenomics for Superior Agronomic Traits. Front. Genet. 2022, 13, 854936. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, X.; Tang, H.; Zhang, Q.; Hua, X.; Ma, X.; Zhu, F.; Jones, T.; Zhu, X.; Bowers, J.; et al. Allele-defined genome of the autopolyploid sugarcane Saccharum spontaneum L. Nat. Genet. 2018, 50, 1565–1573. [Google Scholar] [CrossRef] [PubMed]
- Garsmeur, O.; Droc, G.; Antonise, R.; Grimwood, J.; Potier, B.; Aitken, K.; Jenkins, J.; Martin, G.; Charron, C.; Hervouet, C.; et al. A mosaic monoploid reference sequence for the highly complex genome of sugarcane. Nat. Commun. 2018, 9, 2638. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Chen, F.; Zhang, X.; Li, Z.; Zhao, Y.; Lohaus, R.; Chang, X.; Dong, W.; Ho, S.Y.W.; Liu, X.; et al. The water lily genome and the early evolution of flowering plants. Nature 2020, 577, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Edger, P.P.; Poorten, T.J.; VanBuren, R.; Hardigan, M.A.; Colle, M.; McKain, M.R.; Smith, R.D.; Teresi, S.J.; Nelson, A.D.L.; Wai, C.M.; et al. Origin and evolution of the octoploid strawberry genome. Nat. Genet. 2019, 51, 541–547. [Google Scholar] [CrossRef]
- Zhuang, W.; Chen, H.; Yang, M.; Wang, J.; Pandey, M.K.; Zhang, C.; Chang, W.C.; Zhang, L.; Zhang, X.; Tang, R.; et al. The genome of cultivated peanut provides insight into legume karyotypes, polyploid evolution and crop domestication. Nat. Genet. 2019, 51, 865–876. [Google Scholar] [CrossRef]
- Otto, S.P. The evolutionary consequences of polyploidy. Cell 2007, 131, 452–462. [Google Scholar] [CrossRef]
- One Thousand Plant Transcriptomes, I. One thousand plant transcriptomes and the phylogenomics of green plants. Nature 2019, 574, 679–685. [Google Scholar] [CrossRef]
- Ren, R.; Wang, H.; Guo, C.; Zhang, N.; Zeng, L.; Chen, Y.; Ma, H.; Qi, J. Widespread Whole Genome Duplications Contribute to Genome Complexity and Species Diversity in Angiosperms. Mol. Plant 2018, 11, 414–428. [Google Scholar] [CrossRef]
- Wang, H.; Guo, C.; Ma, H.; Qi, J. Reply to Zwaenepoel et al.: Meeting the Challenges of Detecting Polyploidy Events from Transcriptomic Data. Mol. Plant 2019, 12, 137–140. [Google Scholar] [CrossRef]
- Yoo, M.J.; Szadkowski, E.; Wendel, J.F. Homoeolog expression bias and expression level dominance in allopolyploid cotton. Heredity 2013, 110, 171–180. [Google Scholar] [CrossRef]
- Pfeifer, M.; Kugler, K.G.; Sandve, S.R.; Zhan, B.; Rudi, H.; Hvidsten, T.R.; International Wheat Genome Sequencing, C.; Mayer, K.F.; Olsen, O.A. Genome interplay in the grain transcriptome of hexaploid bread wheat. Science 2014, 345, 1250091. [Google Scholar] [CrossRef]
- Song, Q.; Guan, X.; Chen, Z.J. Dynamic Roles for Small RNAs and DNA Methylation during Ovule and Fiber Development in Allotetraploid Cotton. PLoS Genet. 2015, 11, e1005724. [Google Scholar] [CrossRef]
- Song, Q.; Zhang, T.; Stelly, D.M.; Chen, Z.J. Epigenomic and functional analyses reveal roles of epialleles in the loss of photoperiod sensitivity during domestication of allotetraploid cottons. Genome Biol. 2017, 18, 99. [Google Scholar] [CrossRef]
- Cheng, F.; Sun, C.; Wu, J.; Schnable, J.; Woodhouse, M.R.; Liang, J.; Cai, C.; Freeling, M.; Wang, X. Epigenetic regulation of subgenome dominance following whole genome triplication in Brassica rapa. New Phytol. 2016, 211, 288–299. [Google Scholar] [CrossRef]
- Li, Z.; Wang, M.; Lin, K.; Xie, Y.; Guo, J.; Ye, L.; Zhuang, Y.; Teng, W.; Ran, X.; Tong, Y.; et al. The bread wheat epigenomic map reveals distinct chromatin architectural and evolutionary features of functional genetic elements. Genome Biol. 2019, 20, 139. [Google Scholar] [CrossRef]
- Yuan, Y.; Yang, X.; Feng, M.; Ding, H.; Khan, M.T.; Zhang, J.; Zhang, M. Genome-wide analysis of R2R3-MYB transcription factors family in the autopolyploid Saccharum spontaneum: An exploration of dominance expression and stress response. BMC Genom. 2021, 22, 622. [Google Scholar] [CrossRef]
- Wang, L.; Xie, J.; Hu, J.; Lan, B.; You, C.; Li, F.; Wang, Z.; Wang, H. Comparative epigenomics reveals evolution of duplicated genes in potato and tomato. Plant J. 2018, 93, 460–471. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Xi, Y.; Li, W. BSMAP: Whole genome bisulfite sequence MAPping program. BMC Bioinform. 2009, 10, 232. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Ramirez-Gonzalez, R.H.; Borrill, P.; Lang, D.; Harrington, S.A.; Brinton, J.; Venturini, L.; Davey, M.; Jacobs, J.; van Ex, F.; Pasha, A.; et al. The transcriptional landscape of polyploid wheat. Science 2018, 361, eaar6089. [Google Scholar] [CrossRef]
- Zhang, T.; Hu, Y.; Jiang, W.; Fang, L.; Guan, X.; Chen, J.; Zhang, J.; Saski, C.A.; Scheffler, B.E.; Stelly, D.M.; et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 2015, 33, 531–537. [Google Scholar] [CrossRef]
- Chalhoub, B.; Denoeud, F.; Liu, S.; Parkin, I.A.; Tang, H.; Wang, X.; Chiquet, J.; Belcram, H.; Tong, C.; Samans, B.; et al. Plant genetics. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 2014, 345, 950–953. [Google Scholar] [CrossRef]
- Yang, J.; Liu, D.; Wang, X.; Ji, C.; Cheng, F.; Liu, B.; Hu, Z.; Chen, S.; Pental, D.; Ju, Y.; et al. The genome sequence of allopolyploid Brassica juncea and analysis of differential homoeolog gene expression influencing selection. Nat. Genet. 2016, 48, 1225–1232. [Google Scholar] [CrossRef]
- International Wheat Genome Sequencing, C. A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 2014, 345, 1251788. [Google Scholar] [CrossRef]
- Wang, L.; Shi, Y.; Chang, X.; Jing, S.; Zhang, Q.; You, C.; Yuan, H.; Wang, H. DNA methylome analysis provides evidence that the expansion of the tea genome is linked to TE bursts. Plant Biotechnol. J. 2019, 17, 826–835. [Google Scholar] [CrossRef] [PubMed]
- Niederhuth, C.E.; Bewick, A.J.; Ji, L.; Alabady, M.S.; Kim, K.D.; Li, Q.; Rohr, N.A.; Rambani, A.; Burke, J.M.; Udall, J.A.; et al. Widespread natural variation of DNA methylation within angiosperms. Genome Biol. 2016, 17, 194. [Google Scholar] [CrossRef] [PubMed]
- Crisp, P.A.; Marand, A.P.; Noshay, J.M.; Zhou, P.; Lu, Z.; Schmitz, R.J.; Springer, N.M. Stable unmethylated DNA demarcates expressed genes and their cis-regulatory space in plant genomes. Proc. Natl. Acad. Sci. USA 2020, 117, 23991–24000. [Google Scholar] [CrossRef] [PubMed]
- Espinas, N.A.; Tu, L.N.; Furci, L.; Shimajiri, Y.; Harukawa, Y.; Miura, S.; Takuno, S.; Saze, H. Transcriptional regulation of genes bearing intronic heterochromatin in the rice genome. PLoS Genet. 2020, 16, e1008637. [Google Scholar] [CrossRef]
- Le, T.N.; Miyazaki, Y.; Takuno, S.; Saze, H. Epigenetic regulation of intragenic transposable elements impacts gene transcription in Arabidopsis thaliana. Nucleic Acids Res. 2015, 43, 3911–3921. [Google Scholar] [CrossRef]
- Ossowski, S.; Schneeberger, K.; Lucas-Lledo, J.I.; Warthmann, N.; Clark, R.M.; Shaw, R.G.; Weigel, D.; Lynch, M. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science 2010, 327, 92–94. [Google Scholar] [CrossRef]
- Kiefer, C.; Willing, E.M.; Jiao, W.B.; Sun, H.; Piednoel, M.; Humann, U.; Hartwig, B.; Koch, M.A.; Schneeberger, K. Interspecies association mapping links reduced CG to TG substitution rates to the loss of gene-body methylation. Nat. Plants 2019, 5, 846–855. [Google Scholar] [CrossRef]
- Ong-Abdullah, M.; Ordway, J.M.; Jiang, N.; Ooi, S.E.; Kok, S.Y.; Sarpan, N.; Azimi, N.; Hashim, A.T.; Ishak, Z.; Rosli, S.K.; et al. Loss of Karma transposon methylation underlies the mantled somaclonal variant of oil palm. Nature 2015, 525, 533–537. [Google Scholar] [CrossRef]
- Wang, H.; Beyene, G.; Zhai, J.; Feng, S.; Fahlgren, N.; Taylor, N.J.; Bart, R.; Carrington, J.C.; Jacobsen, S.E.; Ausin, I. CG gene body DNA methylation changes and evolution of duplicated genes in cassava. Proc. Natl. Acad. Sci. USA 2015, 112, 13729–13734. [Google Scholar] [CrossRef]
- Dowen, R.H.; Pelizzola, M.; Schmitz, R.J.; Lister, R.; Dowen, J.M.; Nery, J.R.; Dixon, J.E.; Ecker, J.R. Widespread dynamic DNA methylation in response to biotic stress. Proc. Natl. Acad. Sci. USA 2012, 109, E2183–E2191. [Google Scholar] [CrossRef]
- Quadrana, L.; Almeida, J.; Asis, R.; Duffy, T.; Dominguez, P.G.; Bermudez, L.; Conti, G.; Correa da Silva, J.V.; Peralta, I.E.; Colot, V.; et al. Natural occurring epialleles determine vitamin E accumulation in tomato fruits. Nat. Commun. 2014, 5, 3027. [Google Scholar] [CrossRef]
- Secco, D.; Wang, C.; Shou, H.; Schultz, M.D.; Chiarenza, S.; Nussaume, L.; Ecker, J.R.; Whelan, J.; Lister, R. Stress induced gene expression drives transient DNA methylation changes at adjacent repetitive elements. Elife 2015, 4, e09343. [Google Scholar] [CrossRef]
- Klopfenstein, D.V.; Zhang, L.; Pedersen, B.S.; Ramírez, F.; Warwick Vesztrocy, A.; Naldi, A.; Mungall, C.J.; Yunes, J.M.; Botvinnik, O.; Weigel, M.; et al. GOATOOLS: A Python library for Gene Ontology analyses. Sci. Rep. 2018, 8, 10872. [Google Scholar] [CrossRef]
- Wu, J.; Lin, L.; Xu, M.; Chen, P.; Liu, D.; Sun, Q.; Ran, L.; Wang, Y. Homoeolog expression bias and expression level dominance in resynthesized allopolyploid Brassica napus. BMC Genom. 2018, 19, 586. [Google Scholar] [CrossRef]
- Chodavarapu, R.K.; Feng, S.; Ding, B.; Simon, S.A.; Lopez, D.; Jia, Y.; Wang, G.L.; Meyers, B.C.; Jacobsen, S.E.; Pellegrini, M. Transcriptome and methylome interactions in rice hybrids. Proc. Natl. Acad. Sci. USA 2012, 109, 12040–12045. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.; Chen, Z.J. Epigenetic and developmental regulation in plant polyploids. Curr. Opin. Plant Biol. 2015, 24, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Jiao, W.; Yuan, J.; Jiang, S.; Liu, Y.; Wang, L.; Liu, M.; Zheng, D.; Ye, W.; Wang, X.; Chen, Z.J. Asymmetrical changes of gene expression, small RNAs and chromatin in two resynthesized wheat allotetraploids. Plant J. 2018, 93, 828–842. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Liu, D.; Wu, J.; Zhao, X.; Hao, M.; Geng, S.; Yan, J.; Jiang, X.; Zhang, L.; Wu, J.; et al. mRNA and Small RNA Transcriptomes Reveal Insights into Dynamic Homoeolog Regulation of Allopolyploid Heterosis in Nascent Hexaploid Wheat. Plant Cell 2014, 26, 1878–1900. [Google Scholar] [CrossRef]
- Hu, W.; Ji, C.; Shi, H.; Liang, Z.; Ding, Z.; Ye, J.; Ou, W.; Zhou, G.; Tie, W.; Yan, Y.; et al. Allele-defined Genome Reveals Biallelic Differentiation during Cassava Evolution. Mol. Plant 2021, 14, 851–854. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xue, Y.; Shi, Y.; Qi, Y.; Yu, H.; Zou, C.; Cao, X.; Sun, C.; Chen, B.; Wang, H. Epigenetic and Genetic Contribution for Expression Bias of Homologous Alleles in Polyploid Sugarcane. Agronomy 2022, 12, 2852. https://doi.org/10.3390/agronomy12112852
Xue Y, Shi Y, Qi Y, Yu H, Zou C, Cao X, Sun C, Chen B, Wang H. Epigenetic and Genetic Contribution for Expression Bias of Homologous Alleles in Polyploid Sugarcane. Agronomy. 2022; 12(11):2852. https://doi.org/10.3390/agronomy12112852
Chicago/Turabian StyleXue, Yajie, Yan Shi, Yuchuan Qi, Hang Yu, Chengwu Zou, Xueying Cao, Cuimin Sun, Baoshan Chen, and Haifeng Wang. 2022. "Epigenetic and Genetic Contribution for Expression Bias of Homologous Alleles in Polyploid Sugarcane" Agronomy 12, no. 11: 2852. https://doi.org/10.3390/agronomy12112852
APA StyleXue, Y., Shi, Y., Qi, Y., Yu, H., Zou, C., Cao, X., Sun, C., Chen, B., & Wang, H. (2022). Epigenetic and Genetic Contribution for Expression Bias of Homologous Alleles in Polyploid Sugarcane. Agronomy, 12(11), 2852. https://doi.org/10.3390/agronomy12112852






