Abstract
It is crucial to assess genetically superior parents when developing novel hybrids. This experiment was conducted to find out the diversity of 27 Capsicum annuum mutant lines derived from two varieties.To achieve the objective, 23 morpho-physiological and yield traits were recorded through two planting seasons. Highly significant differences (p < 0.01) were recorded among the studied traits. There was a strong to moderately positive phenotypic association between yield and all other morphological traits except first bifurcation length, stem diameter, pedicle length, flowering date, and maturity date. A higher Genotypic Coefficient of Variation (GCV) and Phenotypic Coefficient of Variation (PCV), combined with moderate to high heritability and high hereditary progress, have been found in the number of fruits per plant, fruit yield per plant, and number of seeds per fruit. High heritability was found in yield characteristics, vis-à-visnumber of seeds per fruit, number of fruits per plant, and indicated high genetic advance. The studied genotypes were divided into six groups after the cluster analysis. Based on the correlation matrix of 23 quantitative characteristics, principal component analysis revealed that the percentage of variation for PC1 and PC2 is 28%and 19%, respectively, andPC1 represents the largest percentage of the overall total variation. The calculated genetic distance also explains the potential of heterosis breeding. The revealed findings might be helpful for breeders to target quantitative characters and the parental lines of C. annuum during the execution of their future breeding programmes for developing high-yielding and climate-resilient chilli varieties.
1. Introduction
Chilli (Capsicum annuum L.) of the genus Capsicum includes about 25 regularly used species with four cultivar groups: Chinense (West Indies chilli), Frutescens (bird chilli), Annuum (hot chilli), and the sweet pepper group [1]. It is said to have originated in South and Central America and belongs to the Solanaceae family. It is a spice crop that can also be used as a vegetable and is widely produced across the world [2,3]. The nutritional and functional benefits of chilli pepper fruits, such as capsaicinoids, carotenoids, antioxidant vitamins, and phenolic components, make them an important vegetable. In addition, hot pepper fruits are utilized as food coloring and flavoring [4]. Capsicum annuum genotypes have a variety of different and intriguing growth and yield characteristics, including fruit size, weight of fruit, fruit colour, pungency, flowering, plant height, and maturity, which may be somehow beneficial for breeding purposes. Many experiments have shown that critical yield traits in C. annuum genotypes have high genetic changeability, heritability, and hereditary advancement [5,6,7,8].
It was reported that there is a wide range of variance in the capacity of chilli genotypes for flowering, setting fruit, yield, and other qualitative characteristics [9,10]. The systematic breeding process includes numerous phases, such as collecting germplasm, analysing genetic diversity, producing genetic variability, implementing selection, and preparing chosen genotypes for commercial distribution [11]. Investigation and improved knowledge of the variability available in a crop population is necessary for efficient and successful breeding research, so that plant breeders may use it for crop development. Furthermore, the effectiveness of any crop development effort is determined not just by the quantity of genetic diversity contained in a crop, but also by the degree of variation heritable from parent to progeny [12]. A large range of diversity in genotypes gives enough opportunity for boosting fruit production and other desired features through systematic breeding. Estimating the genetic diversity inherent in a crop’s germplasm is a prerequisite for having a successful breeding programme [13]. The long-term sustainability of plant populations and their capacity to adapt to changing climatic and environmental circumstances may both be significantly influenced by genetic variety, which is another reason why it is crucial to ensure this. Development of new varietiesis a continuous process to mitigate various demands of growers, so the information aboutparental lines can help the breeders to use the germplasm more confidently.
Several chilli researchers concluded that PCV was higher than GCV for the different characteristics tested [14,15,16]. Fruit yield per plant, seed yield per plant, fruit length, green chilli fruit weight, and fruit production per hectare each had extremely high heritability [17]. Knowledge of genetic distance between parents is vital forbenefiting from transgressive segregation [18,19]. The breeder must have information on the degree of genetic divergence in order to select the proper type of parent for targeted hybridization in heterosis breeding [19,20]. Furthermore, choosing varied parents within a suitable range increases the odds of improving various features in the progeny. One of the fundamental prerequisites for establishing efficient breeding procedures is a careful assessment of the type and extent of variability in the germplasm resource [21]. The genetic variability, correlations, and associations between qualitative and quantitative features, as well as heritability estimates, all play a role in determining the best breeding approach for improving yield and its components in each crop. High heritability together with high hereditary advancement indicated the role of the additive gene for selected characters [22]. Information on genetic distance between parents is also important in order to benefit transgressive segregation [18]. Hence, clustering analysis and genetic distance determination are also essential [23]. The breeder can select the best type of parents for deliberate hybridization in heterosis breeding by having complete knowledge of the nature and degree of genetic difference [24]. The following objectives were pursued in this experiment: (a) to calculate the genetic diversity of 27 chilli genotypes based on their morphological, physiological, and yield traits; and (b) to estimate the genetic variance components, heritability, and genetic advance as a selection criterion for further chilli breeding initiatives.
2. Materials and Methods
2.1. Experiment Location
The experiment was conducted under a rain shelter in the nethouse at the Institute of Tropical Agriculture, Faculty of Agriculture, Universiti Putra Malaysia (UPM), located at 03°00″12.6′N, 101°47″22.4′E. Over the period of two seasons, the evaluation was repeated in a humid tropical climate. The first season was conducted in 2019 and the second was in 2020. The average daily temperature ranged from 19 to 36 °C and relative humidity was recorded between 80–90% during the experimental tenure.
2.2. Plant Materials
A total of 27 selected advanced chilli mutant lines in the M4 generation were used in this study, which were derived from two varieties, viz. Chilli Bangi 3 and Chilli Bangi 5 (Table 1). The advanced mutants were selected based on their excellent agronomic performance.

Table 1.
Selected 27 Capsicum annuum genotypes from mutant lines with gamma source.
2.3. Experimental Design and Layout
The seeds were first sown in seed trays containing peat moss with one or two seeds per cell and were later transplanted after four weeks to prepared polythene pots (17 × 30 cm) filled with cocoa dust, having small holes to drain excess water. The experiment was laid in a randomized complete block design with three replications. Two pots were assigned for each genotype in each replication (54 pots per replication) and were oriented east to west (spaced 75 cm × 150 cm). Seedlings emerged within 3–10 days after sowing and were transplanted 4 weeks after sowing. Fertigation system of cropping was adopted for both irrigation and fertilization. Drip system of irrigation was applied. Throughout the cropping season, intercultural activities including supplemental irrigations and plant protection approaches were carried out as required. Agronomic recommendations were followed.
2.4. Data Collection
2.4.1. Measuring the Morphological, Physiological, and Yield Components
Table 2 contains data on morphological, physiological, and yield parameters that were assessed and reported 90 days after transplantation. The quantitative morphological features obtained in this study include plant height, stem diameter, fruit number, and fruit weight, which could be counted or quantified using particular measuring instruments.

Table 2.
The quantitative characteristics of chosen chilli genotypes with detail description.
2.4.2. Genetic Variance, Heritability, and Advance
An analysis of variance was performed to detect genotype differences and to assess genetic and environmental impacts on several attributes.
- (a)
- Calculation of genotypic variance using following formulae:
- (b)
- Calculation of phenotypic variance using following formula:σ2p = σ2g + MSE
where σ2g is the genotypic variance, σ2p is the phenotypic variance, MSG is the meansquare of genotypes, MSE is mean square of error, and r is number of replications.
- (c)
- Phenotypic and Genotypic Coefficient of Variation (PCV and GCV). Estimates of phenotypic and genotypic coefficient of variation were calculated according to Singh and Choudhary [25] as follows:
σ2p is the phenotypic variance, σ2g is the genotypic variance, and is the mean of the trait. GCV and PCV values were categorized as low (0–10%), moderate (10–20%), and high (20% and above) following Sivasubramanian and Madhavamenon [26].
- (d)
- Broad sense heritability h2B ratio of genetic variance (σ2g) to phenotypic variance (σ2g).
The formula used for broad sense heritability is as follows:
where σ2g is the genotypic variance and σ2p is the phenotypic variance. The heritability percentage was categorized as low (0–30%), moderate (30–60%), and high (≥60%) in accordance with Johnson et al. [27].
- (e)
- Estimated and Expected Genetic Advance. Expected genetic advance (GA) (as percentage of the mean) was calculated using the method of Assefa et al. [28] and selection intensity (K) was assumed to be 5%. Genetic advance was marked as low (0–10%), moderate (10–20%), and high (>20%) by following Johnson et al. [29].
K is a constant which represents the selection intensity. When K is 5%, the value is 2.06. σ2p is phenotypic standard deviation, h2B is the heritability, and is the mean of traits.
2.5. Data Analysis
The 27 accessions were characterised morphologically and agronomically using a randomised complete block design, with four replicates consisting of two pot plants from each accession as the source of variance. One-way ANOVA was used to examine all of the data sets using SAS 9.4 statistical analysis software (North Carolina State University, Raleigh, NC, USA). The significance level was set at >0.05, and the LSD test was used to see if there were any significant differences between the means. A correlation coefficient was also determined. The dendrogram was mapped using SAHN clustering of the UPGMA method through the application of NTSYS 2.1 (Numerical Taxonomy Multivariate Analysis System, Exeter Software, Setauket, NY, USA) software. In addition, principal component analysis (PCA) was employed to generate 2Dvisualisations.
3. Results and Discussion
3.1. Morpho-Physiological and Yield Component
Among the tested genotypes, the results showed that there are highly significant differences (p < 0.01) for all the parameters measured (Table 3).

Table 3.
Analysis of variance (mean squares) of all studied characteristics of 27 chilli genotypes over two seasons.
3.1.1. Growth and Physiological Components
The highest germination percentage was observed for Genotype 5 (94.2%) and the lowest was recorded for Genotype 11 (75.3%). Genotype 9 (84.70 cm) was recorded as the tallest plant while Genotype 24 (66.96 cm) was the shortest one in respect of taken plant height (Table 3). In the case of stem diameter, Genotype 10 and Genotype 26 werethe lowest and the highest, respectively, where the value ranged between 10.58 and 17.84 cm. The highest leaf number was found in Genotype 27 (743.67), whereas for days to first flowering and maturity the Genotype 15 (18.6) andGenotype 27 (64.5 days) was the earliest, respectively.
Genotype 2 (39.63 mm) was found with the longest pedicle; on the contrary, Genotype 5 was the smallest (22.10 mm). Among studied physiological traits, it was found that in the case of relative chlorophyll content Genotype 15 and Genotype 13 werethe highest and lowest, respectively, as shown in Table 4. Within all the observed chilli genotypes, Genotype 16 was found with the highest photosynthesis rate (22.10 µmol CO2 m−2s−1) followed by Genotype 22 and Genotype 7; however, the lowest photosynthesis rate (15.65 µmol CO2 m−2s−1) was found for the Genotype 4. On the other hand, Genotype 15 (0.76 mol H2O m−2s−1) was recorded for the highest stomata conductance, followed by Genotype 13 and Genotype 6, respectively, but the lowest value for this trait was recorded for the Genotype 20 (0.38 mol H2O m−2s−1). For transpiration rate, the highest value was indicated by the Genotype 27 (6.86 mmol H2O m−2s−1) and lowest was found for the Genotype 1 (4.72 mmolH2O m−2s−1).

Table 4.
Mean for morphological, physiological, and yield characteristics of the 27 studied chilli genotypes planted over two seasons.
The differences at the gene level might be the cause behind the variation of the studied chilli genotypes. Ridzuan et al. [30] and Usman et al. [31] also concluded with similar findings after havingconducted experiments with different chilli genotypes. Overtwo different growing seasons most of the values were not significantly different except first bifurcation length, stem diameter, first flowering date, and maturity date (Table 4). Plant growth and development are dependenton physiological processes (e.g., photosynthesis) which in turn follow various factors in the environment in order to proceed optimally [32]. Chlorophyll content, photosynthesis rate, stomata conductance, and transpiration rate were the physiological characters measured in other experiments. Higher chlorophyll content values signify the more prominent dependability of a plant’s chloroplast membranes prompting higher rates of photosynthesis, more dry matter accumulation, and higher productivity [31].
3.1.2. Yield and Yield Contributing Traits
The Genotype 17 produced the highest fruit number per plant with the number 157.2, which was followed by Genotype 9 (154.3) and Genotype 24 (146.7), respectively (Table 4). The lowest fruit number per plant was produced by Genotype 27 (71.0). For fruit length and breadth, Genotype 15 (102.8 mm) and Genotype 4 (22.7 mm) werefound the longest and thickest, respectively. On the other hand, Genotype 18 was recorded as the lowest for both of thesetraits, having the value 70.4mm for length and 12.4mm for breadth. The highest (14.96 g) average green fruit weight was found for Genotype 13, while Genotype 4 was the lowest (9.67 g). In respect of dry fruit weight, Genotype 22 and Genotype 8 werethe highest (1.33 g) and the lowest (0.89 g), respectively. Genotype 5 produced the highest (84.50) seed number in a single fruit, whileGenotype 18 produced the lowest number of seed with the value of 39.50 in a fruit. Genotype 3 and Genotype 13 wereobservedas the highest for hundred seed weight and fruit wall thickness, respectively. On the contrary, Genotype 18 and Genotype 15 were the lowest for these traits, respectively. The highestyield per plant was recorded for the Genotype 2 with the value 1.74 kg. This was followed by Genotype 9 and Genotype 5 with the values 1.63 and 1.62 kg, respectively. However, Genotype 25 was the lowest yielder (0.89 kg) among all the studied genotypes.
No significant difference was found for yield and yield contributing traits whenboth of the seasons were considered (Table 3). A slightly higher yield was found in the second season, which might havehappened due to environmental effect. Ridzuan et al. [33] found similar results when an experiment was done to find out the variability among different chilli genotypes.
3.2. Correlation between Different Traits
Pearson correlation coefficients were calculated to determine the relationships among the traits, with a significant level at p ≤ 0.05 and high significant level at p ≤ 0.01 (Table 5). Yield showed significantly positive correlation with the physiological traits. However, transpiration rate and relative chlorophyll content had low contribution, whereas photosynthesis rate and stomatal conductance had moderate contribution to yield. Among the morphological and growth traits, first bifurcation length, stem diameter, pedicle length flowering and maturity date showed negative correlation with yield. However, apositive correlation was seen in the cases of germination percentage, stem diameter, number of leaves per plant, and primary and secondary branches. Germination percentage, first bifurcation length and days to maturity moderately contributed to total yield per plant, which was also highly significant statistically. Moreover, in respect of yield related traits, fruit number, fruit breadth, number of seed per fruit, and hundred seed weight showedmoderately positive correlation with yield, which was also highly significant. The majority of characteristics do notexist in isolation; rather, they are linked to one another in intricate ways that have an influence on the yield. This relationship could be favourable or unfavourable. Raihana et al. also corroborate this finding [30].

Table 5.
Combine analysis for correlation coefficient among 23 traits.
Considering the plant height, it was observed that plant diameter, leaf number, and other physiological traits were negatively correlated. However, the flowering and maturity date, fruit length, and breadth showed low positive correlation with plant height. On the other hand, days to flowering was highly correlated with maturity date, relative chlorophyll content, photo synthesis rate, stomata conductance, and transpiration rate, and the magnitude of correlation was positive and statistically highly significant. Moreover, in case of days to maturity, highly positive and statistically significant correlation was also found with the studied physiological traits. Fruit number per plant was positively correlated with all other studied traits excluding first bifurcation length, pedicle length, flowering, and maturity date. Moreover, number of leaves showed moderate to high correlation with the physiological traits, which was statistically highly significant also.
3.3. Genetic Analysis, Broad-Sense Heritability, and Genetic Advance
The variance components, environmental variance, genotypic variance (GV) and phenotypic variance (PV), genotypic coefficient of variation (GCV) and phenotypic coefficient of variation (PCV), heritability (h2B), and genetic advance (GA) havebeen presented in Table 6. For all the studied traits, genotypic variance was higher than the phenotypic variance. The phenotypic coefficient of variation (PCV) was greater than the genotypic coefficient of variation (GCV) for all traits, showing that environment had minimal impact on trait expression and that a significant amount of variance was controlled by genotypic composition.

Table 6.
Genetic variance, broad-sense heritability, and genetic advance for 23 traits in 27 Capsicum annuum genotypes from the combined analysis.
High PCVwas recorded for primary branches, number of seeds per fruit, and stomata conductance. Moreover, most of the remaining traits were found with moderate PCV, excluding germination percentage, plant height, maturity date, and hundred seed weight, which were estimated as having a lower PCV value. The GCV value ranged from 6.3 to 24.8, indicating high variability among the traits. The highest GCV value was found for stomata conductance, followed by number of seeds per fruit and number of fruits per plant, signifying the potential to select these traits. Among the yield contributing traits, fruit length and hundred seed weight were found to have a lower GCV value. According to Falconer [34], heritability percentage is considered low when values range from 0 to 30%, moderate when values range from 30 to 60%, and high when values exceed 60%. Broad sense heritability of studied traits was high for most of the traits, except the primary and secondary branch numbers, which showed lower heritability. First bifurcation length, plant height, and hundred seed weight all had moderate heritability. All the yield contributing traits, including total yield per plant, revealed high heritability.
Among the studied 23 traits, the highest genetic advance (49%) was recorded for stomata conductance, followed by the number of seeds per fruit (40%), while the lowest value was recorded for secondary branches (7%), followed by plant height (8.4%). Raihana et al. [31] proposed that heritability estimates combined with genetic advancement are usually superior to heritability alone when it comes to selecting superior individuals. For number of fruits per plant, number of seeds per fruit, and stomata conductance, high heritability was combined with very high genetic advance as a percentage of mean, indicating that these traits were controlled by additive gene action and that standard selection procedures could be effective for the isolation of superior genotypes for these traits. These results are in accordance with results of earlier research by Chattopadhyay et al. [7], Kumar et al. [35], and Agasimani et al. [36] for fruit yield per plant, and Sreelathakumary and Rajamony [37] for number of fruits per plant. High heritability coupled with moderate genetic advance as percent of mean was observed for stem diameter, fruit breadth, fruit dry and fresh weight, transpiration rate, and total yield per plant, indicating the preponderance of additive and non-additive gene action. Further improvement of these traits would be possible through mass selection, progeny selection, and hybridization procedures intending to exploit the additive gene action that was reported by Tembhurne et al. [38] (2010) and Suryakumari et al. [39]. Low heritability was associated with low genetic advance as % of mean and which was observed for primary and secondary branches, indicating the presence of nonadditive gene action for these traits and that their improvement could be achieved through heterosis breeding.
3.4. Clustering and Principal Components Analysis
For selecting the desired parents, estimation of existing diversity among the genotypes through genetic diversity analysis plays avery important role. The summarized data on the degree and nature of genetic variability is essential for choosing the right parent for targeted crosses [22,40].
Based on different recorded traits, the studied 27 genotypes were successfully clustered into four major groups. Geleta et al. [41] also conducted an experiment with twenty-nine diversified genotypes and clustered them based on morphological character. The Euclidian distance was calculated by using the data of different traits, and the unweighted pair group method with arithmetic means (UPGMA) dendrogram was constructed using those values. The dendrogram explains that the genotypes with common trends remain in the same cluster. The genotypes were grouped into four clusters at a 0.27 dissimilarity coefficient. Groups I, II, and III consist of six, seven, and thirteen genotypes, respectively. There was only one genotype in Group IV. Group I had the highest yielding characteristics; Group II had the most fruits per plant; Group III had early flowering and high yielding characteristics; and Group IV had early maturity and larger fruit diameter. Similar observations were reported by Assefa et al. [26] while experimenting with different chilli populations. Figure 1 describes the result of PCA. Gen 15 was the genotype farthest from the centroid. Gen 23 and Gen 21 were more or less close to the centroid. Based on the combined data of the two seasons, PCA further explains cluster analysis, yielding the two-dimensional graphical illustration (Figure 2), showing that most of the genotypes were dispersed at close distances at PC1, while few were dispersed at great distances as revealed by the Eigenvector. The variation percentages of PC1 and PC2 are 28% and 19%, respectively, with PC1 showing the highest of the total variation.
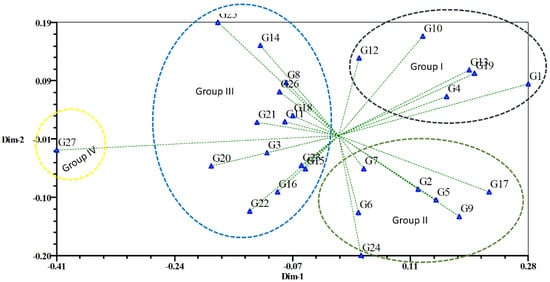
Figure 1.
Principal component analysis showing the relationship among 27 chilli genotypes in a two-dimensional graph.
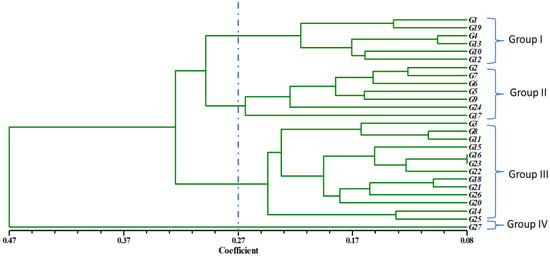
Figure 2.
Relationship among the 27chilli genotypes based on 23 traits using SAHN clustering of UPGMA method.
4. Conclusions
Variability among the base generation of parental lines creates more scope for selecting the targeted genotypes to develop the recombinant type and for heterosis breeding. In the present study, information was gathered about 27 chilli genotypes regarding 23 morphological and yield-related traits. Obviously, this information will pave the way forusing the better ones in various breeding programmes for the improvement of this crop. Hence, estimation of correlation, GCV, PCV, heritability in the broad sense, and genetic advance help to select the genotypes and the selection indices for their exact exploitation. The results of this experiment present an insight into the genetic diversity of the studied chilli population. Considering all the diversity patterns and analysis, the studied genotypes were allocated into four different groups. Group III had the highest (13) number of genotypes, whereas Group IV had only one. The calculated genetic distance also reveals the potential forheterosis breeding. Considering all the information and practical crop conditions, nine genotypes were selected for further hybridization. However, for developing yield and other quality traits, Gen 2, Gen 3, Gen 5, Gen 9, Gen 15, Gen 16, Gen 22, Gen 23, and Gen 27 were selected as better parents, having early flowering, high yielding, and highpungency level characteristics, to design an effective future breeding programme.
Author Contributions
K.M.R.K. and M.Y.R. conceived the research idea; K.M.R.K. carried out the research and initial draft; data analyses by O.Y. and M.A.H.; editing and approval of the final version of the manuscript by A.M., M.F.I., A.R.H., R.R., M.H. and M.F.N.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the research grant title evaluation and selection of crop varieties for utilization to increase yield and quality with grant vot number 6383900, Universiti Putra Malaysia. K. M. Rezaul Karim would like to express his appreciation to the National Agricultural Technology Program (NATP II) of the Bangladesh Agricultural Research Council (BARC) for PhD fellowship financial support.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors pay thanks to the Universiti Putra Malaysia (UPM) and the Bangladesh Sugarcrop Research Institute (BSRI), People’s Republic of Bangladesh, for their support.
Conflicts of Interest
The authors have no conflict of interest to publish this manuscript.
References
- Nsebiyera, V.; Logose, M.; Ochwo-Ssemakula, M.; Sseruwagi, P.; Gibson, P.; Ojiewo, C.O. Morphological characterization of local and exotic hot pepper (Capsicum annuum L.) collections in Uganda. Bioremediation Biodivers. Bioavailab. 2013, 7, 22–32. [Google Scholar]
- Dias, G.B.; Gomes, V.M.; Moraes, T.M.S.; Zottich, U.P.; Rabelo, G.R.; Carvalho, A.O.; Moulin, M.; Goncalves, L.S.A.; Rodrigues, R.; da Cunha, M. Characterization of Capsicum species using anatomical and molecular data. Gene. Mol. Res. 2013, 12, 6488–6501. [Google Scholar] [CrossRef]
- Wahyuni, Y.; Ballester, A.R.; Tikunov, Y.; De Vos, R.C.; Pelgrom, K.T.; Maharijaya, A.; Sudarmonowati, E.; Bino, R.J.; Bovy, A.G. Metabolomics and molecular marker analysis to explore pepper (Capsicum sp.) biodiversity. Metabolomics 2013, 9, 130–144. [Google Scholar] [CrossRef] [PubMed]
- Virga, G.; Licata, M.; Consentino, B.B.; Tuttolomondo, T.; Sabatino, L.; Leto, C.; La Bella, S. Agro-morphological characterization of sicilian chili pepper accessions for ornamental purposes. Plants 2020, 9, 1400. [Google Scholar] [CrossRef] [PubMed]
- Sreelathakumary, I.; Rajamony, L. Variability, heritability and genetic advance in chilli (Capsicum annuum L.). J. Trop. Agri. 2006, 42, 35–37. [Google Scholar]
- Singh, Y.; Sharma, M.; Sharma, A. Genetic variation, association of characters and their direct and indirect contributions for improvement in chilli peppers. Int. J. Veg. Sci. 2009, 15, 340–368. [Google Scholar] [CrossRef]
- Chattopadhyay, A.; Sharangi, A.B.; Dai, N.; Dutta, S. Diversity of genetic resources and genetic association analysis of green and dry chillies of Eastern India. Chil. J. Agric. Res. 2011, 71, 350–356. [Google Scholar] [CrossRef]
- Pandit, M.K.; Adhikary, S. Variability and heritability estimates in some reproductive characters and yield in chilli (Capsicum annuum L.). Int. J. Plant Soil Sci. 2014, 3, 845–853. [Google Scholar] [CrossRef]
- Maurya, A.K.; Kushwaha, M.L.; Jain, V.K.; Singh, N. Evaluation of chilli (Capsicum annuum L.) genotypes for yield and performance against diseases. Prog. Res. Intl. J. 2016, 11, 4615–4617. [Google Scholar]
- Rani, P.U. Fruit seed weight and seed attributes on the quality characteristics in chilli. Madras Agric. J. 1996, 83, 259–264. [Google Scholar]
- Syukur, M.; Sujiprihati, S.; Yunianti, R. Estimation of genetic parameter for quantitative characters of pepper (Capsicum annuum L.). J. Trop. Crop Sci. 2012, 1, 4–8. [Google Scholar] [CrossRef]
- Bello, O.B.; Ige, S.A.; Azeez, M.A.; Afolabi, M.S.; Abdulmaliq, S.Y.; Mahamood, J. Heritability and genetic advance for yield and its component character in chilli. Intl. J. Plant Res. 2014, 2, 138–145. [Google Scholar]
- Parkash, C. Estimation of genetic variability and implications of direct effects of different traits on leaf yield in bathua (Chenopodium album). Indian J. Agri. Sci. 2012, 82, 71–74. [Google Scholar]
- Kadwey, S.; Ashwini, D.; Sunil, P. Genotypes performance and genetic variability studies in Hot Chilli. Indian J. Agric. Res. 2016, 50, 56–60. [Google Scholar]
- Gupta, A.M.; Singh, D.; Kumar, A. Genetic variability, genetic advance and correlation in chilli. Indian J. Agric. Sci. 2009, 79, 221–223. [Google Scholar]
- Bendale, V.W.; Palsuledesai, M.R.; Bhave, S.G.; Sawant, S.S.; Desai, S.S. Genetic evaluation of some economic traits in chilli. Crop Res. 2006, 31, 401–403. [Google Scholar]
- Bharadwaj, D.N.; Singh, S.K.; Singh, H.L. Genetic variability an association of component characters for yield in chilli. Intl. J. Plant Sci. 2007, 2, 93–96. [Google Scholar]
- Lahbib, K.; Bnejdi, F.; Gazzah, E.I. Genetic diversity evaluation of pepper in Tunisia based on morphologic characters. Afr. J. Agric. Res. 2012, 7, 3413–3417. [Google Scholar]
- Khodadadi, M.; Fotokian, M.H.; Miransari, M. Genetic diversity of wheat genotypes based on cluster and principal component analyses for breeding strategies. Aust. J. Crop Sci. 2011, 5, 17–24. [Google Scholar]
- Farhad, M.; Hasanuzzaman, M.; Biswas, B.K.; Arifuzzaman, M.; Islam, M.M. Genetic divergence in chilli. Bangladesh Res. Pub. J. 2010, 3, 1045–1051. [Google Scholar]
- Krishna, U.C.; Madalageri, M.B.; Patil, M.P.; Ravindra, M.; Kotlkal, Y.K. Variability studies in green chilli (Capsicum annuum L.). Karnataka J. Agric. Sci. 2007, 20, 102–104. [Google Scholar]
- Yatung, T.; Dubey, R.K.; Singh, V.; Upadhyay, G. Genetic diversity of chilli (Capsicum annuum L.) genotypes of India based on morphochemical traits. Aust. J. Crop Sci 2014, 8, 97–102. [Google Scholar]
- Hoque, M.N.; Rahman, L. Estimation of Euclidean distance for different morpho-physiological characters in some wild and cultivated rice genotypes (Oryza sativa L.). J. Biol. Sci. 2007, 7, 86–88. [Google Scholar] [CrossRef][Green Version]
- Sen, N.; Biswas, K.; Sinha, S.N. Assessment of genetic divergence through cluster analysis of chilli varieties. J. Biosci. 2021, 46, 52. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.K.; Choudhary, B.D. Biometrical Methods in Quantitative Genetic Analysis; Kalyani Publishers: New Delhi, India, 1977. [Google Scholar]
- Sivasubramanian, S.; Madhavamenon, P. Genotypic and phenotypic variability in rice. Madras Agric. J. 1973, 60, 1093–1096. [Google Scholar]
- Johnson, H.W.; Comstock, R.E.; Harvey, P.H. Genotypic and phenotypic correlations in corn and their implications in selection. Agron. J. 1951, 43, 282–287. [Google Scholar]
- Assefa, K.; Ketema, S.; Tefera, H.; Nguyen, H.T.; Blum, A.; Ayele, M.; Bai, G.; Simane, B.; Kefyalew, T. Diversity among germplasm lines of the Ethiopian cereal tef [Eragrostistef (Zucc.) Trotter]. Euphytica 1999, 106, 87–97. [Google Scholar] [CrossRef]
- Johnson, H.W.; Robinson, H.F.; Comstock, R.E. Estimation of genetic and environmental variability in soybeans. Agron. J. 1955, 47, 314–318. [Google Scholar] [CrossRef]
- Ridzuan, R.B. Development of Anthracnose Resistant Chili Varieties through Marker-assisted Pedigree Selection. Ph.D. Thesis, Universiti Putra Malaysia, Serdang, Malaysia, 2018. [Google Scholar]
- Usman, M.G.; Rafii, M.Y.; Ismail, M.R.; Malek, M.; Latif, M.A. Capsaicin and dihydrocapsaicin determination in chili pepper genotypes using ultra-fast liquid chromatography. Molecules 2014, 19, 6474–6488. [Google Scholar] [CrossRef] [PubMed]
- George, A. Principles of Plant Genetics and Breeding; Backwell Publishing: Victoria, Australia, 2007; pp. 246–248. [Google Scholar]
- Ridzuan, R.; Rafii, M.Y.; Ismail, S.I.; Mohammad Yusoff, M.; Miah, G.; Usman, M. Breeding for anthracnose disease resistance in chili: Progress and prospects. Int. J. Mol. Sci. 2018, 19, 3122. [Google Scholar] [CrossRef] [PubMed]
- Falconer, D.S. Introduction to Quantitative Genetics. Pearson Education: New Delhi, India, 1996; ISBN 8131727408. [Google Scholar]
- Kumar, D.; Bahadur, V.S.; Rangare, D.; Singh, D. Genetic variability, heritability and correlation studies in chilli (Capsicum annuum L.). HortFlora Res. Spectrum 2012, 1, 248–252. [Google Scholar]
- Agasimani, S.; Kumar, H.D. Genetic variability, heritability and genetic advance for yield and its components in byadgikaddichilli (Capsicumannuum L.) accessions. BIOINFOLET-A Q. J. Life Sci. 2013, 10, 50–53. [Google Scholar]
- Sreelathakumary, I.; Rajamony, L. Variability, heritability and correlation studies in chilli (Capsicum spp.) under shade. Indian J. Hortic. 2002, 59, 77–83. [Google Scholar]
- Tembhurne, B.V.; Kuchanur, P.H. Varietal performance, genetic variability and correlation studies in chilli (Capsicum annuum L.). Karnataka J. Agric. Sci. 2010, 21, 541–543. [Google Scholar]
- Surya Kumari, S.; Uma Jyothi, K.; Srihari, D.; Siva Sankar, A.; Ravi Sankar, C. Variability and genetic divergence in paprika (Capsicum annuum L.). J. Spices Aromat. Crops 2011, 19, 71–75. [Google Scholar]
- Guerra, E.P.; Destro, D.; Miranda, L.A.; Montalván, R. Parent selection for intercrossing in food type soybean through multivariate genetic divergence. Acta Sci. Agron. 1999, 21, 429–437. [Google Scholar]
- Geleta, L.F.; Labuschagne, M.T.; Viljoen, C.D. Genetic variability in pepper (Capsicum annuum L.) estimated by morphological data and amplified fragment length polymorphism markers. Biodivers. Conserv. 2005, 14, 2361–2375. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).