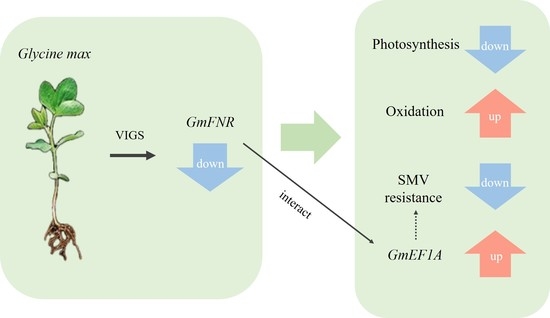
Functional Analysis of A Soybean Ferredoxin-NADP Reductase (FNR) Gene in Response to Soybean Mosaic Virus
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Genetic Materials and SMV Isolate
2.2. Cloning and Sequence Analysis
2.3. RNA Isolation and qRT-PCR Analysis
2.4. Construction of VIGS Plasmids
2.5. Synthesis of In Vitro Transcripts and Inoculation of Plants
2.6. Silencing of GmFNRs in Soybean Plants
2.7. Determination of Chlorophyll
2.8. H2O2 Content and CAT Activity
2.9. Detection of SMV Accumulation
2.10. Yeast-Two-Hybrid Assay
2.11. Bimolecular Fluorescence Complementation (BiFC) Assay
3. Results
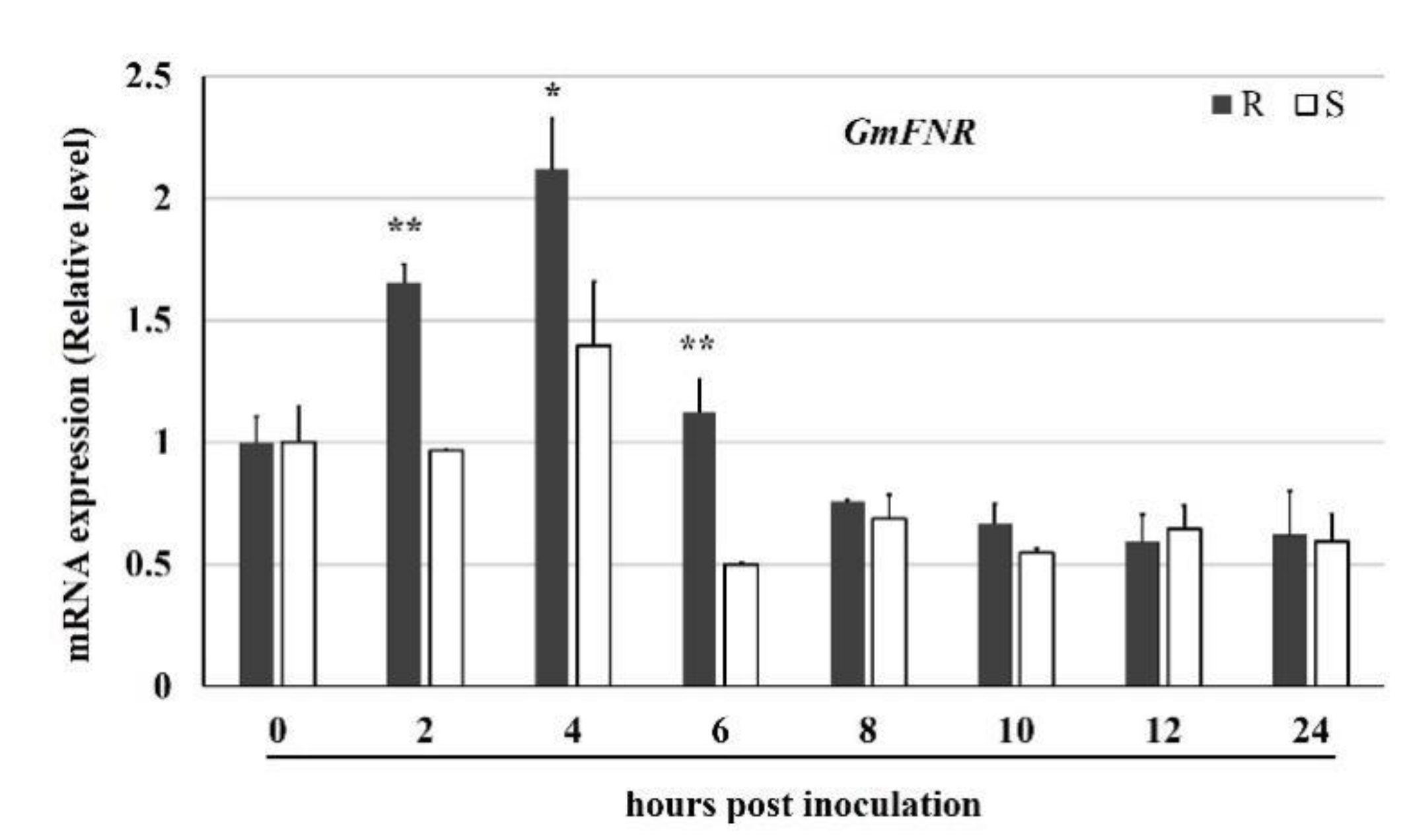
3.1. Sequence and Expression Analyses
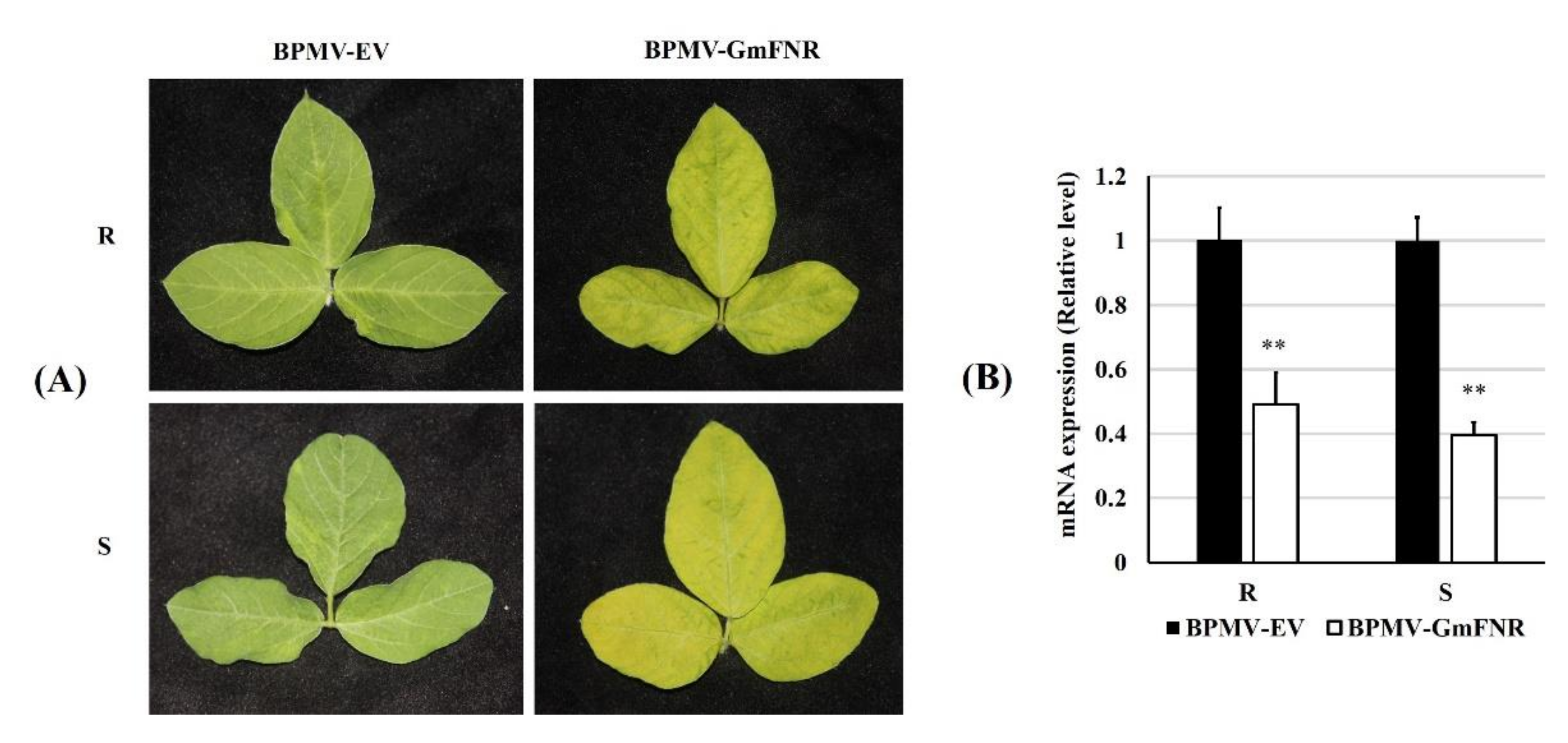
3.2. Silencing of GmFNR in Resistant and Susceptible Lines
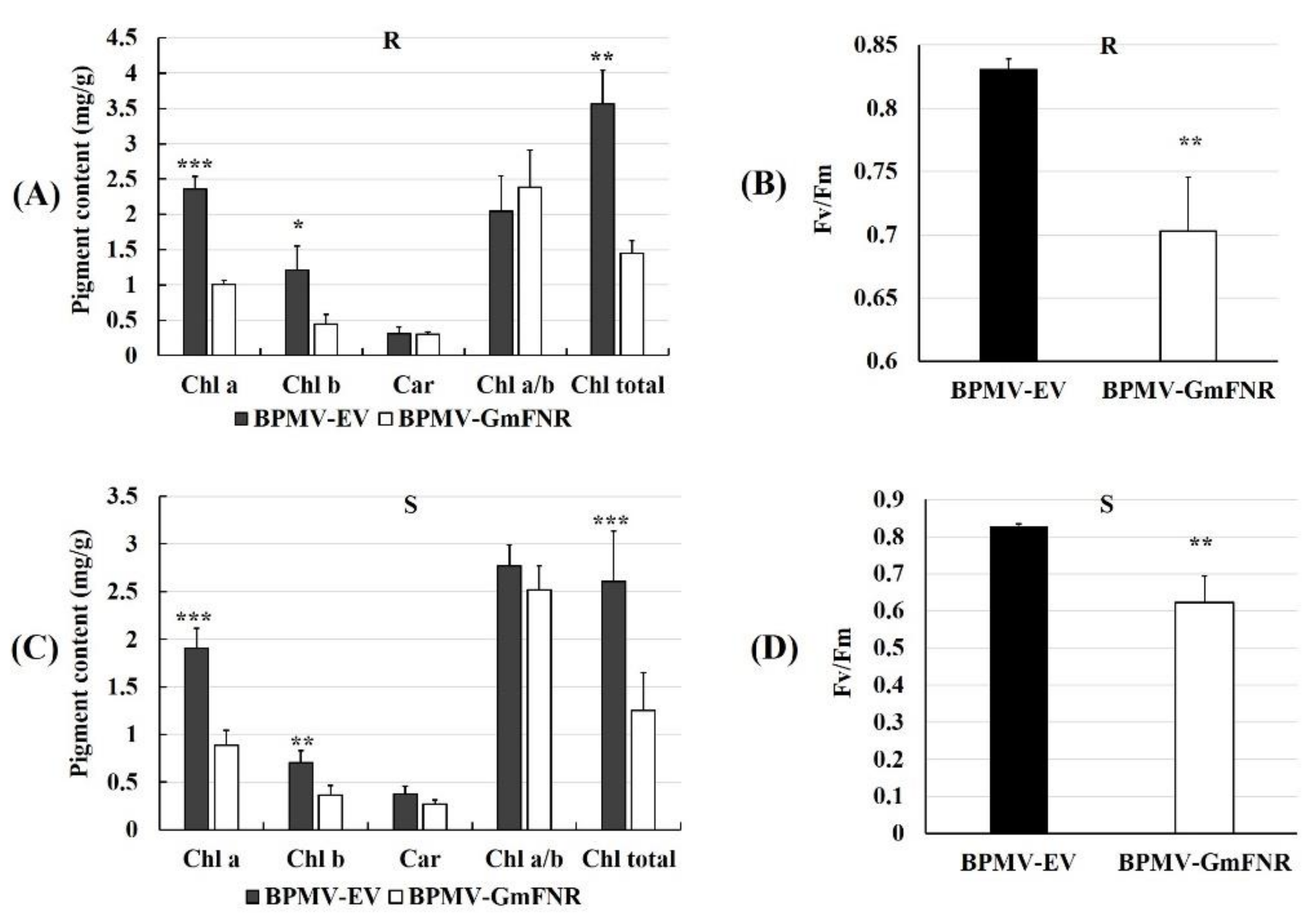
3.3. Photosynthetic Capacity, ROS Production, and CAT Activity
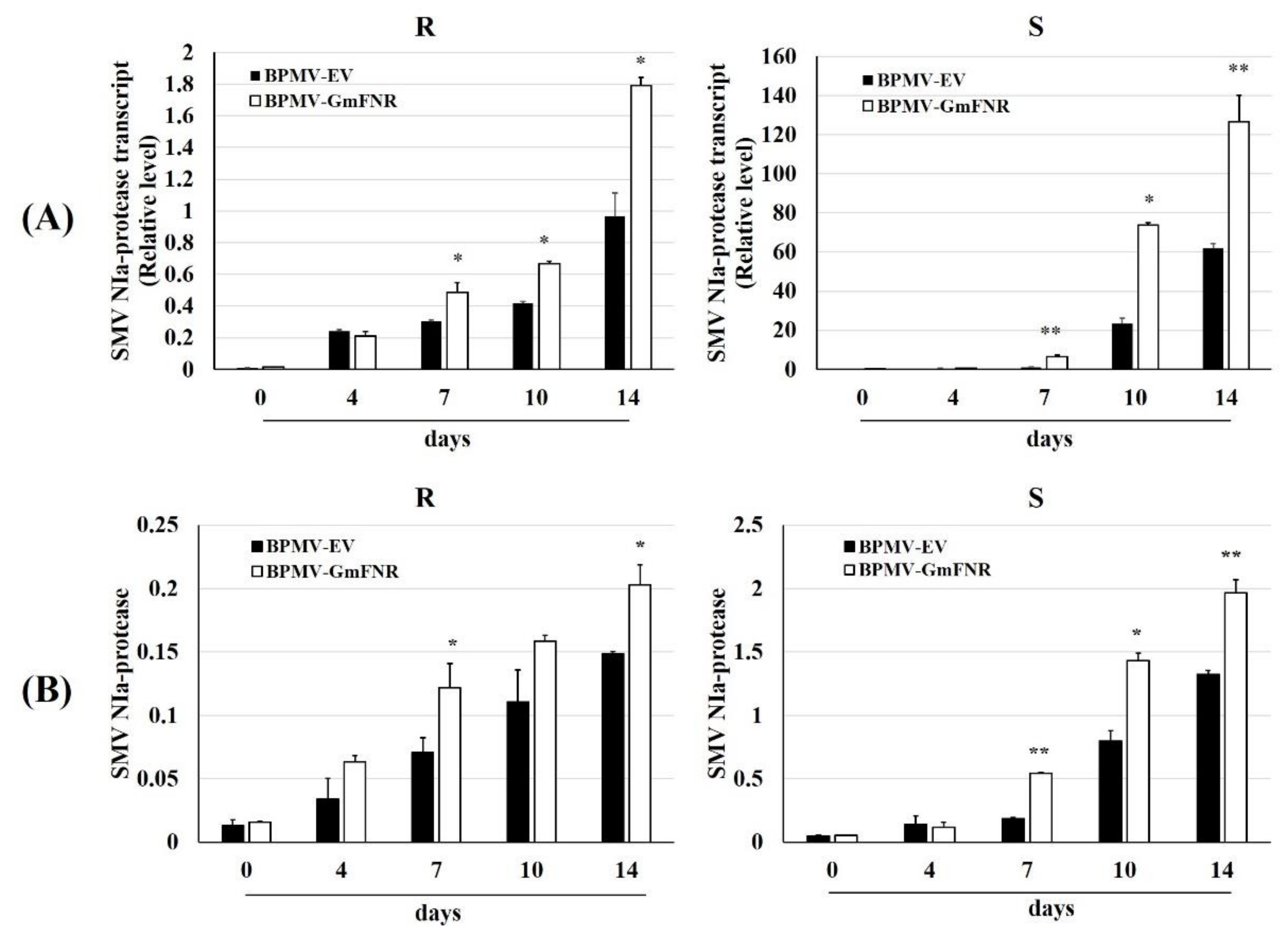
3.4. Silencing Plants Response to SMV Infection
3.5. Proteins Interacting with GmFNR
3.6. The Interaction between GmFNR and EF1A
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Hill, J. Soybean mosaic. In Compendium of Soybean Diseases, 4th ed.; Hartman, G.L., Sinclair, J.B., Rupe, J.C., Eds.; American Phytopathological Society: St. Paul, MN, USA, 2007; pp. 70–71. [Google Scholar]
- Liao, L.; Chen, P.; Buss, G.R.; Yang, Q.; Tolin, S.A. Inheritance and allelism of resistance to soybean mosaic virus in Zao18 soybean from China. J. Hered. 2002, 93, 447–452. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Yang, Q.H.; Zhi, H.J.; Gai, J.Y. Identification and distribution of soybean mosaic virus strains in southern China. Plant Dis. 2010, 94, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Karthikeyan, A.; Li, K.; Jiang, H.; Ren, R.; Li, C.; Zhi, H.; Chen, S.; Gai, J. Inheritance, fine-mapping, and candidate gene analyses of resistance to soybean mosaic virus strain SC5 in soybean. Mol. Genet. Genom. 2017, 292, 811–822. [Google Scholar] [CrossRef] [PubMed]
- Karthikeyan, A.; Li, K.; Li, C.; Yin, J.; Li, N.; Yang, Y.; Song, Y.; Ren, R.; Zhi, H.; Gai, J. Fine-mapping and identifying candidate genes conferring resistance to Soybean mosaic virus strain SC20 in soybean. Theor. Appl. Genet. 2018, 131, 461–476. [Google Scholar] [CrossRef]
- Gao, L.; Sun, S.; Li, K.; Wang, L.; Hou, W.; Wu, C.; Zhi, H.; Han, T. Spatio-temporal characterisation of changes in the resistance of widely grown soybean cultivars to Soybean mosaic virus across a century of breeding in China. Crop Pasture Sci. 2018, 69, 395–405. [Google Scholar] [CrossRef]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.; Song, Q.; Thelen, J.; Cheng, J.; et al. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef]
- Yamagishi, N.; Yoshikawa, N. Virus-induced gene silencing in soybean seeds and the emergence stage of soybean plants with Apple latent spherical virus vectors. Plant Mol. Biol. 2009, 71, 15–24. [Google Scholar] [CrossRef]
- Zhang, C.; Whitham, S.A.; Hill, J.H. Virus-induced gene silencing in soybean and common bean. Methods Mol Biol. 2013, 975, 149–156. [Google Scholar]
- Kim, K.H.; Lim, S.; Kang, Y.J.; Yoon, M.Y.; Nam, M.; Jun, T.H.; Seo, M.J.; Baek, S.B.; Lee, J.H.; Moon, J.K.; et al. Optimization of a Virus-Induced Gene Silencing System with Soybean yellow common mosaic virus for Gene Function Studies in Soybeans. Plant Pathol. 2016, 32, 112–122. [Google Scholar] [CrossRef]
- Zhao, F.; Lim, S.; Igori, D.; Yoo, R.H.; Kwon, S.Y.; Moon, J.S. Development of tobacco ringspot virus-based vectors for foreign gene expression and virus-induced gene silencing in a variety of plants. Virology 2016, 492, 166–178. [Google Scholar] [CrossRef]
- Zhang, C.; Grosic, S.; Whitham, S.A.; Hill, J.H. The requirement of multiple defense genes in soybean Rsv1-mediated extreme resistance to soybean mosaic virus. Mol. Plant Microbe Interact. 2012, 25, 1307–1313. [Google Scholar] [CrossRef]
- Luan, H.; Niu, H.; Luo, J.; Zhi, H. Soybean Cytochrome b5 Is a Restriction Factor for Soybean Mosaic Virus. Viruses 2019, 11, 546. [Google Scholar] [CrossRef]
- Kozuleva, M.; Goss, T.; Twachtmann, M.; Rudi, K.; Trapka, J.; Selinski, J.; Ivanov, B.; Garapati, P.; Steinhoff, H.-J.; Hase, T.; et al. Ferredoxin: NADP (H) oxidoreductase abundance and location influences redox poise and stress tolerance. Plant. Physiol. 2016, 172, 1480–1493. [Google Scholar] [CrossRef]
- Ma, N. Cellular Ultrastructure Analysis and Proteomics Study in Soybean Infected by Soybean Mosaic Virus; Nanjing Agriculture University: Nanjing, China, 2013. [Google Scholar]
- Zheng, G. Fine Mapping, Expression Analysis and Breeding Application of Resistance Genes to Soybean Mosaic Virus in Qihuang No.1; Nanjing Agriculture University: Nanjing, China, 2012. [Google Scholar]
- Costache, M.A.; Campeanu, G.; Neata, G. Studies concerning the extraction of chlorophyll and total carotenoids from vegetables. Rom. Biotechnol. Lett. 2012, 17, 7702–7708. [Google Scholar]
- Waadt, R.; Kudla, J.; Waadt, R.; Kudla, J. In planta visualization of protein interactions using bimolecular fluorescence complementation (BiFC). In Cold Spring Harbor Protocols 2008; Cold Spring Harbor Laboratory: Huntington, NY, USA, 2008. [Google Scholar] [CrossRef]
- Larson, R.L.; Wintermantel, W.M.; Hill, A.; Fortis, L.; Nunez, A. Proteome changes in sugar beet in response to Beet necrotic yellow vein virus. Physiol. Mol. Plant Pathol. 2008, 72, 62–72. [Google Scholar] [CrossRef]
- Wu, L.; Han, Z.; Wang, S.; Wang, X.; Sun, A.; Zu, X.; Chen, Y. Comparative proteomic analysis of the plant–virus interaction in resistant and susceptible ecotypes of maize infected with sugarcane mosaic virus. J. Proteom. 2013, 89, 124–140. [Google Scholar] [CrossRef]
- Li, K.; Xu, C.; Zhang, J. Proteome profile of maize (Zea Mays, L.) leaf tissue at the flowering stage after long-term adjustment to rice black-streaked dwarf virus infection. Gene 2011, 485, 106–113. [Google Scholar] [CrossRef]
- Zhao, J.; Liu, Q.; Zhang, H.; Jia, Q.; Hong, Y.; Liu, Y. The rubisco small subunit is involved in tobamovirus movement and Tm-22-mediated extreme resistance. Plant. Physiol. 2013, 161, 374–383. [Google Scholar] [CrossRef]
- Dube, A.; Bisaillon, M.; Perreault, J.P. Identification of proteins from Prunus persica that interact with peach latent mosaic viroid. J. Virol. 2009, 83, 12057–12067. [Google Scholar] [CrossRef]
- Kaido, M.; Abe, K.; Mine, A.; Hyodo, K.; Taniguchi, T.; Taniguchi, H.; Mise, K.; Okuno, T. GAPDH-A recruits a plant virus movement protein to cortical virus replication complexes to facilitate viral cell-to-cell movement. PLoS Pathog 2014, 10, e1004505. [Google Scholar] [CrossRef]
- Mekhalfi, M.; Puppo, C.; Avilan, L.; Lebrun, R.; Mansuelle, P.; Maberly, S.C.; Gontero, B. Glyceraldehyde-3-phosphate dehydrogenase is regulated by ferredoxin-NADP reductase in the diatom A sterionella Formosa. New Phytol. 2014, 203, 414–423. [Google Scholar] [CrossRef]
- Souza, P.F.; Garcia-Ruiz, H.; Carvalho, F.E. What proteomics can reveal about the plant–virus interactions? Photosynthesis-related proteins on the spotlight. Theor. Exp. Plant Physiol. 2019, 31, 227–248. [Google Scholar] [CrossRef]
- Zhao, J.; Zhang, X.; Hong, Y.; Liu, Y. Chloroplast in plant-virus interaction. Front. Microbiol. 2016, 7, 1565. [Google Scholar] [CrossRef]
- Abbink, T.E.; Peart, J.R.; Mos, T.N.; Baulcombe, D.C.; Bol, J.F.; Linthorst, H.J. Silencing of a gene encoding a protein component of the oxygen-evolving complex of photosystem II enhances virus replication in plants. Virology 2002, 295, 307–319. [Google Scholar] [CrossRef][Green Version]
- Kong, L.; Wu, J.; Lu, L.; Xu, Y.; Zhou, X. Interaction between Rice stripe virus disease-specific protein and host PsbP enhances virus symptoms. Mol. Plant 2014, 7, 691–708. [Google Scholar] [CrossRef]
- Geng, C.; Yan, Z.Y.; Cheng, D.J.; Liu, J.; Tian, Y.-P.; Zhu, C.X.; Wang, H.-Y.; Li, X.-D. Tobacco vein banding mosaic virus 6K2 protein hijacks NbPsbO1 for virus replication. Sci. Rep. 2017, 7, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Brizard, J.P.; Carapito, C.; Delalande, F.; Van Dorsselaer, A.; Brugidou, C. Proteome analysis of plant-virus interactome: Comprehensive data for virus multiplication inside their hosts. Mol. Cell. Proteom. 2006, 5, 2279–2297. [Google Scholar] [CrossRef]
- Cilia, M.; Peter, K.A.; Bereman, M.S.; Howe, K.; Fish, T.; Smith, D.; Gildow, F.; MacCoss, M.J.; Thannhauser, T.W.; Gray, S.M. Discovery and targeted LC-MS/MS of purified polerovirus reveals differences in the virus-host interactome associated with altered aphid transmission. PLoS ONE 2012, 7, e48177. [Google Scholar] [CrossRef]
- Giesler, L.J.; Ghabrial, S.A.; Hunt, T.E.; Hill, J.H. Bean pod mottle virus: A threat to US soybean production. Plant Dis. 2002, 86, 1280–1289. [Google Scholar] [CrossRef][Green Version]
- Foyer, C.H.; Shigeoka, S. Understanding oxidative stress and antioxidant functions to enhance photosynthesis. Plant Physiol. 2011, 155, 93–100. [Google Scholar] [CrossRef]
- Kidou, S.; Ejiri, S. Isolation, characterization and mRNA expression of four cDNAs encoding translation elongation factor 1A from rice (Oryza sativa L.). Plant Mol. Biol. 1998, 36, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Prod’Homme, D.; Jakubiec, A.; Tournier, V.; Drugeon, G.; Jupin, I. Targeting of the turnip yellow mosaic virus 66K replication protein to the chloroplast envelope is mediated by the 140K protein. J. Virol. 2003, 77, 9124–9135. [Google Scholar] [CrossRef] [PubMed]
- Torrance, L.; Cowan, G.H.; Gillespie, T.; Ziegler, A.; Lacomme, C. Barley stripe mosaic virus-encoded proteins triple-gene block 2 and γb localize to chloroplasts in virus-infected monocot and dicot plants, revealing hitherto-unknown roles in virus replication. J. Gen. Virol. 2006, 87, 2403–2411. [Google Scholar] [CrossRef] [PubMed]
- Thivierge, K.; Cotton, S.; Dufresne, P.J.; Mathieu, I.; Beauchemin, C.; Ide, C.; Fortin, M.G.; Laliberté, J.-F. Eukaryotic elongation factor 1A interacts with Turnip mosaic virus RNA-dependent RNA polymerase and VPg-Pro in virus-induced vesicles. Virology 2008, 377, 216–225. [Google Scholar] [CrossRef]
- Yamaji, Y.; Kobayashi, T.; Hamada, K.; Sakurai, K.; Yoshii, A.; Suzuki, M.; Namba, S.; Hibi, T. In vivo interaction between Tobacco mosaic virus RNA-dependent RNA polymerase and host translation elongation factor 1A. Virology 2006, 347, 100–108. [Google Scholar] [CrossRef]
- Luan, H.; Shine, M.; Cui, X.; Chen, X.; Ma, N.; Kachroo, P.; Zhi, H.; Kachroo, A. The potyviral P3 protein targets eukaryotic elongation factor 1A to promote the unfolded protein response and viral pathogenesis. Plant Physiol. 2016, 172, 221–234. [Google Scholar] [CrossRef]



| Genes | Forward Primers | Reverse Primers | Tm (℃) | PCR Size (bp) |
|---|---|---|---|---|
| GmFNRa | CTGGATCC CAAGAGAAAAGTCCTG | TATGGCCA TGACACCATAATGTCAT | 58.0 | 204 |
| GmFNRb | ATGGCTGCTGCGGTTA | ATAGACTTCGACATTCCATTG | 53.0 | 1086 |
| GmEF1Ab | ATGGGTAAGGAAAAGACTCACATCA | CTTCTTCTTGGCAGCGGC | 58.0 | 1341 |
| GmFNRc | CCCAAGACCCCTTACAT | TGTCAATACCATCTGGAATT | 54.0 | 154 |
| GmEF1Ac | AGCGTGGTTTTGTTGCATCC | CACAGCAAAACGACCAAGGG | 60.0 | 317 |
| Tubulinc | GGAGTTCACAGAGGCAGAG | CACTTACGCATCACATAGCA | 60.0 | 189 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, Y.; Karthikeyan, A.; Yang, Y.; Ma, N.; Yin, J.; Yuan, Y.; Wang, L.; Zhi, H. Functional Analysis of A Soybean Ferredoxin-NADP Reductase (FNR) Gene in Response to Soybean Mosaic Virus. Agronomy 2021, 11, 1592. https://doi.org/10.3390/agronomy11081592
Shen Y, Karthikeyan A, Yang Y, Ma N, Yin J, Yuan Y, Wang L, Zhi H. Functional Analysis of A Soybean Ferredoxin-NADP Reductase (FNR) Gene in Response to Soybean Mosaic Virus. Agronomy. 2021; 11(8):1592. https://doi.org/10.3390/agronomy11081592
Chicago/Turabian StyleShen, Yingchao, Adhimoolam Karthikeyan, Yunhua Yang, Na Ma, Jinlong Yin, Yuan Yuan, Liqun Wang, and Haijian Zhi. 2021. "Functional Analysis of A Soybean Ferredoxin-NADP Reductase (FNR) Gene in Response to Soybean Mosaic Virus" Agronomy 11, no. 8: 1592. https://doi.org/10.3390/agronomy11081592
APA StyleShen, Y., Karthikeyan, A., Yang, Y., Ma, N., Yin, J., Yuan, Y., Wang, L., & Zhi, H. (2021). Functional Analysis of A Soybean Ferredoxin-NADP Reductase (FNR) Gene in Response to Soybean Mosaic Virus. Agronomy, 11(8), 1592. https://doi.org/10.3390/agronomy11081592