Identification and Genomic Characterization of Pathogenic Bacillus altitudinis from Common Pear Trees in Morocco
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plants Sampling and Bacterial Strains Isolation
2.2. Pathogenicity Test
2.2.1. Plant Material
2.2.2. Preparation of Bacterial Inoculum
2.2.3. Pathogenicity Tests
2.3. Infiltration Method
2.4. Spraying Method
2.5. Disease Assessment
2.6. Molecular Identification
2.7. Genomic DNA Extraction and Whole Genome Sequencing
2.8. Bioinformatics
2.8.1. Genome Assembly and Annotation
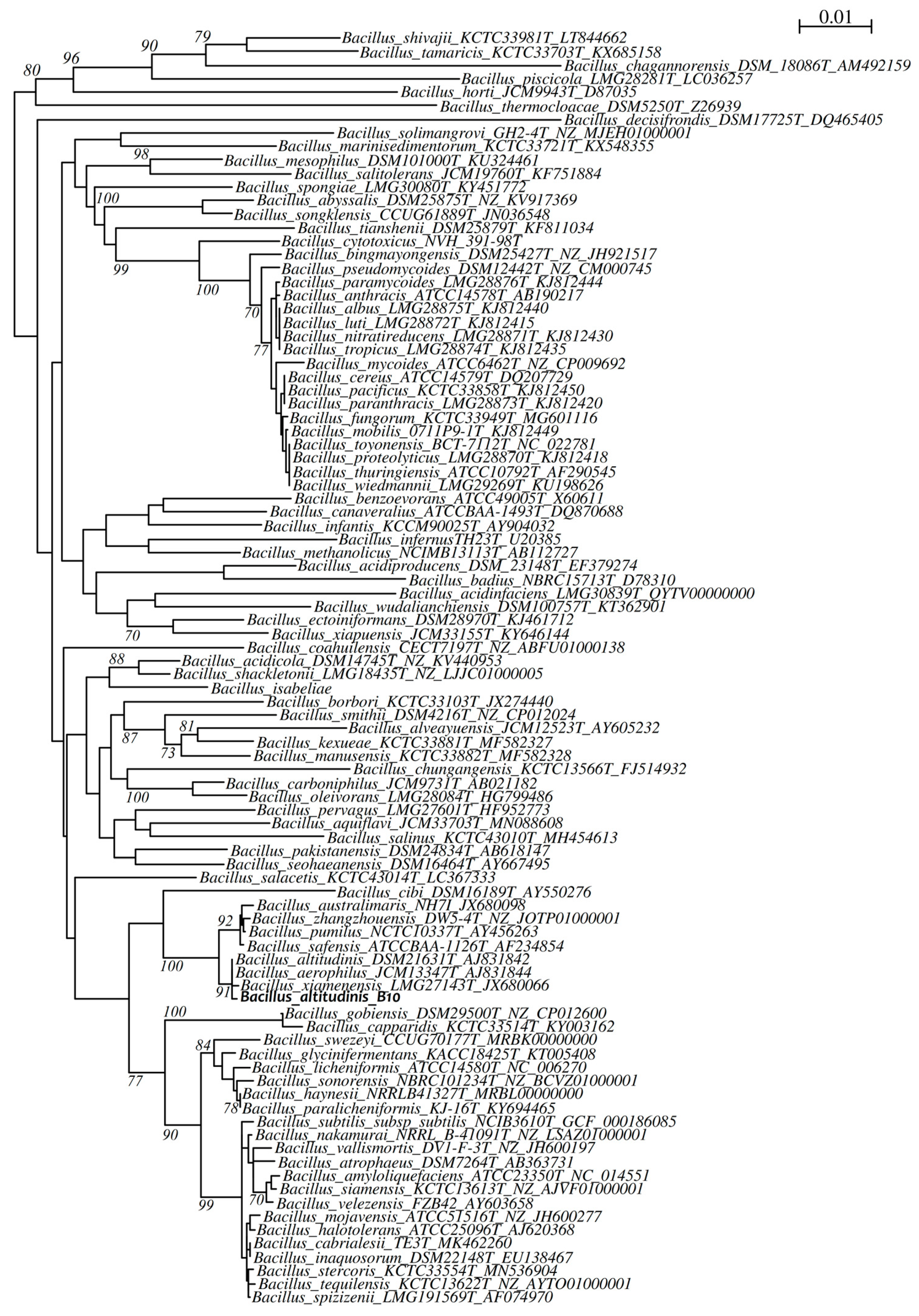
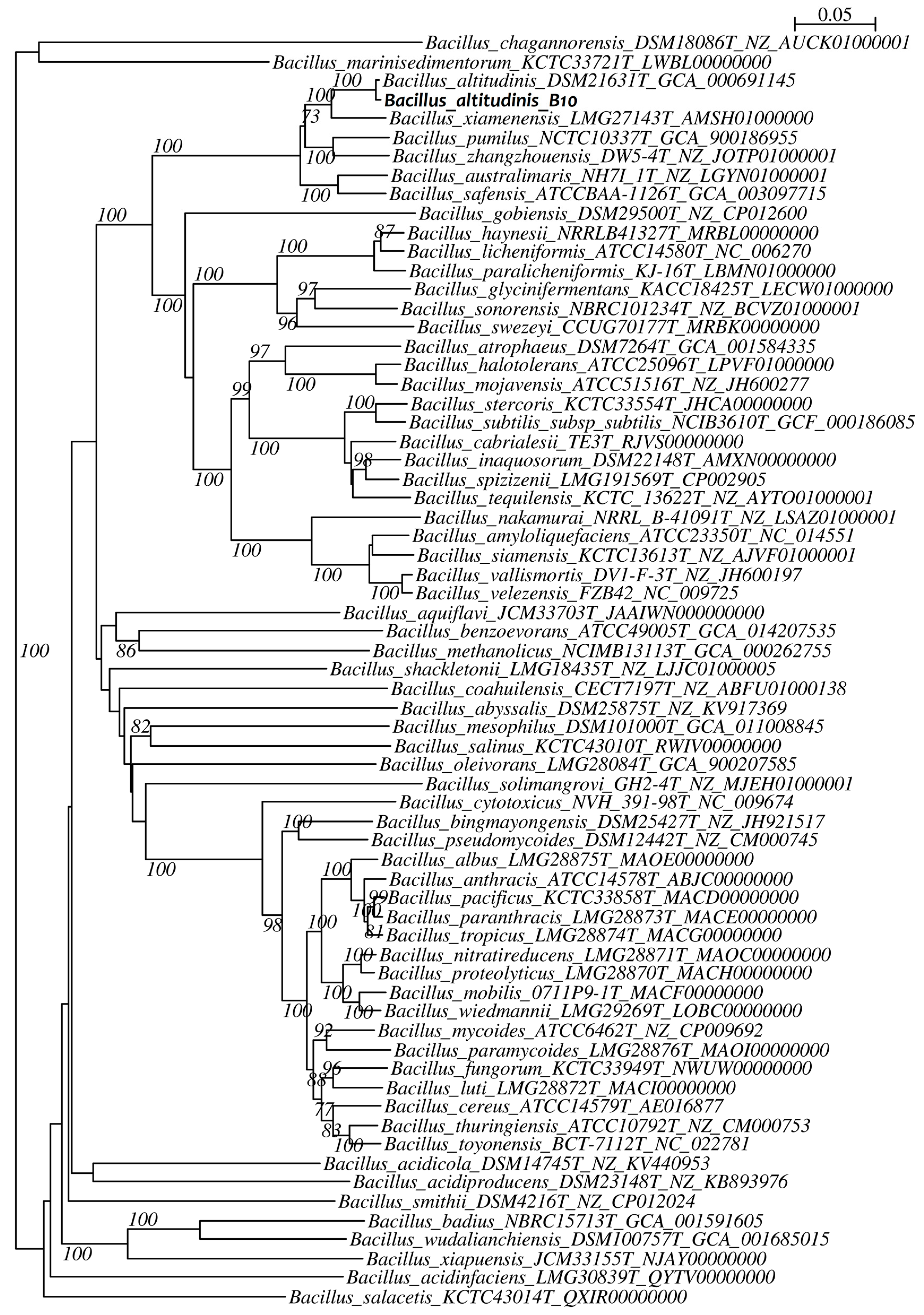
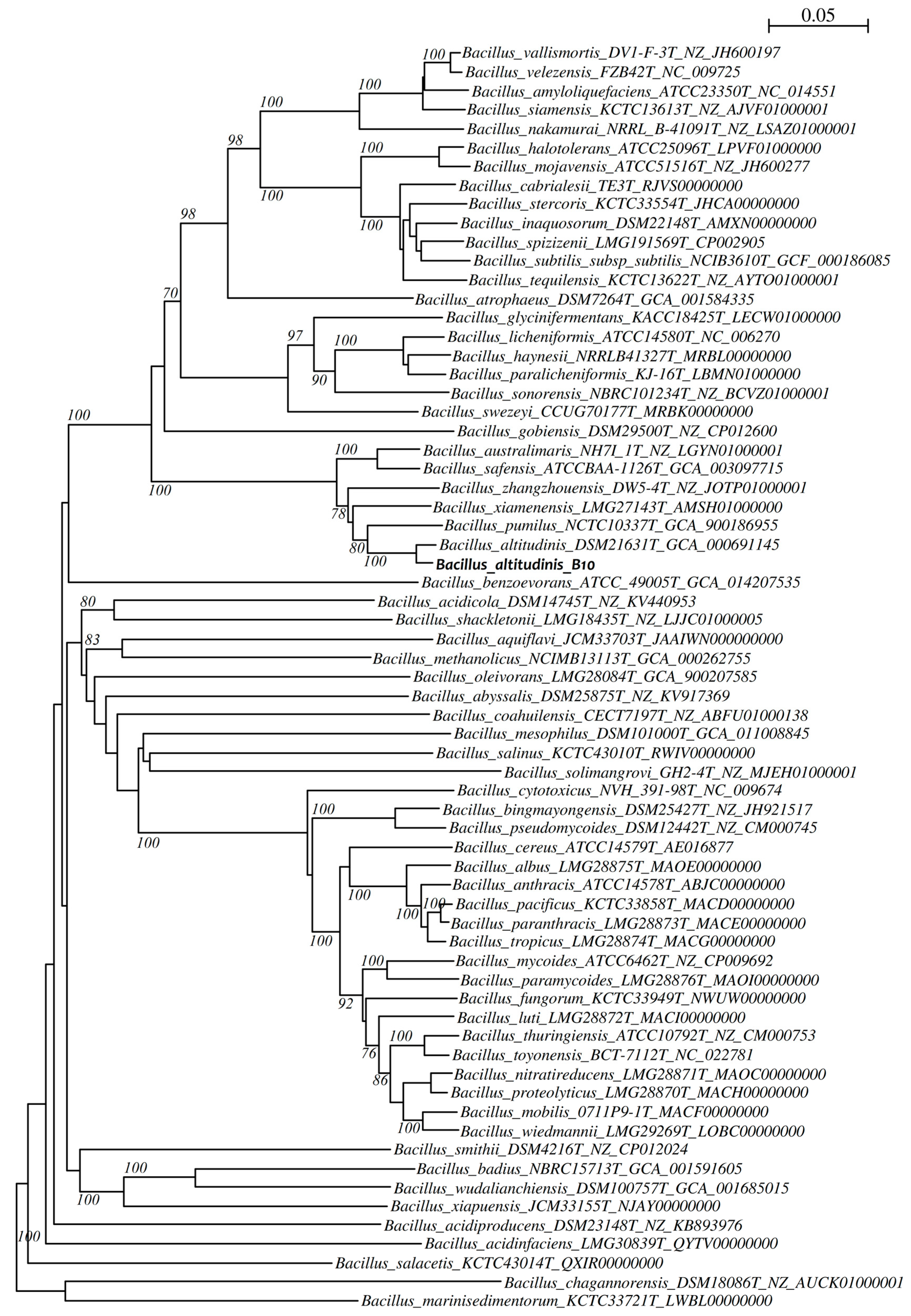
2.8.2. Phylogenetic Analysis
2.8.3. Genome Indexes
2.8.4. Identification of Carbohydrate-Active Enzymes
3. Results
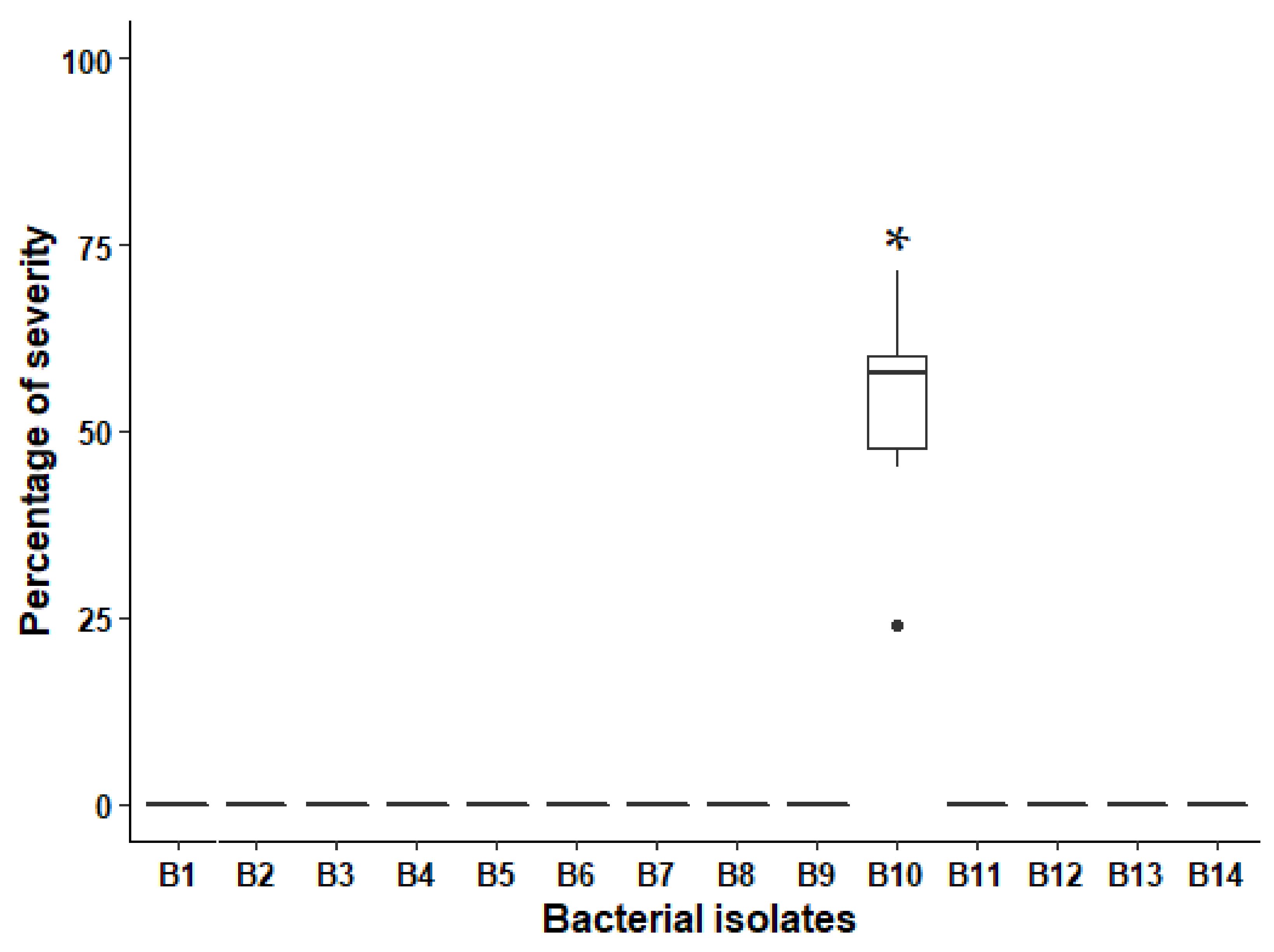
3.1. Pathogenicity Tests
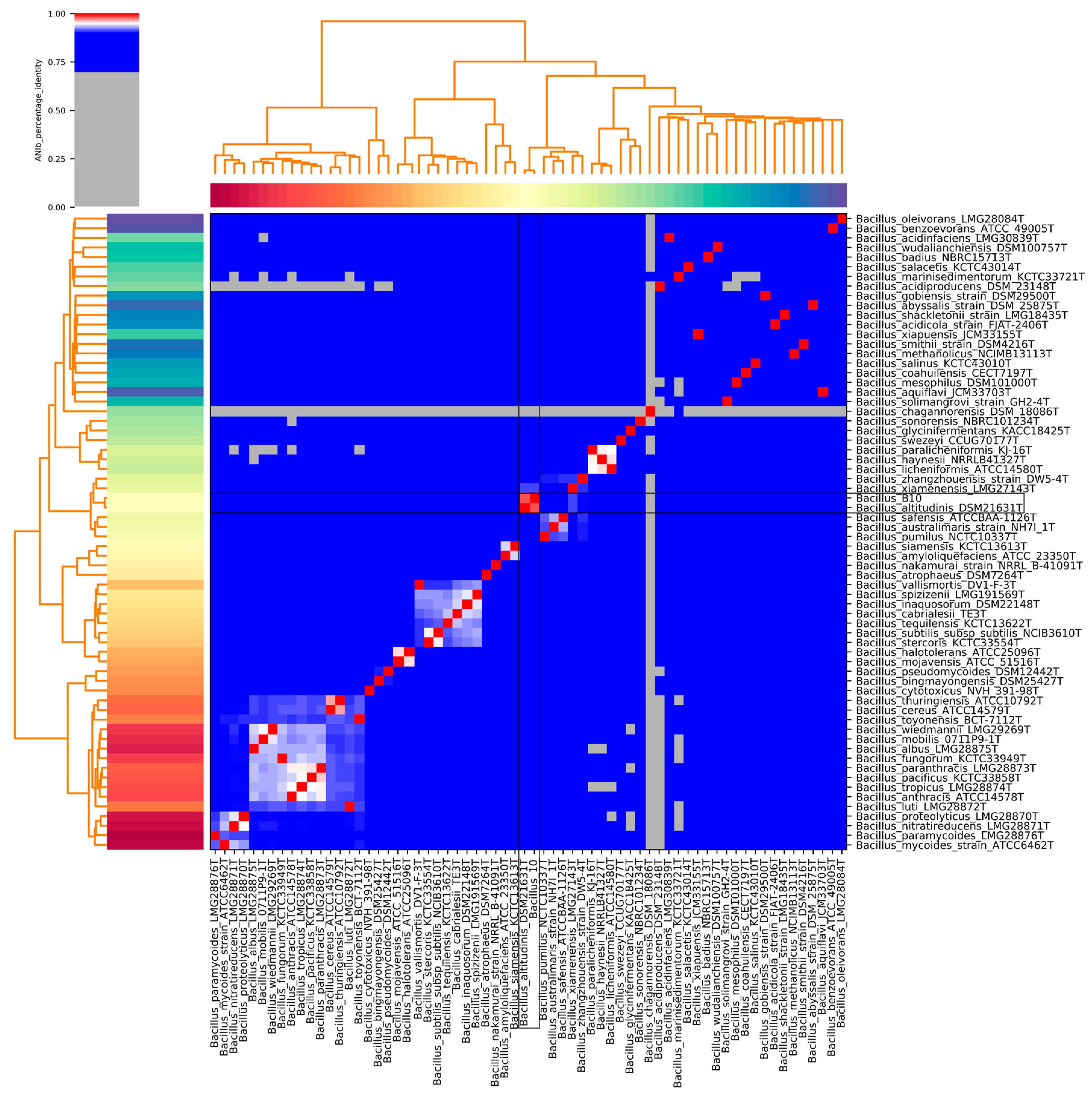
3.2. Taxonomic Identification of the Bacterial Isolate B10
3.3. CAZyme Profiling
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ikinci, A.; Bolat, I.; Ercisli, S.; Kodad, O. Influence of rootstocks on growth, yield, fruit quality and leaf mineral element contents of pear cv. ‘Santa Maria’ in semi-arid conditions. Biol. Res. 2014, 16, 47–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- FAO. Word Pear Production. Food and Agriculture Organisation of the United Nations Crop Statistics, 2013. Available online: http://www.fao.org/faostat/en/#data (accessed on 24 April 2018).
- Gonsalves, P.E. As frutas e seus benefícios. In Fructas Que Curam, 2nd ed.; MG: São Paulo, Brazil, 2002; Volume 1, pp. 131–166. [Google Scholar]
- FAO. Pear production in Morocco. Food and Agriculture Organisation of the United Nations Crop Statistics, 2016. Available online: http://www.fao.org/faostat/en/#data (accessed on 30 April 2018).
- Blancard, D.; Allard, E.; Brest, P. La stemphyliose du poirier ou “macules brunes”. Phytoma 1989, 406, 35–37. [Google Scholar]
- Vilardell, P. Stemphylium vesicarium en plantaciones de peral. Frutic. Prof. 1988, 18, 51–55. [Google Scholar]
- Leu, L.S.; Su, C.C. Isolation, cultivation, and pathogenicity of Xylella fastidiosa, the causal bacterium of pear leaf scorch disease in Taiwan. Plant Dis. 1993, 77, 642–646. [Google Scholar] [CrossRef]
- Elbanna, K.; Elnaggar, S.; Bakeer, A. Characterization of Bacillus altitudinis as a New Causative Agent of Bacterial Soft Rot. J. Phytopathol. 2014, 162, 712–722. [Google Scholar] [CrossRef]
- Kim, W.G.; Hong, S.K.; Park, Y.S. Occurrence of Anthracnose on Fruits of Asian Pear Tree Caused by Colletotrichum acutatum. Mycobiology 2007, 35, 238–240. [Google Scholar] [CrossRef] [Green Version]
- Chatterjee, A. Fire Blight: The Disease and its Causative Agent, Erwinia amylovora. Eur. J. Plant Pathol. 2001, 107, 569. [Google Scholar] [CrossRef]
- Momol, M.T.; Aldwinckle, H.S. Genetic diversity and host range of Erwinia amylovora. In The Disease and Its Causative Agent, Erwinia amylovora; Vanneste, J.L., Ed.; CABI: Wallingford, UK, 2000. [Google Scholar]
- Ramos, L.S.; Lehman, B.L.; Peter, K.A.; McNellis, T.W. Mutation of the Erwinia amylovora argD Gene Causes Arginine Auxotrophy, Nonpathogenicity in Apples, and Reduced Virulence in Pears. Appl. Environ. Microbiol. 2014, 80, 6739–6749. [Google Scholar] [CrossRef] [Green Version]
- Achbani, E.H. Feu bactérien sur Poirier au Maroc. Première apparition de cette brûlure bactérienne sur poirier dans la région de Meknès. Phytoma La Défense Végétaux 2007, 606, 26–28. [Google Scholar]
- Kotan, R.; Sahin, F.; Ala, A. Identification and pathogenicity of bacteria isolated from pome fruit trees in the Eastern Anatolia region of Turkey. J. Plant Dis. Prot. 2005, 113, 8–13. [Google Scholar]
- Brown, K.L. Control of bacterial spores. Br. Med. Bull. 2000, 56, 158–171. [Google Scholar] [CrossRef] [Green Version]
- Turner, J.T.; Backman, P.A. Factors relating to peanut yield increases after seed treatment with Bacillus subtilis. Plant Dis. 1991, 75, 347–353. [Google Scholar] [CrossRef]
- Harsh, P.B.; Ray, F.; Jorge, M.V. Biocontrol of Bacillus subtilis against Infection of Arabidopsis Roots by Pseudomonas syringae is facilitated by Biofilm Formation and Surfactin Production. Plant Physiol. 2004, 134, 307–319. [Google Scholar]
- Bradbury, F. Guide to Plant Pathogenic Bacteria; CAB International: Wallingford, UK, 1986. [Google Scholar]
- Bathily, H.; Babana, A.H.; Samaké, F. Bacillus pumilus a new pathogen on potato tubers in Mali. Afr. J. Microbiol. Res. 2010, 4, 2067–2071. [Google Scholar]
- Galal, A.A.; El Bana, A.A.; Janse, J. Bacillus pumilus, a new pathogen on mango plants. Egypt. J. Phytopathol. 2006, 34, 17–29. [Google Scholar]
- Saleh, O.I.; Huang, P.I.; Huang, J.S. Bacillus pumilus a cause bacterial Blotch of Immature Balady Peach Egypt. J. Phytopathol. 1997, 145, 447–453. [Google Scholar] [CrossRef]
- Li, B.; Qiu, W.; Tan, Q.M.; Su, T.; Fang, Y.; Xie, G.L. Association of a Bacillus species with Leaf and Twig Dieback of Asian pear (Pyrus pyrifolia) in China. J. Plant Pathol. 2009, 91, 705–708. [Google Scholar]
- Vaerewijck, M.J.; De Vos, P.; Lebbe, L.; Scheldeman, P.; Hoste, B.; Heyndrickx, M. Occurrence of Bacillus sporothermodurans and other aerobic spore-forming species in feed concentrate for dairy cattle. J. Appl. Microbiol. 2001, 91, 1074–1084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adiguzel, A.; Ozkan, H.; Baris, O.; Inan, K.; Gulluce, M.; Sahin, F. Identification and characterization of thermophilic bacteria isolated from hot springs in Turkey. J. Microbiol. Methods 2009, 79, 321–328. [Google Scholar] [CrossRef]
- Rintaro, S.; Yousuke, O.; Naoki, K.; Masaru, T. Computer analysis of potential stem structures of rRNA operons in various prokaryote genomes. Gene 2000, 259, 217–222. [Google Scholar]
- Peng, Q.; Yuan, Y.; Gao, M. Bacillus pumilus, a Novel Ginger Rhizome Rot Pathogen in China. Plant Dis. 2013, 97, 1308–1315. [Google Scholar] [CrossRef] [Green Version]
- Cesbron, S.; Paulin, J.P.; Tharaud, M.; Barny, M.A.; Brisset, M.N. The alternative sigma factor HrpL negatively modulates the flagellar system in the phytopathogenic bacterium Erwinia amylovora under hrp-inducing conditions. FEMS Microbiol. Lett. 2006, 257, 221–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patil, A.G.; Ambadkar, C.V.; Kashid, V.S.; Navgire, K.D. Standardization of methods for pathogenicity of pomegranate bacterial blight caused by Xanthomonas axonopodis pv. Punicae. J. Pharmacogn. Phytochem. 2017, 6, 1763–1765. [Google Scholar]
- Nash, A.F.; Gardner, R.G. Tomato early blight resistance in a breeding line derived from Lycopersicon hirsutum PI 126445. Plant Dis. 1988, 72, 206–209. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://www.R-project.org/ (accessed on 10 May 2021).
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 10 May 2021).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [Green Version]
- Mikheenko, A.; Prjibelski, A.; Saveliev, V.; Antipov, D.; Gurevich, A. Versatile genome assembly evaluation with QUAST-LG. Bioinformatics 2018, 34, i142–i150. [Google Scholar] [CrossRef]
- Bushnell, B. BBMap: A Fast, Accurate, Splice-Aware Aligner; Lawrence Berkeley National Lab. (LBNL): Berkeley, CA, USA, 2014. [Google Scholar]
- Tatusova, T.; Dicuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Pritchard, L. PYANI: Python Module for Average Nucleotide Identity Analyses. 2017. Available online: https://github.com/widdowquinn/pyani/releases/tag/v0.2.7 (accessed on 14 December 2020).
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Goker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Yohe, T.; Huang, L.; Entwistle, S.; Wu, P.; Yang, Z.; Busk, P.K.; Xu, Y.; Yin, Y. dbCAN2: A meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018, 46, W95–W101. [Google Scholar] [CrossRef] [Green Version]
- Rawlings, N.D.; Barrett, A.J.; Finn, R. Twenty years of the MEROPS database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2016, 44, D343–D350. [Google Scholar] [CrossRef] [Green Version]
- Logan, N.A.; Halket, G. Developments in the taxonomy of aerobic, endospores-forming bacteria. In Endospore-Forming Soil Bacteria; Logan, N., Vos, P., Eds.; Springer: Berlin/Heidelberg, Germany, 2011; Volume 27, pp. 1–29. [Google Scholar]
- Collier, F.A.; Elliot, S.L.; Ellis, R.J. Spatial variation in Bacillus thuringiensis/cereus populations within the phyllosphere of broad-leaved dock (Rumex obtusifolius) and surrounding habitats. FEMS Microbiol. Ecol. 2005, 54, 417–425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srinivasan, R.; Karaoz, U.; Volegova, M.; MacKichan, J.; Kato-Maeda, M.; Miller, S.; Nadarajan, R.; Brodie, E.L.; Lynch, S.V. Use of 16S rRNA gene for identification of a broad range of clinically relevant bacterial pathogens. PLoS ONE 2015, 10, e0117617. [Google Scholar] [CrossRef] [PubMed]
- Espariz, M.; Zuljan, F.A.; Esteban, L.; Magni, C. Taxonomic identity resolution of highly phylogenetically related strains and selection of phylogenetic markers by using genome-scale methods: The Bacillus pumilus group case. PLoS ONE 2016, 11, e0163098. [Google Scholar] [CrossRef]
- Gundlach, J.; Mehne, F.M.P.; Herzberg, C.; Kampf, J.; Valerius, O.; Kaever, V.; Stülke, J. An essential poison: Synthesis and degradation of cyclic di-AMP in Bacillus subtilis. J. Bacteriol. 2015, 197, 3265–3274. [Google Scholar] [CrossRef] [Green Version]
- Chiu, W.F.; Di, Y.P.; Choue, Y.Y.; Si, F.J. Some bacteria causing dacay of Chinese cabbage in storage. Acta Phytopathologica Sin. 1964, 7, 127–134. [Google Scholar]
- Sunar, K.; Dey, P.; Chakraborty, U.; Chakraborty, B. Biocontrol efficacy and plant growth promoting activity of Bacillus altitudinis isolated from Darjeeling hills, India. J. Basic Microbiol. 2015, 55, 91–104. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Zhou, D.; Chen, X.; Zhang, J.; Huang, H.; Wei, L. Isolation and characterization of Bacillus altitudinis JSCX-1 as a new potential biocontrol agent against Phytophthora sojae in soybean (Glycine max (L.) Merr). Plant Soil 2017, 416, 53–66. [Google Scholar] [CrossRef] [Green Version]
- Kumaravel, S.; Thankappan, S.; Raghupathi, S.; Uthandi, S. Draft genome sequence of plant growth-promoting and drought-tolerant Bacillus altitudinis FD48, isolated from rice phylloplane. Genome Announc. 2018, 6, e00019-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumvinit, A.; Akarapisan, A. Characterization of blackleg and soft rot from potato in northern Thailand. J. Phytopathol. 2019, 167, 655–666. [Google Scholar] [CrossRef]
- Liu, Q.; Li, D.; Hu, H.; Hu, H.; Huang, Q.; Zhang, Y.; Zhou, F.; Song, P.; Guan, Y.; Bu, R.; et al. First report of Bacillus altitudinis causing seed rot of pomegranate in China. Australas. Plant Pathol. 2021, 50, 427–429. [Google Scholar] [CrossRef]


| Parameter | Data for Strain | |
|---|---|---|
| Bacillus altitudinis B10-bis | Bacillus altitudinis B10 | |
| Location | Greenhouse | Field |
| DNA library prep kit | DNA Prep kit (Illumina) (2 × 300) | DNA Prep kit (Illumina) (2 × 300) |
| Sequencing metrics | ||
| No. Of total reads | 2,030,980 | 2,169,478 |
| Mean coverage (×) | 123 | 136 |
| Accession no. | ||
| GenBank | na | JAGSNB000000000 |
| Genomic data | ||
| Genome size (bp) | 3,649,248 | 3,648,564 |
| G + C content (%) | 41.27 | 41.27 |
| No. of contigs | 44 | 48 |
| N50 (bp) | 293,183 | 293,183 |
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | |
|---|---|---|---|---|---|---|---|
| 1 | - | 83.8 | 44 | 39.2 | 36.4 | 36.3 | 35.9 |
| 2 | 98.1 | - | 44 | 39.2 | 36.4 | 36.1 | 35.8 |
| 3 | 91.3 | 91.3 | - | 42.2 | 37.6 | 36.7 | 37.2 |
| 4 | 89.8 | 89.8 | 90.9 | - | 42.3 | 41.7 | 41.6 |
| 5 | 88.9 | 88.8 | 89.3 | 90.8 | - | 45.1 | 52 |
| 6 | 88.7 | 88.7 | 88.9 | 90.6 | 91.6 | - | 43.5 |
| 7 | 88.6 | 88.6 | 89.1 | 90.6 | 93.3 | 91.2 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lemjiber, N.; Naamani, K.; Merieau, A.; Dihazi, A.; Zhar, N.; Jediyi, H.; Boukerb, A.M. Identification and Genomic Characterization of Pathogenic Bacillus altitudinis from Common Pear Trees in Morocco. Agronomy 2021, 11, 1344. https://doi.org/10.3390/agronomy11071344
Lemjiber N, Naamani K, Merieau A, Dihazi A, Zhar N, Jediyi H, Boukerb AM. Identification and Genomic Characterization of Pathogenic Bacillus altitudinis from Common Pear Trees in Morocco. Agronomy. 2021; 11(7):1344. https://doi.org/10.3390/agronomy11071344
Chicago/Turabian StyleLemjiber, Naima, Khalid Naamani, Annabelle Merieau, Abdelhi Dihazi, Nawal Zhar, Hicham Jediyi, and Amine M. Boukerb. 2021. "Identification and Genomic Characterization of Pathogenic Bacillus altitudinis from Common Pear Trees in Morocco" Agronomy 11, no. 7: 1344. https://doi.org/10.3390/agronomy11071344
APA StyleLemjiber, N., Naamani, K., Merieau, A., Dihazi, A., Zhar, N., Jediyi, H., & Boukerb, A. M. (2021). Identification and Genomic Characterization of Pathogenic Bacillus altitudinis from Common Pear Trees in Morocco. Agronomy, 11(7), 1344. https://doi.org/10.3390/agronomy11071344






