Using Plant-Based Preparations to Protect Common Bean against Halo Blight Disease: The Potential of Nettle to Trigger the Immune System
Abstract
:1. Introduction
2. Materials and Methods
2.1. PBP Sampling and Processing
2.2. Bioassays of PBPs against Pseudomonas syringae pv. phaseolicola
2.3. Experimental Design and PBP Application
2.4. H2O2 Content Measurement and Activity Assays of Antioxidant Enzymes
2.5. Gene Expression Analysis
2.6. Yield Assessment
2.7. Infection Assays
2.8. Statistical Analysis and Data Representation
3. Results
3.1. Screening of PBPs against Halo Blight Disease
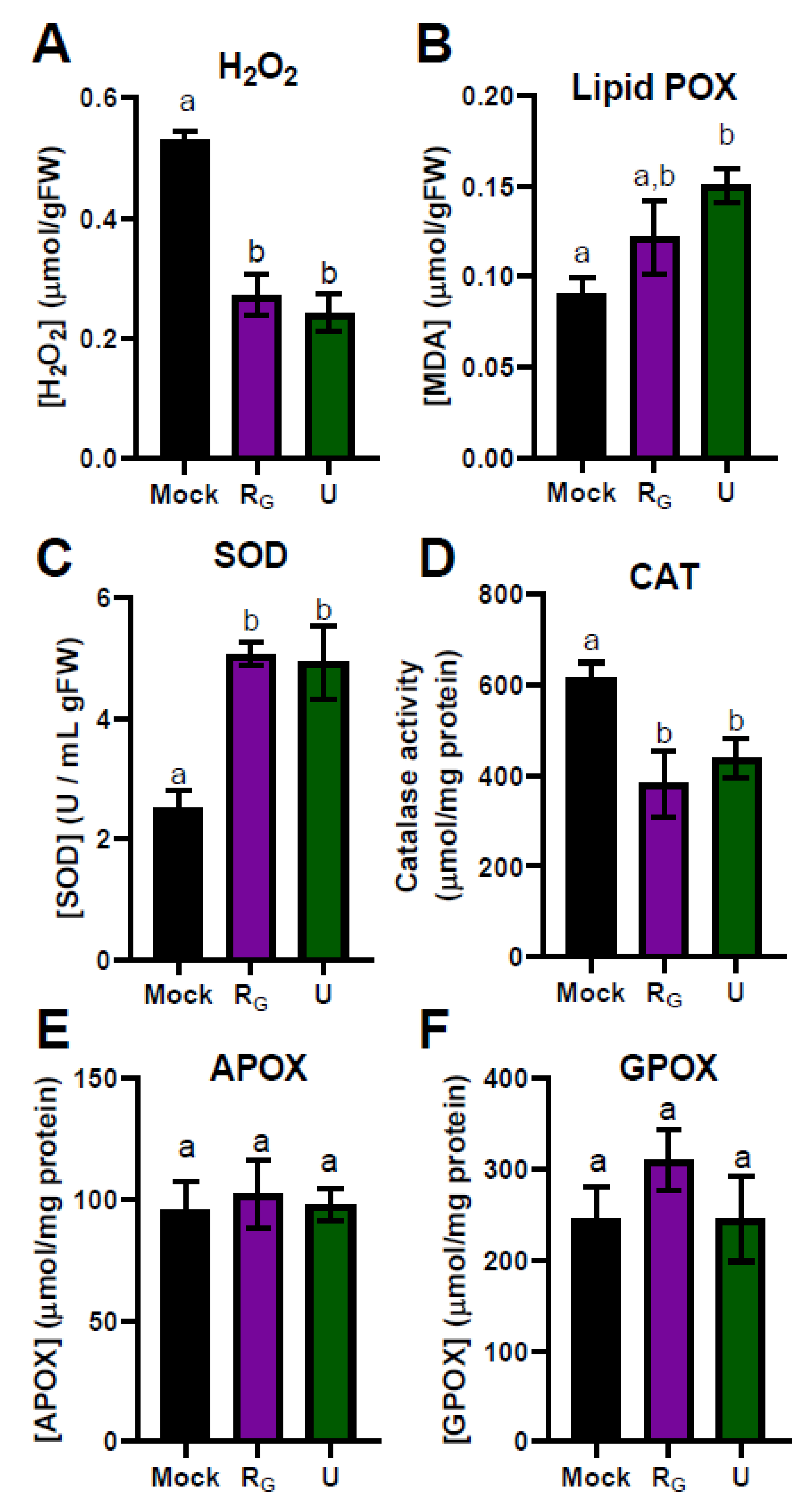
3.2. Effect of the Application of Nettle and Grapevine Pomace PBPs on the Physiological Redox State of Common Bean
3.3. U-PBP Increased Expression of Some Defense-Related Genes
3.4. Yield Did Not Vary after Treatments
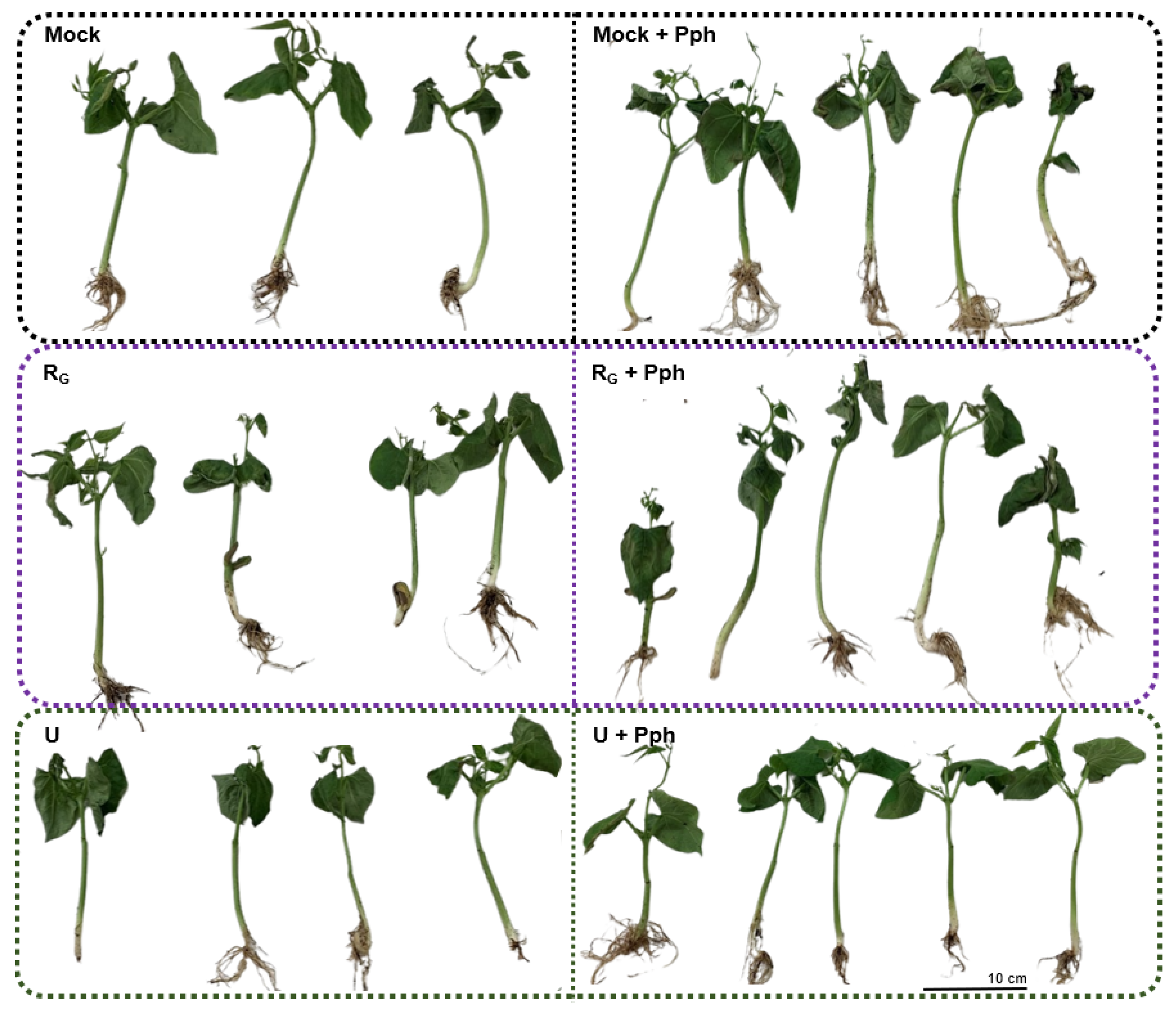
3.5. Common Bean Showed Less Symptoms of Halo Blight Disease after Application of U-PBP
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- González, A.M.; Godoy, L.; Santalla, M. Dissection of Resistance Genes to Pseudomonas syringae pv. phaseolicola in UI3 Common Bean Cultivar. Int. J. Mol. Sci. 2017, 18, 2503. [Google Scholar] [CrossRef] [Green Version]
- Arnold, D.L.; Lovell, H.C.; Jackson, R.W.; Mansfield, J.W. Pseudomonas syringae pv. phaseolicola: From “has bean” to supermodel. Mol. Plant Pathol. 2011, 12, 617–627. [Google Scholar] [CrossRef]
- Asensio-S.-Manzanera, M.C.; Asensio, C.; Singh, S.P. Gamete selection for resistance to common and halo bacterial blights in dry bean intergene pool populations. Crop Sci. 2006, 46, 131–135. [Google Scholar] [CrossRef]
- Zhang, S.; Fu, Y.; Mersha, Z.; Pernezny, K. Assessment of copper resistance in Pseudomonas syringae pv. phaseolicola, the pathogen of halo blight on snap bean. Crop Prot. 2017, 98, 8–15. [Google Scholar] [CrossRef]
- Monteagudo, A.B.; Rodiño, A.P.; Lema, M.; De La Fuente, M.; Santalla, M.; De Ron, A.M.; Singh, S.P. Resistance to infection by fungal, bacterial, and viral pathogens in a common bean core collection from the Iberian Peninsula. HortScience 2006, 41, 319–322. [Google Scholar] [CrossRef] [Green Version]
- De la Rubia, A.G.; Centeno, M.L.; Moreno-González, V.; De Castro, M.; García-Angulo, P. Perception and first defense responses against Pseudomonas syringae pv. phaseolicola in Phaseolus vulgaris L.: Identification of Wall-Associated Kinase receptors. Phytopathology 2021, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Aguilar, K.; Ramírez-Carrasco, G.; Hernández-Chávez, J.L.; Barraza, A.; Alvarez-Venegas, R. Use of BABA and INA As Activators of a Primed State in the Common Bean (Phaseolus vulgaris L.). Front. Plant Sci. 2016, 7, 653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramírez-Carrasco, G.; Martínez-Aguilar, K.; Alvarez-Venegas, R. Transgenerational Defense Priming for Crop Protection against Plant Pathogens: A Hypothesis. Front. Plant Sci. 2017, 8, 696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De la Rubia, A.G.; Mélida, H.; Centeno, M.L.; Encina, A.; García-Angulo, P. Immune Priming Triggers Cell Wall Remodeling and Increased Resistance to Halo Blight Disease in Common Bean. Plants 2021, 10, 1514. [Google Scholar] [CrossRef]
- Kauss, H.; Theisinger-Hinkel, E.; Mindermann, R.; Conrath, U. Dichloroisonicotinic and salicylic acid, inducers of systemic acquired resistance, enhance fungal elicitor responses in parsley cells. Plant J. 1992, 2, 655–660. [Google Scholar] [CrossRef]
- Görlach, J.; Volrath, S.; Knauf-Beiter, G.; Hengy, G.; Beckhove, U.; Kogel, K.H.; Oostendorp, M.; Staub, T.; Ward, E.; Kessmann, H.; et al. Benzothiadiazole, a novel class of inducers of systemic acquired resistance, activates gene expression and disease resistance in wheat. Plant Cell 1996, 8, 629–643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heil, M.; Hilpert, A.; Kaiser, W.; Linsenmair, K.E. Reduced growth and seed set following chemical induction of pathogen defence: Does systemic acquired resistance (SAR) incur allocation costs? J. Ecol. 2000, 88, 645–654. [Google Scholar] [CrossRef]
- Boller, T.; Felix, G. A Renaissance of Elicitors: Perception of Microbe-Associated Molecular Patterns and Danger Signals by Pattern-Recognition Receptors. Annu. Rev. Plant Biol. 2009, 60, 379–406. [Google Scholar] [CrossRef] [PubMed]
- Lotze, M.T.; Zeh, H.J.; Rubartelli, A.; Sparvero, L.J.; Amoscato, A.A.; Washburn, N.R.; DeVera, M.E.; Liang, X.; Tör, M.; Billiar, T. The grateful dead: Damage-associated molecular pattern molecules and reduction/oxidation regulate immunity. Immunol. Rev. 2007, 220, 60–81. [Google Scholar] [CrossRef]
- Hou, S.; Liu, Z.; Shen, H.; Wu, D. Damage-Associated Molecular Pattern-Triggered Immunity in Plants. Front. Plant Sci. 2019, 10, 646. [Google Scholar] [CrossRef] [PubMed]
- Voxeur, A.; Habrylo, O.; Guénin, S.; Miart, F.; Soulié, M.C.; Rihouey, C.; Pau-Roblot, C.; Domon, J.M.; Gutierrez, L.; Pelloux, J.; et al. Oligogalacturonide production upon Arabidopsis thaliana-Botrytis cinerea interaction. Proc. Natl. Acad. Sci. USA 2019, 116, 19743–19752. [Google Scholar] [CrossRef] [Green Version]
- Mélida, H.; Bacete, L.; Ruprecht, C.; Rebaque, D.; Del Hierro, I.; López, G.; Brunner, F.; Pfrengle, F.; Molina, A. Arabinoxylan-Oligosaccharides Act as Damage Associated Molecular Patterns in Plants Regulating Disease Resistance. Front. Plant Sci. 2020, 11, 1210. [Google Scholar] [CrossRef]
- Mélida, H.; Sopeña-Torres, S.; Bacete, L.; Garrido-Arandia, M.; Jordá, L.; López, G.; Muñoz-Barrios, A.; Pacios, L.F.; Molina, A. Non-branched β-1,3-glucan oligosaccharides trigger immune responses in Arabidopsis. Plant J. 2018, 93, 34–49. [Google Scholar] [CrossRef] [Green Version]
- Rebaque, D.; del Hierro, I.; López, G.; Bacete, L.; Vilaplana, F.; Dallabernardina, P.; Pfrengle, F.; Jordá, L.; Sánchez-Vallet, A.; Pérez, R.; et al. Cell wall-derived mixed-linked β-1,3/1,4-glucans trigger immune responses and disease resistance in plants. Plant J. 2021, 106, 601–615. [Google Scholar] [CrossRef]
- Couto, D.; Zipfel, C. Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 2016, 16, 537–552. [Google Scholar] [CrossRef]
- Smirnoff, N.; Arnaud, D. Hydrogen peroxide metabolism and functions in plants. N. Phytol. 2019, 221, 1197–1214. [Google Scholar] [CrossRef]
- López-Moral, A.; Agustí-Brisach, C.; Trapero, A. Plant Biostimulants: New Insights into the Biological Control of Verticillium Wilt of Olive. Front. Plant Sci. 2021, 12, 782. [Google Scholar] [CrossRef]
- Rakoczy-Lelek, R.; Smoleń, S.; Grzanka, M.; Ambroziak, K.; Pitala, J.; Skoczylas, Ł.; Liszka-Skoczylas, M.; Kardasz, H. Effectiveness of Foliar Biofortification of Carrot With Iodine and Selenium in a Field Condition. Front. Plant Sci. 2021, 12, 656283. [Google Scholar] [CrossRef] [PubMed]
- Qiu, H.; Su, L.; Wang, H.; Zhang, Z. Chitosan elicitation of saponin accumulation in Psammosilene tunicoides hairy roots by modulating antioxidant activity, nitric oxide production and differential gene expression. Plant Physiol. Biochem. 2021, 166, 115–127. [Google Scholar] [CrossRef]
- Shahrajabian, M.H.; Chaski, C.; Polyzos, N.; Tzortzakis, N.; Petropoulos, S.A. Sustainable Agriculture Systems in Vegetable Production Using Chitin and Chitosan as Plant Biostimulants. Biomolecules 2021, 11, 819. [Google Scholar] [CrossRef]
- Barna, D.; Kisvarga, S.; Kovács, S.; Csatári, G.; Tóth, I.O.; Fári, M.G.; Alshaal, T.; Bákonyi, N. Raw and Fermented Alfalfa Brown Juice Induces Changes in the Germination and Development of French Marigold (Tagetes patula L.) Plants. Plants 2021, 10, 1076. [Google Scholar] [CrossRef]
- El Boukhari, M.E.M.; Barakate, M.; Choumani, N.; Bouhia, Y.; Lyamlouli, K. Ulva lactuca Extract and Fractions as Seed Priming Agents Mitigate Salinity Stress in Tomato Seedlings. Plants 2021, 10, 1104. [Google Scholar] [CrossRef]
- Zaynab, M.; Fatima, M.; Abbas, S.; Sharif, Y.; Umair, M.; Zafar, M.H.; Bahadar, K. Role of secondary metabolites in plant defense against pathogens. Microb. Pathog. 2018, 124, 198–202. [Google Scholar] [CrossRef]
- Kadoglidou, K.; Chatzopoulou, P.; Maloupa, E.; Kalaitzidis, A.; Ghoghoberidze, S.; Katsantonis, D. Mentha and oregano soil amendment induces enhancement of tomato tolerance against soilborne diseases, yield and quality. Agronomy 2020, 10, 406. [Google Scholar] [CrossRef] [Green Version]
- Latique, S.; Mrid, R.B.; Kabach, I.; Kchikich, A.; Sammama, H.; Yasri, A.; Nhiri, M.; El Kaoua, M.; Douira, A.; Selmaoui, K. Foliar Application of Ulva rigida Water Extracts Improves Salinity Tolerance in Wheat (Triticum durum L.). Agronomy 2021, 11, 265. [Google Scholar] [CrossRef]
- Yusoff, S.F.; Haron, F.F.; Asib, N.; Mohamed, M.T.M.; Ismail, S.I. Development of Vernonia amygdalina Leaf Extract Emulsion Formulations in Controlling Gray Mold Disease on Tomato (Lycopersicon esculentum Mill.). Agronomy 2021, 11, 373. [Google Scholar] [CrossRef]
- Bella, E.L.; Baglieri, A.; Rovetto, E.I.; Stevanato, P.; Puglisi, I. Foliar spray application of Chlorella vulgaris extract: Effect on the growth of lettuce seedlings. Agronomy 2021, 11, 308. [Google Scholar] [CrossRef]
- Abdel Latef, A.A.H.; Zaid, A.; Alwaleed, E.A. Influences of Priming on Selected Physiological Attributes and Protein Pattern Responses of Salinized Wheat with Extracts of Hormophysa cuneiformis and Actinotrichia fragilis. Agronomy 2021, 11, 545. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Largo-Gosens, A.; de Castro, M.; Alonso-Simón, A.; García-Angulo, P.; Acebes, J.L.; Encina, A.; Álvarez, J.M. Quinclorac-habituation of bean (Phaseolus vulgaris) cultured cells is related to an increase in their antioxidant capacity. Plant Physiol. Biochem. 2016, 107, 257–263. [Google Scholar] [CrossRef]
- Bradford, M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Cheeseman, J.M. Hydrogen peroxide concentrations in leaves under natural conditions. J. Exp. Bot. 2006, 57, 2435–2444. [Google Scholar] [CrossRef] [Green Version]
- Buege, J.A.; Aust, S.D. Microsomal Lipid Peroxidation. Methods Enzymol. 1978, 52, 302–310. [Google Scholar] [CrossRef]
- Droillard, M.J.; Paulin, A.; Massot, J.C. Free radical production, catalase and superoxide dismutase activities and membrane integrity during senescence of petals of cut carnations (Dianthus caryophyllus). Physiol. Plant. 1987, 71, 197–202. [Google Scholar] [CrossRef]
- Hossain, M.A.; Asada, K. Inactivation of ascorbate peroxidase in spinach chloroplasts on dark addition of hydrogen peroxide: Its protection by ascorbate. Plant Cell Physiol. 1984, 25, 1285–1295. [Google Scholar] [CrossRef]
- Ádám, A.L.; Bestwick, C.S.; Barna, B.; Mansfield, J.W. Enzymes regulating the accumulation of active oxygen species during the hypersensitive reaction of bean to Pseudomonas syringae pv. phaseolicola. Planta 1995, 197, 240–249. [Google Scholar] [CrossRef]
- Borges, A.; Tsai, S.M.; Caldas, D.G.G. Validation of reference genes for RT-qPCR normalization in common bean during biotic and abiotic stresses. Plant Cell Rep. 2012, 31, 827–838. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Guerrero-González, M.L.; Rodríguez-Kessler, M.; Rodríguez-Guerra, R.; González-Chavira, M.; Simpson, J.; Sanchez, F.; Jiménez-Bremont, J.F. Differential expression of Phaseolus vulgaris genes induced during the interaction with Rhizoctonia solani. Plant Cell Rep. 2011, 30, 1465–1473. [Google Scholar] [CrossRef] [PubMed]
- Mayo, S.; Gutiérrez, S.; Malmierca, M.G.; Lorenzana, A.; Campelo, M.P.; Hermosa, R.; Casquero, P.A. Influence of Rhizoctonia solani and Trichoderma spp. In growth of bean (Phaseolus vulgaris L.) and in the induction of plant defense-related genes. Front. Plant Sci. 2015, 6, 685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, P.; Jha, A.B.; Dubey, R.S.; Pessarakli, M. Reactive Oxygen Species, Oxidative Damage, and Antioxidative Defense Mechanism in Plants under Stressful Conditions. J. Bot. 2012, 2012, 217037. [Google Scholar] [CrossRef] [Green Version]
- Pedre, B.; Young, D.; Charlier, D.; Mourenza, Á.; Rosado, L.A.; Marcos-Pascual, L.; Wahni, K.; Martens, E.; de la Rubia, A.G.; Belousov, V.V.; et al. Structural snapshots of OxyR reveal the peroxidatic mechanism of H2O2 sensing. Proc. Natl. Acad. Sci. USA 2018, 115, E11623–E11632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breen, S.; Williams, S.J.; Outram, M.; Kobe, B.; Solomon, P.S. Emerging Insights into the Functions of Pathogenesis-Related Protein 1. Trends Plant Sci. 2017, 22, 871–879. [Google Scholar] [CrossRef]
- Krofta, K.; Houška, M.; Mikyška, A.; Čermák, P. Changes in the composition of hop secondary metabolites induced by high hydrostatic pressure. J. Inst. Brew. 2018, 124, 158–172. [Google Scholar] [CrossRef]
- Flythe, M.D. The antimicrobial effects of hops (Humulus lupulus L.) on ruminal hyper ammonia-producing bacteria. Lett. Appl. Microbiol. 2009, 48, 712–717. [Google Scholar] [CrossRef]
- Pallag, A.; Filip, G.A.; Olteanu, D.; Clichici, S.; Baldea, I.; Jurca, T.; Micle, O.; Vicas, L.; Marian, E.; Soriţău, O.; et al. Equisetum arvense L. extract induces antibacterial activity and modulates oxidative stress, inflammation, and apoptosis in endothelial vascular cells exposed to hyperosmotic stress. Oxid. Med. Cell. Longev. 2018, 2018, 3060525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rogozhin, E.A.; Vasilchenko, A.S.; Barashkova, A.S.; Smirnov, A.N.; Zavriev, S.K.; Demushkin, V.P. Peptide extracts from seven medicinal plants discovered to inhibit oomycete phytophthora infestans, a causative agent of potato late blight disease. Plants 2020, 9, 1294. [Google Scholar] [CrossRef] [PubMed]
- Augspole, I.; Duma, M.; Ozola, B.; Cinkmanis, I. Phenolic profile of fresh and frozen netthle, goutweed, dandelion ad chickweed leaves. Foodbalt 2017, 36–39. [Google Scholar] [CrossRef]
- Kregiel, D.; Pawlikowska, E.; Antolak, H. Urtica spp.: Ordinary plants with extraordinary properties. Molecules 2018, 23, 1664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furiga, A.; Lonvaud-Funel, A.; Badet, C. In vitro study of antioxidant capacity and antibacterial activity on oral anaerobes of a grape seed extract. Food Chem. 2009, 113, 1037–1040. [Google Scholar] [CrossRef]
- Costa, J.R.; Amorim, M.; Vilas-Boas, A.; Tonon, R.V.; Cabral, L.M.C.; Pastrana, L.; Pintado, M. Impact of: In vitro gastrointestinal digestion on the chemical composition, bioactive properties, and cytotoxicity of Vitis vinifera L. cv. Syrah grape pomace extract. Food Funct. 2019, 10, 1856–1869. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, D.A.; Salvador, A.A.; Smânia, A.; Smânia, E.F.A.; Maraschin, M.; Ferreira, S.R.S. Antimicrobial activity and composition profile of grape (Vitis vinifera) pomace extracts obtained by supercritical fluids. J. Biotechnol. 2013, 164, 423–432. [Google Scholar] [CrossRef] [PubMed]
- Al-Mousawi, A.H.; Al-kaabi, S.J.; Albaghdadi, A.J.H.; Almulla, A.F.; Raheem, A.; Algon, A.A.A. Effect of Black Grape Seed Extract (Vitis vinifera) on Biofilm Formation of Methicillin-Resistant Staphylococcus aureus and Staphylococcus haemolyticus. Curr. Microbiol. 2020, 77, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Waszczak, C.; Carmody, M.; Kangasjärvi, J. Reactive Oxygen Species in Plant Signaling. Annu. Rev. Plant Biol. 2018, 69, 209–236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dat, J.; Vandenabeele, S.; Vranová, E.; Van Montagu, M.; Inzé, D.; Van Breusegem, F. Dual action of the active oxygen species during plant stress responses. Cell. Mol. Life Sci. 2000, 57, 779–795. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. PPB 2010, 48, 909–930. [Google Scholar] [CrossRef]
- Mittler, R. ROS Are Good. Trends Plant Sci. 2017, 22, 11–19. [Google Scholar] [CrossRef] [Green Version]
- Leonard, S.S.; Xia, C.; Jiang, B.H.; Stinefelt, B.; Klandorf, H.; Harris, G.K.; Shi, X. Resveratrol scavenges reactive oxygen species and effects radical-induced cellular responses. Biochem. Biophys. Res. Commun. 2003, 309, 1017–1026. [Google Scholar] [CrossRef] [PubMed]
- Bakshi, M.; Oelmüller, R. Wrky transcription factors jack of many trades in plants. Plant Signal. Behav. 2014, 9, e27700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kapos, P.; Devendrakumar, K.T.; Li, X. Plant NLRs: From discovery to application. Plant Sci. 2019, 279, 3–18. [Google Scholar] [CrossRef]
- Van Loon, L.C.; Van Strien, E.A. The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol. Mol. Plant Pathol. 1999, 55, 85–97. [Google Scholar] [CrossRef]
- Betsuyaku, S.; Katou, S.; Takebayashi, Y.; Sakakibara, H.; Nomura, N.; Fukuda, H. Salicylic Acid and Jasmonic Acid Pathways are Activated in Spatially Different Domains around the Infection Site during Effector-Triggered Immunity in Arabidopsis thaliana. Plant Cell Physiol. 2018, 59, 8–16. [Google Scholar] [CrossRef]
- Van Loon, L.C.; Rep, M.; Pieterse, C.M.J. Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 2006, 44, 135–162. [Google Scholar] [CrossRef] [Green Version]
- Abo-Zaid, G.A.; Matar, S.M.; Abdelkhalek, A. Induction of plant resistance against tobacco mosaic virus using the biocontrol agent streptomyces cellulosae isolate actino 48. Agronomy 2020, 10, 1620. [Google Scholar] [CrossRef]
- Huot, B.; Yao, J.; Montgomery, B.L.; He, S.Y. Growth-defense tradeoffs in plants: A balancing act to optimize fitness. Mol. Plant 2014, 7, 1267–1287. [Google Scholar] [CrossRef] [Green Version]
- Ning, Y.; Liu, W.; Wang, G.L. Balancing Immunity and Yield in Crop Plants. Trends Plant Sci. 2017, 22, 1069–1079. [Google Scholar] [CrossRef] [PubMed]
- Lawton, K.A.; Friedrich, L.; Hunt, M.; Weymann, K.; Delaney, T.; Kessmann, H.; Staub, T.; Ryals, J. Benzothiadiazole induces disease resistance in Arabidopsis by activation of the systemic acquired resistance signal transduction pathway. Plant J. 1996, 10, 71–82. [Google Scholar] [CrossRef] [PubMed]
- Maricic, B.; Radman, S.; Romic, M.; Perkovic, J.; Major, N.; Urlic, B.; Palcic, I.; Ban, D.; Zoric, Z.; Ban, S.G. Stinging Nettle (Urtica dioica L.) as an Aqueous Plant-Based Extract Fertilizer in Green Bean (Phaseolus vulgaris L.) Sustainable Agriculture. Sustainability 2021, 13, 4042. [Google Scholar] [CrossRef]





| Gene Name | Sequence (5′-3′) Fw/Rev | Amplicon Length | Primer Efficiency | Annealing Temperature | Functional Annotation | Reference or NCBI Accesion |
|---|---|---|---|---|---|---|
| Ukn1 | ATTCCCATCATGCAGCAAAG/ AGATCCCTCCAGGTCAATCC | 192 bp | 97.3% | 62.1 °C | Unknown | [42] |
| PR1 | TGGTCCTAACGGAGGATCAC/ TGGCTTTTCCAGCTTTGAGT | 170 bp | 94.0% | 63.4 °C | Pathogenesis related 1 | [44] |
| MAPKK | TTCTACATGGGCAGGTTTCC/ GGGAGATGAAATCCCTGAAG | 132 bp | 92.3% | 60.6 °C | Mitogen activated protein kinase kinase | [45] |
| WRKY33 | TTTCACAGGACAGGTTCCAGC/ CCTTTGACAGAAATGACTGAAGGA | 161 bp | 93.8% | 63.8 °C | WRKY transcription factor | [45] |
| RIN4 | GTTCAGTGTTGCTGCTTTGC/ CACACACCTCATGACCATACAC | 117 bp | 92.2% | 63.6 °C | RPM1 interacting protein 4 | [6] |
| PAL1 | TGAGAGAGGAGTTGGGCACT TTCCACTCTCCAAGGCATTC | 135 bp | 99.4% | 62.9 °C | Phenylalanine ammonia-lyase | [45] |
| Mock | U | RG | ||||
|---|---|---|---|---|---|---|
| Pods/Plant | 3.889 | ±0.964 ns | 4.467 | ±0.576 ns | 3.154 | ±0.470 ns |
| Yield (g/Plant) | 3.481 | ±1.273 ns | 3.473 | ±0.874 ns | 3.360 | ±0.658 ns |
| nº seeds/Plant | 11.333 | ±3.167 ns | 11.800 | ±1.598 ns | 12.615 | ±2.407 ns |
| Pod length (cm) | 12.470 | ±0.440 ns | 12.520 | ±0.300 ns | 11.810 | ±0.430 ns |
| Pod weight (g) | 4.352 | ±0.646 ns | 5.662 | ±0.423 ns | 4.536 | ±0.446 ns |
| nº seeds/Pod | 3.110 | ±0.390 ns | 2.900 | ±0.280 ns | 3.620 | ±0.360 ns |
| Chlorophyll (µg/mL/g FW) | 206.564 | ±28.337 ns | 224.632 | ±22.415 ns | 218.675 | ±18.057 ns |
| Maximum Quantum Yield | 0.803 | ±0.015 ns | 0.801 | ±0.008 ns | 0.806 | ±0.016 ns |
| Effective Quantum Yield | 0.526 | ±0.037 ns | 0.490 | ±0.031 ns | 0.541 | ±0.027 ns |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De la Rubia, A.G.; De Castro, M.; Medina-Lozano, I.; García-Angulo, P. Using Plant-Based Preparations to Protect Common Bean against Halo Blight Disease: The Potential of Nettle to Trigger the Immune System. Agronomy 2022, 12, 63. https://doi.org/10.3390/agronomy12010063
De la Rubia AG, De Castro M, Medina-Lozano I, García-Angulo P. Using Plant-Based Preparations to Protect Common Bean against Halo Blight Disease: The Potential of Nettle to Trigger the Immune System. Agronomy. 2022; 12(1):63. https://doi.org/10.3390/agronomy12010063
Chicago/Turabian StyleDe la Rubia, Alfonso Gonzalo, María De Castro, Inés Medina-Lozano, and Penélope García-Angulo. 2022. "Using Plant-Based Preparations to Protect Common Bean against Halo Blight Disease: The Potential of Nettle to Trigger the Immune System" Agronomy 12, no. 1: 63. https://doi.org/10.3390/agronomy12010063
APA StyleDe la Rubia, A. G., De Castro, M., Medina-Lozano, I., & García-Angulo, P. (2022). Using Plant-Based Preparations to Protect Common Bean against Halo Blight Disease: The Potential of Nettle to Trigger the Immune System. Agronomy, 12(1), 63. https://doi.org/10.3390/agronomy12010063







