Integrative Transcriptomic and Metabolomic Analysis at Organ Scale Reveals Gene Modules Involved in the Responses to Suboptimal Nitrogen Supply in Tomato
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Treatments
2.2. Analysis of Growth-Related Parameters
2.3. Photosynthetic Determinations
2.4. Metabolomic Analyses
2.5. Transcriptomic Analysis
2.6. Differential Expression and Gene Co-Expression Network Analyses
2.7. Yield Determinations
2.8. Statistical Analyses
3. Results
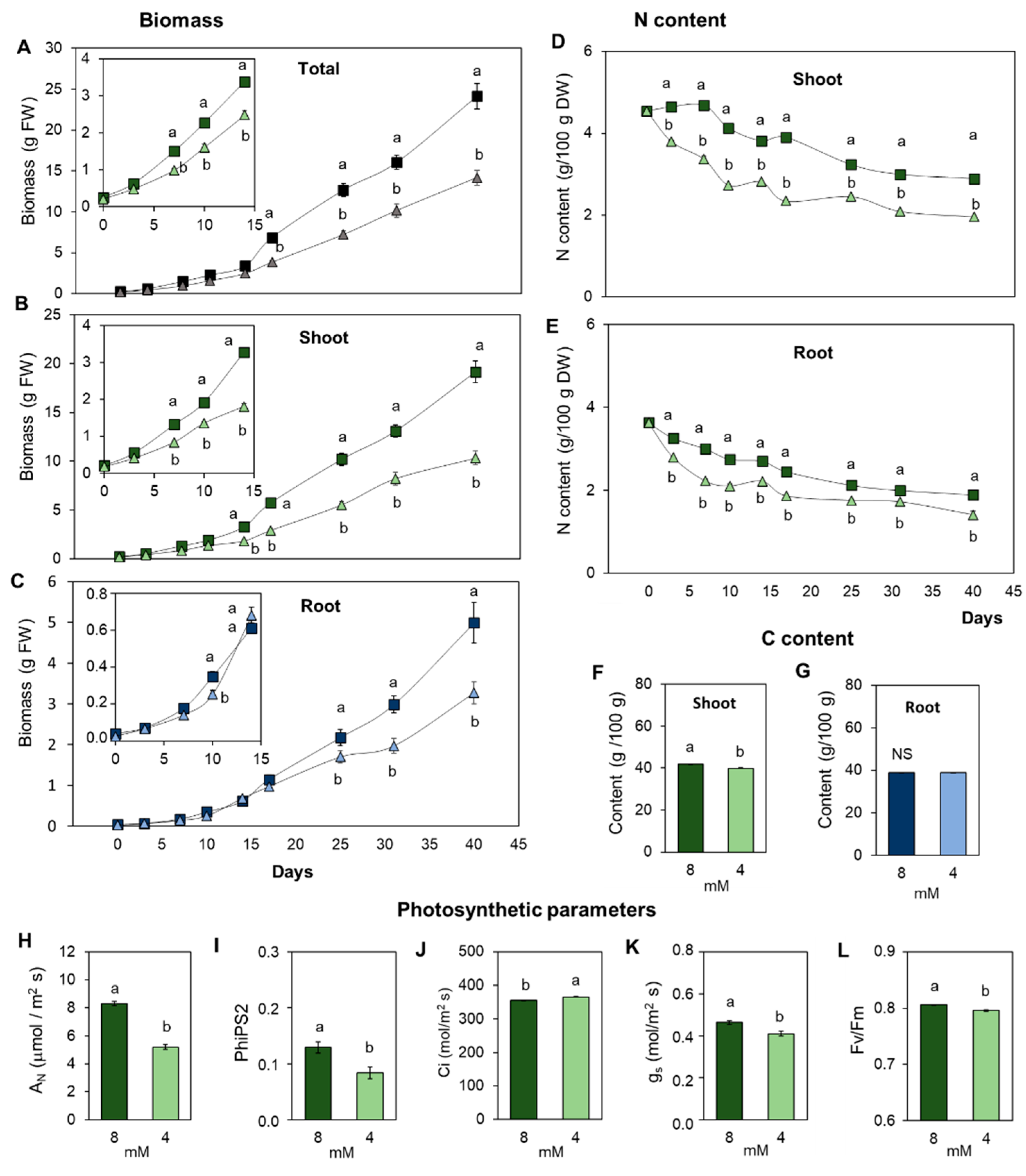
3.1. Suboptimal Nitrogen Supply Promotes Different Root and Shoot Growth Responses in Tomato
3.2. The Content of Essential Elements in the Plant Is Differentially Affected in Leaves and Roots by the Limitation in N Supply
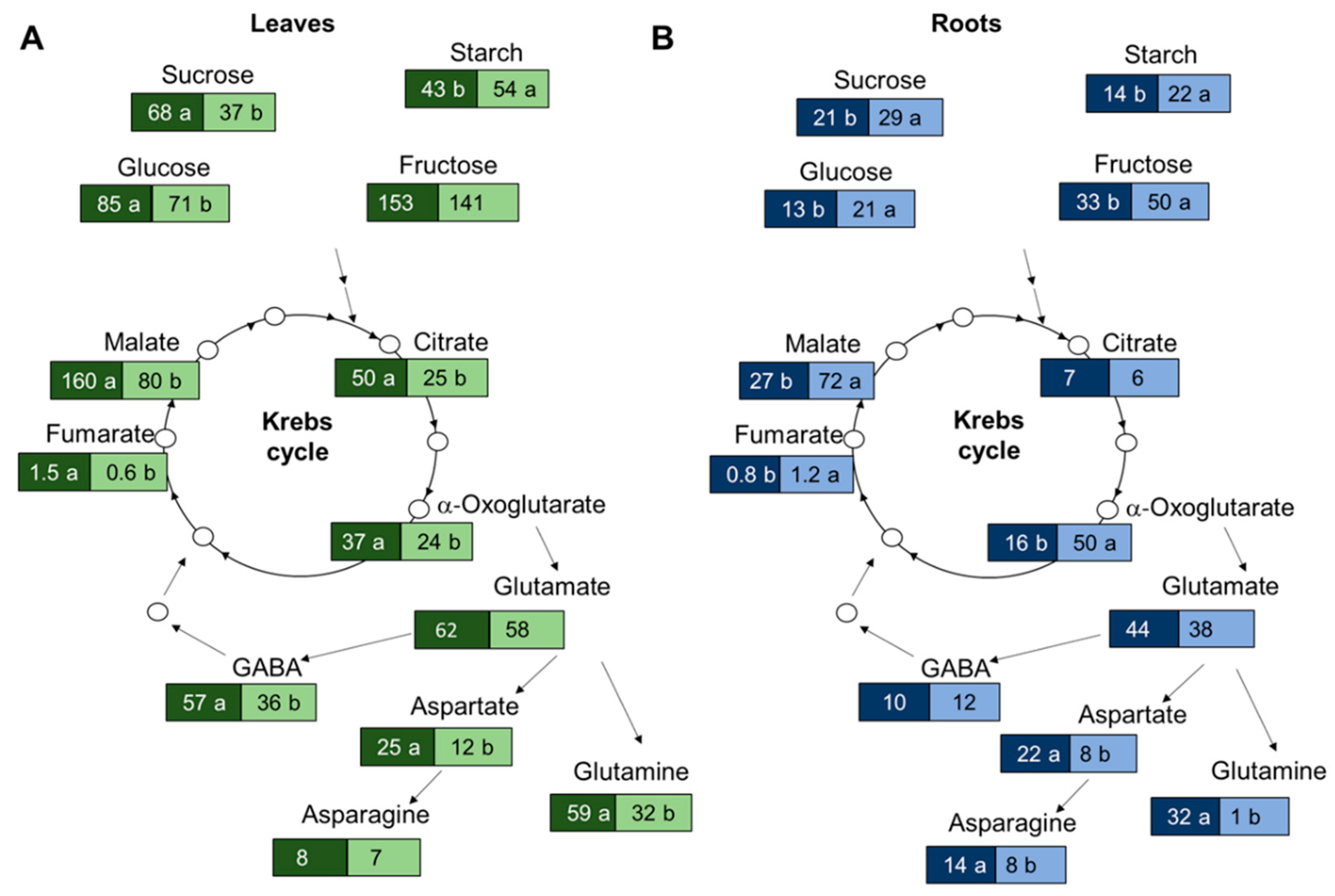
3.3. Suboptimal N Supply Promotes Different Changes in Shoot and Root C/N Metabolism
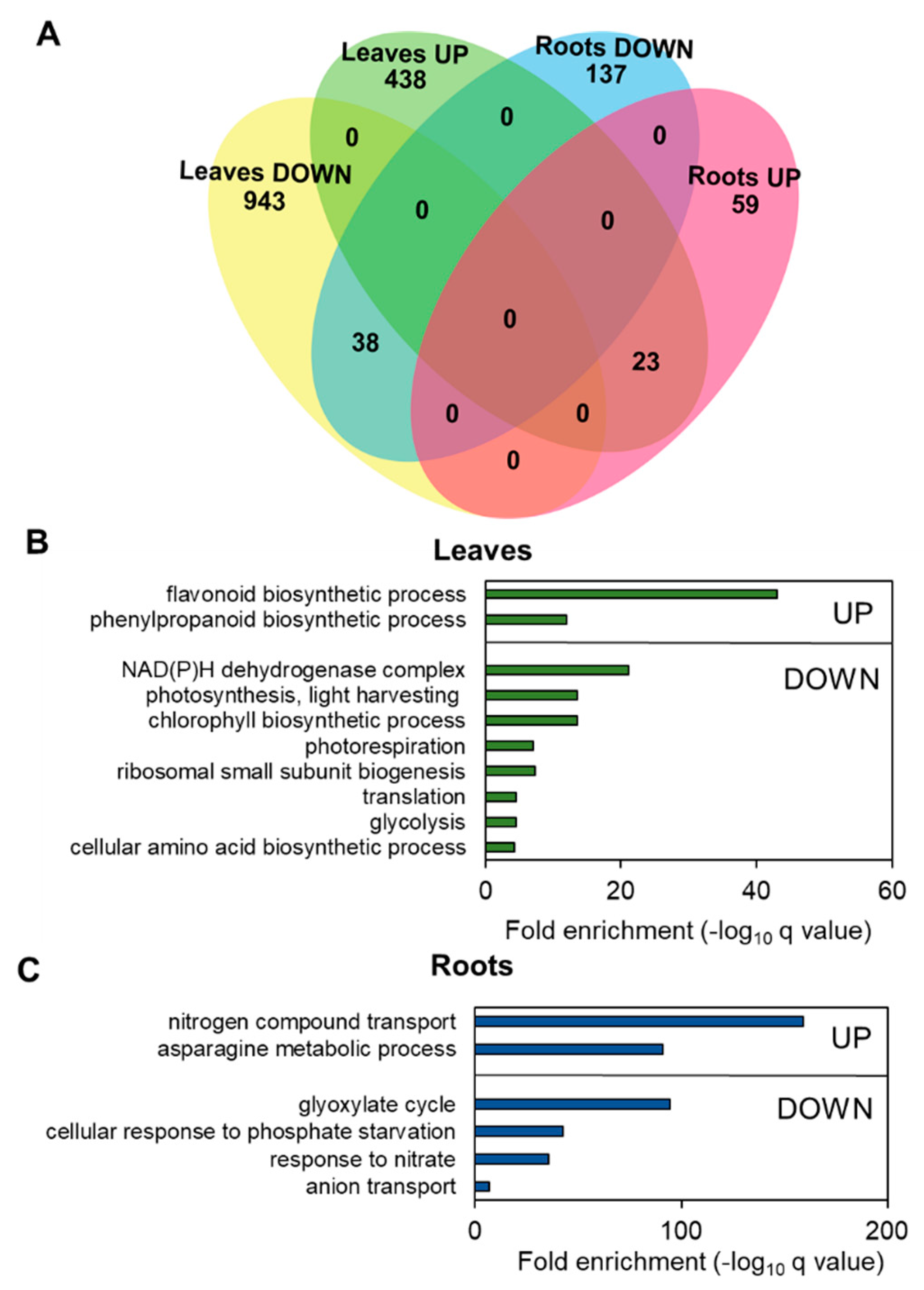
3.4. Nitrogen Limitation Promotes Specific Changes in Leaf and Root Transcriptomes
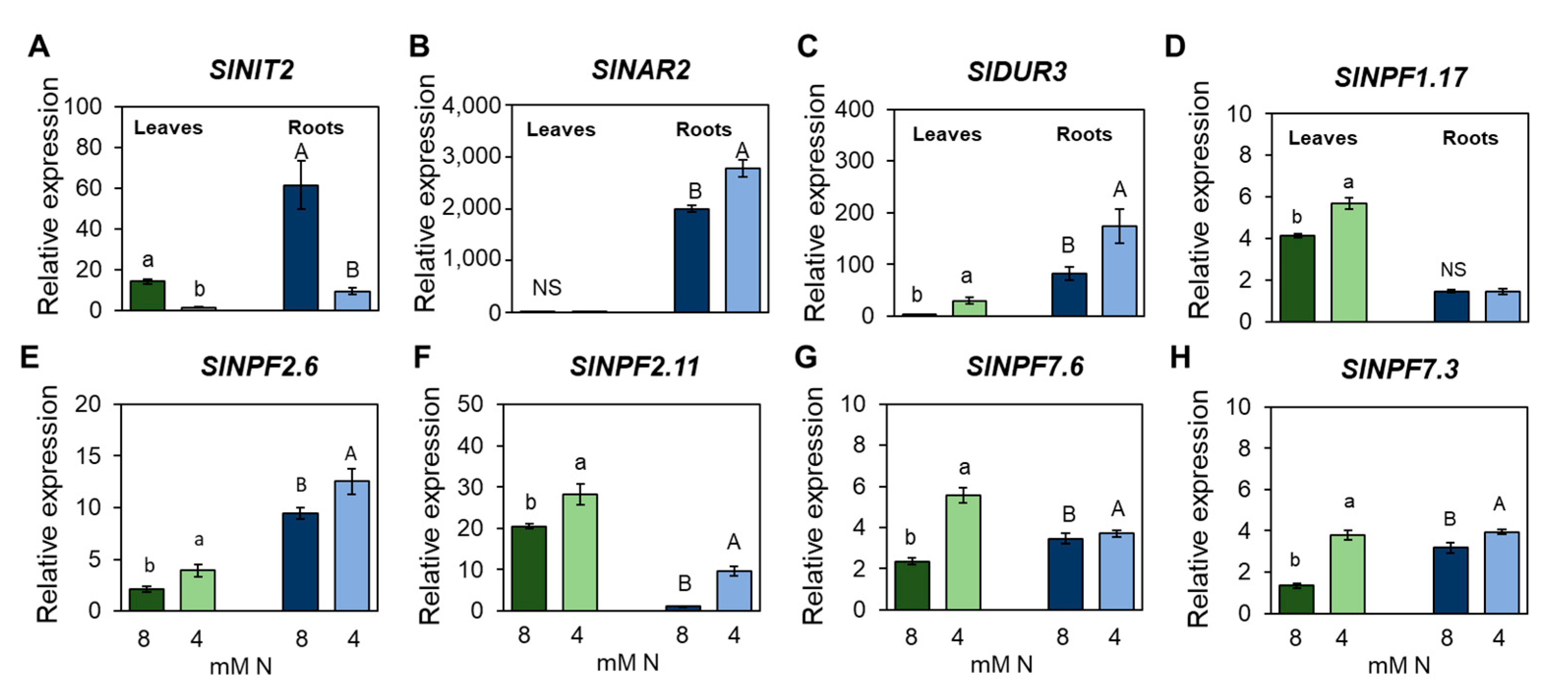
3.5. Identification of Transcription Factors Involved in the Responses to Nitrogen Limitation at Organ Scale
3.6. Transcriptomic and Metabolomic Dynamics in Roots and Leaves under N Limitation Conditions
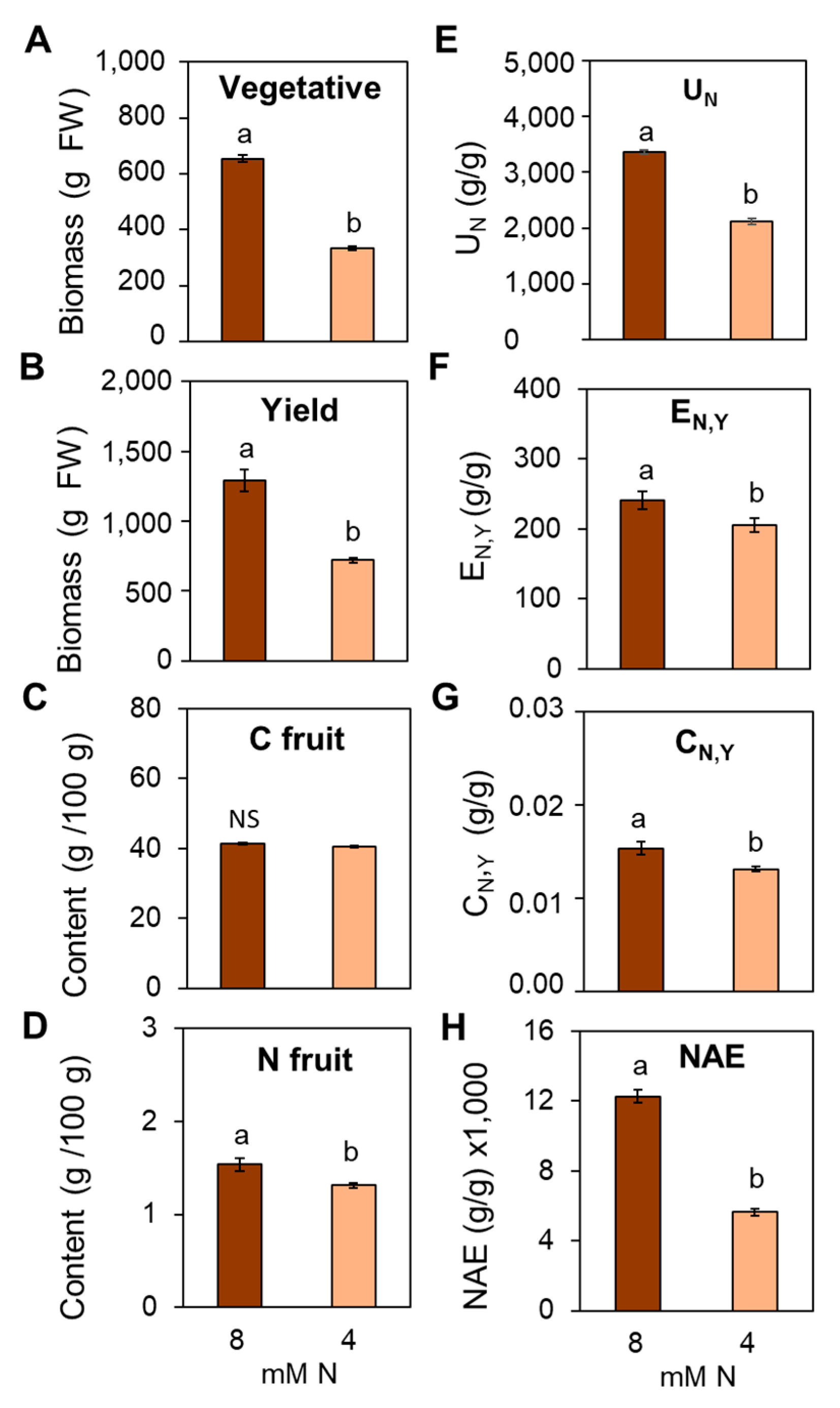
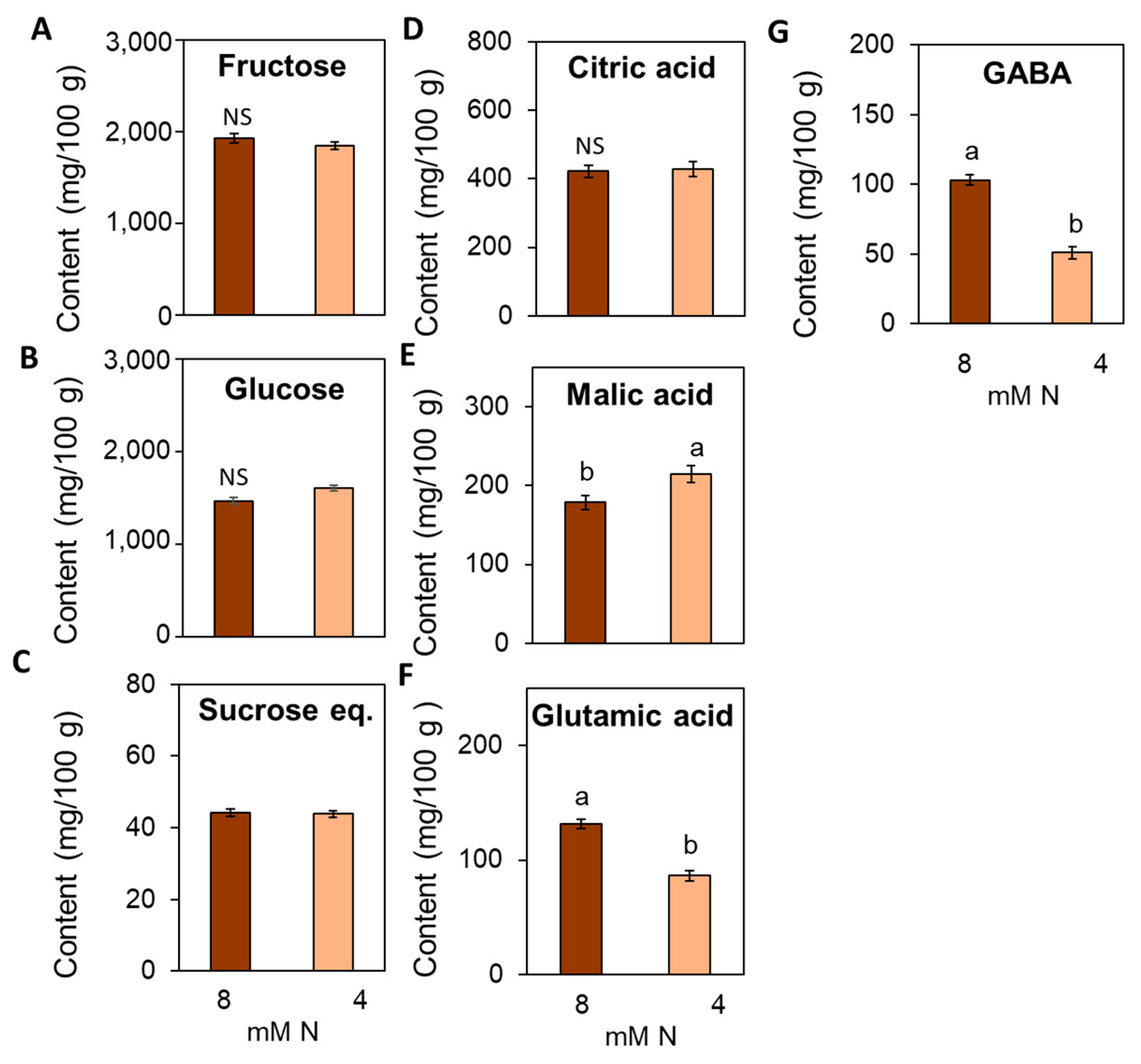
3.7. Suboptimal N Supply Promotes Significant Changes in Yield and Fruit Quality in Tomato
4. Discussion
4.1. Organ-Specific Adaptation of the Primary Metabolism to N Availability in Tomato
4.2. Long-Term Expression Analyses Identified Several Categories Involved in N Responses in Tomato
4.3. The Transcriptomic Analysis Revealed a Group of Candidate Transcription Factors Involved in Responses to N Deficiency
4.4. Impact of Suboptimal N Supply on Yield and Fruit Quality
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Olson, R.A.; Kurtz, L.T. Crop Nitrogen Requirements, Utilization, and Fertilization. In Nitrogen in Agricultural Soils; Stevenson, F.J., Ed.; American Society of Agronomy: Madison, WI, USA, 1982; Volume 22, pp. 567–604. [Google Scholar]
- Blumenthal, J.M.; Baltensperger, D.D.; Cassman, K.G.; Mason, S.C.; Pavlista, A.D. Importance and Effect of Nitrogen on Crop Quality and Health. In Nitrogen in the Environment; Hatfield, J.L., Follett, R.F., Eds.; Elsevier: Amsterdam, The Netherlands, 2008; Volume 3, pp. 51–70. [Google Scholar] [CrossRef] [Green Version]
- Kraiser, T.; Gras, D.E.; Gutierrez, A.G.; González, B.; Gutiérrez, R.A. A Holistic View of Nitrogen Acquisition in Plants. J. Exp. Bot. 2011, 62, 1455–1466. [Google Scholar] [CrossRef]
- Robertson, G.P.; Vitousek, P.M. Nitrogen in Agriculture: Balancing the Cost of an Essential Resource. Annu. Rev. Environ. Resour. 2009, 34, 97–125. [Google Scholar] [CrossRef] [Green Version]
- Food and Agriculture Organization of The United Nations FAO. Statistics Database; Food and Agriculture Organization of The United Nations: Rome, Italy; Available online: http//www.fao.org/faostat/en/#data (accessed on 7 January 2021).
- Mogollón, J.M.; Lassaletta, L.; Beusen, A.; van Grinsven, H.J.M.; Westhoek, H.; Bouwman, A.F. Assessing Future Reactive Nitrogen Inputs into Global Croplands Based on the Shared Socioeconomic Pathways. Environ. Res. Lett. 2018, 13, 044008. [Google Scholar] [CrossRef]
- Li, H.; Hu, B.; Chu, C. Nitrogen Use Efficiency in Crops: Lessons from Arabidopsis and Rice. J. Exp. Bot. 2017, 68, 2477–2488. [Google Scholar] [CrossRef]
- Smil, V. Nitrogen in Crop Production: An Account of Global Flows. Glob. Biogeochem. Cycles 1999, 13, 647–662. [Google Scholar] [CrossRef] [Green Version]
- van Bueren, E.T.L.; Struik, P.C. Diverse Concepts of Breeding for Nitrogen Use Efficiency. A Review. Agron. Sustain. Dev. 2017, 37, 50. [Google Scholar] [CrossRef] [Green Version]
- Frusciante, L.; Carli, P.; Ercolano, M.R.; Pernice, R.; Di Matteo, A.; Fogliano, V.; Pellegrini, N. Antioxidant Nutritional Quality of Tomato. Mol. Nutr. Food Res. 2007, 51, 609–617. [Google Scholar] [CrossRef] [PubMed]
- Nicola, S.; Tibaldi, G.; Fontana, E. Tomato Production Systems and Their Application to the Tropics. In Proceedings of the International Symposium on Tomato in the Tropics, Villa de Leyva, Colombia, 9–13 September 2008; Fischer, G., Magnitskiy, S., Nicola, S., Eds.; ISHS: Leuven, Belgium, 2009. [Google Scholar]
- Zotarelli, L.; Scholberg, J.M.; Dukes, M.D.; Muñoz-Carpena, R.; Icerman, J. Tomato Yield, Biomass Accumulation, Root Distribution and Irrigation Water Use Efficiency on a Sandy Soil, As Affected by Nitrogen Rate and Irrigation Scheduling. Agric. Water Manag. 2009, 96, 23–34. [Google Scholar] [CrossRef]
- Elia, A.; Conversa, G. Agronomic and Physiological Responses of a Tomato Crop to Nitrogen Input. Eur. J. Agron. 2012, 40, 64–74. [Google Scholar] [CrossRef]
- Ronga, D.; Parisi, M.; Pentangelo, A.; Mori, M.; Di Mola, I. Effects of Nitrogen Management on Biomass Production and Dry Matter Distribution of Processing Tomato Cropped in Southern Italy. Agronomy 2019, 9, 855. [Google Scholar] [CrossRef] [Green Version]
- Bénard, C.; Gautier, H.; Bourgaud, F.; Grasselly, D.; Navez, B.; Caris-Veyrat, C.; Weiss, M.; Génard, M. Effects of Low Nitrogen Supply on Tomato (Solanum lycopersicum) Fruit Yield and Quality with Special Emphasis on Sugars, Acids, Ascorbate, Carotenoids, and Phenolic Compounds. J. Agric. Food Chem. 2009, 57, 4112–4123. [Google Scholar] [CrossRef] [PubMed]
- Truffault, V.; Ristorto, M.; Brajeul, E.; Vercambre, G.; Gautier, H. To Stop Nitrogen Overdose in Soilless Tomato Crop: A Way to Promote Fruit Quality Without Affecting Fruit Yield. Agronomy 2019, 9, 80. [Google Scholar] [CrossRef] [Green Version]
- Hirel, B.; Tetu, T.; Lea, P.J.; Dubois, F. Improving Nitrogen Use Efficiency in Crops for Sustainable Agriculture. Sustainability 2011, 3, 1452–1485. [Google Scholar] [CrossRef]
- Abenavoli, M.R.; Longo, C.; Lupini, A.; Miller, T.; Araniti, F.; Mercati, F.; Princi, M.P.; Sunseri, F. Phenotyping Two Tomato Genotypes with Different Nitrogen Use Efficiency. Plant Physiol. Biochem. 2016, 107, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Gu, F.; Chen, J.; Yang, H.; Jiang, J.; Du, T.; Zhang, J. Assessing the Response of Yield and Comprehensive Fruit Quality of Tomato Grown in Greenhouse to Deficit Irrigation and Nitrogen Application Strategies. Agric. Water Manag. 2015, 161, 9–19. [Google Scholar] [CrossRef]
- Kant, S. Understanding Nitrate Uptake, Signaling and Remobilisation for Improving Plant Nitrogen Use Efficiency. Semin. Cell Dev. Biol. 2018, 74, 89–96. [Google Scholar] [CrossRef]
- Iqbal, A.; Qiang, D.; Alamzeb, M.; Xiangru, W.; Huiping, G.; Hengheng, Z.; Nianchang, P.; Xiling, Z.; Meizhen, S. Untangling the Molecular Mechanisms and Functions of Nitrate to Improve Nitrogen Use Efficiency. J. Sci. Food Agric. 2020, 100, 904–914. [Google Scholar] [CrossRef]
- Tegeder, M.; Masclaux-Daubresse, C. Source and Sink Mechanisms of Nitrogen Transport and Use. New Phytol. 2018, 217, 35–53. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.-H.; Köhler, B.; Cao, F.-Q.; Liu, L.-H. Molecular and Physiological Aspects of Urea Transport in Higher Plants. Plant Sci. 2008, 175, 467–477. [Google Scholar] [CrossRef]
- Yoneyama, T.; Suzuki, A. Light-independent nitrogen assimilation in plant leaves: Nitrate incorporation into glutamine, glutamate, aspartate, and asparagine traced by n-15. Plants 2020, 9, 1303. [Google Scholar] [CrossRef]
- McAllister, C.H.; Beatty, P.H.; Good, A.G. Engineering Nitrogen Use Efficient Crop Plants: The Current Status. Plant Biotechnol. J. 2012, 10, 1011–1025. [Google Scholar] [CrossRef]
- Plett, D.; Garnett, T.; Okamoto, M. Molecular Genetics to Discover and Improve Nitrogen Use Efficiency in Crop Plants. In Plant Macronutrient Use Efficiency; Hossain, M.A., Kamiya, T., Burritt, D.J., Phan Tran, L.-S., Fujiwara, T., Eds.; Elsevier: Amsterdam, The Netherlands, 2017; Volume 6, pp. 93–122. [Google Scholar]
- Lupini, A.; Princi, M.P.; Araniti, F.; Miller, T.; Sunseri, F.; Abenavoli, M.R. Physiological and Molecular Responses in Tomato under Different Forms of N Nutrition. J. Plant Physiol. 2017, 216, 17–25. [Google Scholar] [CrossRef]
- Lauter, F.R.; Ninnemann, O.; Bucher, M.; Riesmeier, J.W.; Frommer, W. Preferential Expression of an Ammonium Transporter and of Two Putative Nitrate Transporters in Root Hairs of Tomato. Proc. Natl. Acad. Sci. USA 1996, 93, 8139–8144. [Google Scholar] [CrossRef] [Green Version]
- Ono, F.; Frommer, W.B.; Wirén, N. Coordinated Diurnal Regulation of Low- and High-Affinity Nitrate Transporters in Tomato. Plant Biol. 2000, 2, 17–23. [Google Scholar] [CrossRef]
- Wang, Y.-H.; Garvin, D.F.; Kochian, L.V. Nitrate-Induced Genes in Tomato Roots. Array Analysis Reveals Novel Genes That May Play a Role in Nitrogen Nutrition. Plant Physiol. 2001, 127, 345–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, Y.; Yi, H.; Bao, J.; Gong, J. LeNRT2.3 Functions in Nitrate Acquisition and Long-Distance Transport in Tomato. FEBS Lett. 2015, 589, 1072–1079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forde, B.G. Nitrogen Signalling Pathways Shaping Root System Architecture: An Update. Curr. Opin. Plant Biol. 2014, 21, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Masclaux-Daubresse, C.; Daniel-Vedele, F.; Dechorgnat, J.; Chardon, F.; Gaufichon, L.; Suzuki, A. Nitrogen Uptake, Assimilation and Remobilization in Plants: Challenges for Sustainable and Productive Agriculture. Ann. Bot. 2010, 105, 1141–1157. [Google Scholar] [CrossRef] [Green Version]
- Scheible, W.-R.; Morcuende, R.; Czechowski, T.; Fritz, C.; Osuna, D.; Palacios-Rojas, N.; Schindelasch, D.; Thimm, O.; Udvardi, M.K.; Stitt, M. Genome-Wide Reprogramming of Primary and Secondary Metabolism, Protein Synthesis, Cellular Growth Processes, and the Regulatory Infrastructure of Arabidopsis in Response to Nitrogen. Plant Physiol. 2004, 136, 2483–2499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krapp, A.; Berthomé, R.; Orsel, M.; Mercey-Boutet, S.; Yu, A.; Castaings, L.; Elftieh, S.; Major, H.; Renou, J.-P.; Daniel-Vedele, F. Arabidopsis Roots and Shoots Show Distinct Temporal Adaptation Patterns Toward Nitrogen Starvation. Plant Physiol. 2011, 157, 1255–1282. [Google Scholar] [CrossRef] [Green Version]
- Vidal, E.; Álvarez, J.M.; Moyano, T.C.; Gutiérrez, R.A. Transcriptional Networks in the Nitrate Response of Arabidopsis Thaliana. Curr. Opin. Plant Biol. 2015, 27, 125–132. [Google Scholar] [CrossRef]
- Wang, Y.-Y.; Cheng, Y.H.; Chen, K.-E.; Tsay, Y.-F. Nitrate Transport, Signaling, and Use Efficiency. Annu. Rev. Plant Biol. 2018, 69, 85–122. [Google Scholar] [CrossRef]
- Vidal, E.A.; Alvarez, J.M.; Araus, V.; Riveras, E.; Brooks, M.D.; Krouk, G.; Ruffel, S.; Lejay, L.; Crawford, N.M.; Coruzzi, G.M.; et al. Nitrate in 2020: Thirty Years from Transport to Signaling Networks. Plant Cell 2020, 32, 2094–2119. [Google Scholar] [CrossRef] [PubMed]
- Gaudinier, A.; Rodriguez-Medina, J.; Zhang, L.; Olson, A.; Liseron-Monfils, C.; Bågman, A.-M.; Foret, J.; Abbitt, S.; Tang, M.; Li, B.; et al. Transcriptional Regulation of Nitrogen-Associated Metabolism and Growth. Nat. Cell Biol. 2018, 563, 259–264. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Quan, S.; Qi, S.; Xu, N.; Wang, Y. Novel Aspects of Nitrate Regulation in Arabidopsis. Front. Plant Sci. 2020, 11. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, Y.; Wu, W. Potassium and Phosphorus Transport and Signaling in Plants. J. Integr. Plant Biol. 2021, 63, 34–52. [Google Scholar] [CrossRef]
- Varala, K.; Marshall-Colón, A.; Cirrone, J.; Brooks, M.D.; Pasquino, A.V.; Léran, S.; Mittal, S.; Rock, T.M.; Edwards, M.B.; Kim, G.J.; et al. Temporal Transcriptional Logic of Dynamic Regulatory Networks Underlying Nitrogen Signaling and Use in Plants. Proc. Natl. Acad. Sci. USA 2018, 115, 6494–6499. [Google Scholar] [CrossRef] [Green Version]
- Brooks, M.D.; Cirrone, J.; Pasquino, A.V.; Alvarez, J.M.; Swift, J.; Mittal, S.; Juang, C.-L.; Varala, K.; Gutiérrez, R.A.; Krouk, G.; et al. Network Walking Charts Transcriptional Dynamics of Nitrogen Signaling by Integrating Validated and Predicted Genome-Wide Interactions. Nat. Commun. 2019, 10, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Hu, B.; Chu, C. Towards Understanding the Hierarchical Nitrogen Signalling Network in Plants. Curr. Opin. Plant Biol. 2020, 55, 60–65. [Google Scholar] [CrossRef]
- Luo, J.; Havé, M.; Clément, G.; Tellier, F.; Balliau, T.; Launay-Avon, A.; Guérard, F.; Zivy, M.; Masclaux-Daubresse, C. Integrating Multiple Omics to Identify Common and Specific Molecular Changes Occurring in Arabidopsis under Chronic Nitrate and Sulfate Limitations. J. Exp. Bot. 2020, 71, 6471–6490. [Google Scholar] [CrossRef] [PubMed]
- Balazadeh, S.; Schildhauer, J.; Araújo, W.L.; Munné-Bosch, S.; Fernie, A.R.; Proost, S.; Humbeck, K.; Mueller-Roeber, B. Reversal of Senescence by N Resupply to N-Starved Arabidopsis Thaliana: Transcriptomic and Metabolomic Consequences. J. Exp. Bot. 2014, 65, 3975–3992. [Google Scholar] [CrossRef] [Green Version]
- Curci, P.L.; Cigliano, R.A.; Zuluaga, D.L.; Janni, M.; Sanseverino, W.; Sonnante, G. Transcriptomic Response of Durum Wheat to Nitrogen Starvation. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef]
- Gupta, S.; Yadav, B.; Raj, U.; Freilich, S.; Varadwaj, P.K. Transcriptomic Analysis of Soil Grown T. Aestivum Cv. Root to Reveal the Changes in Expression of Genes in Response to Multiple Nutrients Deficiency. Front. Plant Sci. 2017, 8, 1025. [Google Scholar] [CrossRef] [Green Version]
- Shin, S.-Y.; Jeong, J.S.; Lim, J.Y.; Kim, T.; Park, J.H.; Kim, J.-K.; Shin, C. Transcriptomic Analyses of Rice (Oryza Sativa) Genes and Non-Coding RNAs under Nitrogen Starvation Using Multiple Omics Technologies. BMC Genom. 2018, 19, 1–20. [Google Scholar] [CrossRef]
- Su, H.; Zhang, X.; He, Y.; Li, L.; Wang, Y.; Hong, G.; Xu, P. Transcriptomic Analysis Reveals the Molecular Adaptation of Three Major Secondary Metabolic Pathways to Multiple Macronutrient Starvation in Tea (Camellia sinensis). Genes 2020, 11, 241. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Ding, G.; Yang, N.; White, P.J.; Ye, X.; Cai, H.; Lu, J.; Shi, L.; Xu, F. Comparative Genome and Transcriptome Analysis Unravels Key Factors of Nitrogen Use Efficiency in Brassica Napus L. Plant Cell Environ. 2020, 43, 712–731. [Google Scholar] [CrossRef]
- Gálvez, J.H.; Tai, H.H.; Lagüe, M.; Zebarth, B.J.; Strömvik, M.V. The Nitrogen Responsive Transcriptome in Potato (Solanum Tuberosum L.) Reveals Significant Gene Regulatory Motifs. Sci. Rep. 2016, 6, 26090. [Google Scholar] [CrossRef] [Green Version]
- Tiwari, J.K.; Buckseth, T.; Zinta, R.; Saraswati, A.; Singh, R.K.; Rawat, S.; Dua, V.K.; Chakrabarti, S.K. Transcriptome Analysis of Potato Shoots, Roots and Stolons under Nitrogen Stress. Sci. Rep. 2020, 10, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Wang, Y.; Zhao, Y.; Zhang, Y.; Zhang, J.; Ma, H.; Han, Y. Transcriptome Analysis Reveals Nitrogen Deficiency Induced Alterations in Leaf and Root of Three Cultivars of Potato (Solanum tuberosum L.). PLoS ONE 2020, 15, e0240662. [Google Scholar] [CrossRef]
- Canales, J.; Uribe, F.; Henríquez-Valencia, C.; Lovazzano, C.; Medina, J.; Vidal, E.A. Transcriptomic Analysis at Organ and Time Scale Reveals Gene Regulatory Networks Controlling the Sulfate Starvation Response of Solanum Lycopersicum. BMC Plant Biol. 2020, 20, 1–17. [Google Scholar] [CrossRef]
- Hoagland, D.R.; Arnon, D.I. The Water-Culture Method for Growing Plants without Soil; Californian Agricultural Experimental Station; Circular No. 347; University of California: Berkeley, CA, USA, 1950. [Google Scholar]
- Dumas, Y.; Leoni, C.; Portas, C.A.M.; Bièche, B. Influence of water and nitrogen availability on yield and quality of processing tomato in the european union countries. Acta Hortic. 1993, 376, 185–192. [Google Scholar] [CrossRef]
- Bruggink, G. A Comparative Analysis of the Influence of Light on Growth of Young Tomato and Carnation Plants. Sci. Hortic. 1992, 51, 71–81. [Google Scholar] [CrossRef]
- Renau-Morata, B.; Molina, R.V.; Carrillo, L.; Cebolla-Cornejo, J.; Sánchez-Perales, M.; Pollmann, S.; Domínguez-Figueroa, J.; Corrales, A.R.; Flexas, J.; Vicente-Carbajosa, J.; et al. Ectopic Expression of CDF3 Genes in Tomato Enhances Biomass Production and Yield under Salinity Stress Conditions. Front. Plant Sci. 2017, 8, 660. [Google Scholar] [CrossRef] [Green Version]
- Cebolla-Cornejo, J.; Valcárcel, M.; Herrero-Martínez, J.M.; Roselló, S.; Nuez, F. High Efficiency Joint CZE Determination of Sugars and Acids in Vegetables and Fruits. Electrophoresis 2012, 33, 2416–2423. [Google Scholar] [CrossRef] [PubMed]
- Galiana-Balaguer, L.; Ibáñez, G.; Cebolla-Cornejo, J.; Roselló, S. Evaluation of Germplasm in Solanum Section Lycopersicon for Tomato Taste Improvement. Turk. J. Agric. For. 2018, 42, 309–321. [Google Scholar] [CrossRef]
- Martí, R.; Valcárcel, M.; Herrero-Martínez, J.M.; Cebolla-Cornejo, J.; Roselló, S. Fast Simultaneous Determination of Prominent Polyphenols in Vegetables and Fruits by Reversed Phase Liquid Chromatography Using a Fused-Core Column. Food Chem. 2015, 169, 169–179. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. Stringtie enables improved reconstruction of a transcriptome from rna-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Tian, T.; Liu, Y.; Yan, H.; You, Q.; Yi, X.; Du, Z.; Xu, W.; Su, Z. AgriGO v2.0: A GO Analysis Toolkit for the Agricultural Community, 2017 Update. Nucleic Acids Res. 2017, 45, W122–W129. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R Package for Weighted Correlation Network Analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [Green Version]
- Lemoine, G.G.; Scott-Boyer, M.-P.; Ambroise, B.; Périn, O.; Droit, A. GWENA: Gene Co-Expression Networks Analysis and Extended Modules Characterization in a Single Bioconductor Package. BMC Bioinform. 2021, 22, 267. [Google Scholar] [CrossRef] [PubMed]
- Weih, M. A Calculation Tool for Analyzing Nitrogen Use Efficiency in Annual and Perennial Crops. Agronomy 2014, 4, 470–477. [Google Scholar] [CrossRef] [Green Version]
- Domínguez-Figueroa, J.; Carrillo, L.; Renau-Morata, B.; Yang, L.; Molina, R.-V.; Marino, D.; Canales, J.; Weih, M.; Vicente-Carbajosa, J.; Nebauer, S.G.; et al. The Arabidopsis Transcription Factor CDF3 Is Involved in Nitrogen Responses and Improves Nitrogen Use Efficiency in Tomato. Front. Plant Sci. 2020, 11. [Google Scholar] [CrossRef]
- Wahle, E.A.; Masiunas, J.B. Population Density and Nitrogen Fertility Effects on Tomato Growth and Yield. HortScience 2003, 38, 367–372. [Google Scholar] [CrossRef] [Green Version]
- Coruzzi, G.; Bush, D. Nitrogen and Carbon Nutrient and Metabolite Signaling in Plants. Plant Physiol. 2001, 125, 61–64. [Google Scholar] [CrossRef] [Green Version]
- Foyer, C.H.; Parry, M.; Noctor, G. Markers and Signals Associated with Nitrogen Assimilation in Higher Plants. J. Exp. Bot. 2003, 54, 585–593. [Google Scholar] [CrossRef] [PubMed]
- Bouain, N.; Krouk, G.; Lacombe, B.; Rouached, H. Getting to the Root of Plant Mineral Nutrition: Combinatorial Nutrient Stresses Reveal Emergent Properties. Trends Plant Sci. 2019, 24, 542–552. [Google Scholar] [CrossRef]
- Riedell, W.E.; Pikul, J.L.; Jaradat, A.A.; Schumacher, T.E. Crop Rotation and Nitrogen Input Effects on Soil Fertility, Maize Mineral Nutrition, Yield, and Seed Composition. Agron. J. 2009, 101, 870–879. [Google Scholar] [CrossRef] [Green Version]
- Nunes-Nesi, A.; Fernie, A.R.; Stitt, M. Metabolic and Signaling Aspects Underpinning the Regulation of Plant Carbon Nitrogen Interactions. Mol. Plant 2010, 3, 973–996. [Google Scholar] [CrossRef]
- Tschoep, H.; Gibon, Y.; Carillo, P.; Armengaud, P.; Szecowka, M.; Nunes-Nesi, A.; Fernie, A.R.; Koehl, K.; Stitt, M. Adjustment of Growth and Central Metabolism to a Mild But Sustained Nitrogen-Limitation InArabidopsis. Plant Cell Environ. 2009, 32, 300–318. [Google Scholar] [CrossRef]
- Chen, K.-E.; Chen, H.-Y.; Tseng, C.-S.; Tsay, Y.-F. Improving Nitrogen Use Efficiency by Manipulating Nitrate Remobilization in Plants. Nat. Plants 2020, 6, 1126–1135. [Google Scholar] [CrossRef] [PubMed]
- Xuan, W.; Beeckman, T.; Xu, G. Plant Nitrogen Nutrition: Sensing and Signaling. Curr. Opin. Plant Biol. 2017, 39, 57–65. [Google Scholar] [CrossRef]
- Jiang, L.; Ball, G.; Hodgman, C.; Coules, A.; Zhao, H.; Lu, C. Analysis of Gene Regulatory Networks of Maize in Response to Nitrogen. Genes 2018, 9, 151. [Google Scholar] [CrossRef] [Green Version]
- Zuluaga, D.L.; Sonnante, G. The Use of Nitrogen and Its Regulation in Cereals: Structural Genes, Transcription Factors, and the Role of MiRNAs. Plants 2019, 8, 294. [Google Scholar] [CrossRef] [Green Version]
- Ueda, Y.; Ohtsuki, N.; Kadota, K.; Tezuka, A.; Nagano, A.J.; Kadowaki, T.; Kim, Y.; Miyao, M.; Yanagisawa, S. Gene Regulatory Network and Its Constituent Transcription Factors That Control nitrogen-deficiency Responses in Rice. New Phytol. 2020, 227, 1434–1452. [Google Scholar] [CrossRef] [PubMed]
- Schwacke, R.; Soto, G.Y.P.; Krause, K.; Bolger, A.M.; Arsova, B.; Hallab, A.; Gruden, K.; Stitt, M.; Bolger, M.; Usadel, B. MapMan4: A Refined Protein Classification and Annotation Framework Applicable to Multi-Omics Data Analysis. Mol. Plant 2019, 12, 879–892. [Google Scholar] [CrossRef] [PubMed]
- Martí, R.; Roselló, S.; Cebolla-Cornejo, J. Tomato As a Source of Carotenoids and Polyphenols Targeted to Cancer Prevention. Cancers 2016, 8, 58. [Google Scholar] [CrossRef]
- Zheng, Z.-L. Carbon and Nitrogen Nutrient Balance Signaling in Plants. Plant Signal. Behav. 2009, 4, 584–591. [Google Scholar] [CrossRef] [Green Version]
- Vega, A.; O’Brien, J.A.; Gutiérrez, R.A. Nitrate and Hormonal Signaling Crosstalk for Plant Growth and Development. Curr. Opin. Plant Biol. 2019, 52, 155–163. [Google Scholar] [CrossRef]
- Kant, S.; Bi, Y.-M.; Rothstein, S.J. Understanding Plant Response to Nitrogen Limitation for the Improvement of Crop Nitrogen Use Efficiency. J. Exp. Bot. 2010, 62, 1499–1509. [Google Scholar] [CrossRef] [Green Version]
- Sung, J.; Lee, S.; Lee, Y.; Ha, S.; Song, B.; Kim, T.; Waters, B.; Krishnan, H.B. Metabolomic Profiling from Leaves and Roots of Tomato (Solanum Lycopersicum L.) Plants Grown under Nitrogen, Phosphorus or Potassium-Deficient Condition. Plant Sci. 2015, 241, 55–64. [Google Scholar] [CrossRef]
- Xin, W.; Zhang, L.; Zhang, W.; Gao, J.; Yi, J.; Zhen, X.; Li, Z.; Zhao, Y.; Peng, C.; Zhao, C. An Integrated Analysis of the Rice Transcriptome and Metabolome Reveals Differential Regulation of Carbon and Nitrogen Metabolism in Response to Nitrogen Availability. Int. J. Mol. Sci. 2019, 20, 2349. [Google Scholar] [CrossRef] [Green Version]
- Guidi, L.; Lorefice, G.; Pardossi, A.; Malorgio, F.; Tognoni, F.; Soldatini, G. Growth and Photosynthesis of Lycopersicon Esculentum (L.) Plants As Affected by Nitrogen Deficiency. Biol. Plant. 1997, 39, 235–244. [Google Scholar] [CrossRef]
- Hikosaka, K. Interspecific Difference in the photosynthesis nitrogen relationship: Patterns, Physiological Causes, and Ecological Importance. J. Plant Res. 2004, 117, 481–494. [Google Scholar] [CrossRef]
- Rao, I.M.; Terry, N. Photosynthetic Adaptation to Nutrient Stress. In Probing Photosynthesis; Yunus, M., Pathre, U., Mohanty, P., Eds.; Taylor & Francis: London, UK, 2000; Volume 21, pp. 379–397. [Google Scholar]
- Reuscher, S.; Akiyama, M.; Yasuda, T.; Makino, H.; Aoki, K.; Shibata, D.; Shiratake, K. The Sugar Transporter Inventory of Tomato: Genome-Wide Identification and Expression Analysis. Plant Cell Physiol. 2014, 55, 1123–1141. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Ruan, Y.-L. Shoot–root Carbon Allocation, Sugar Signalling and Their Coupling with Nitrogen Uptake and Assimilation. Funct. Plant Biol. 2016, 43, 105–113. [Google Scholar] [CrossRef]
- Valle, E.M.; Boggio, S.B.; Heldt, H.W. Free Amino Acid Composition of Phloem Sap and Growing Fruit of Lycopersicon Esculentum. Plant Cell Physiol. 1998, 39, 458–461. [Google Scholar] [CrossRef] [Green Version]
- Fritz, C.; Palacios-Rojas, N.; Feil, R.; Stitt, M. Regulation of Secondary Metabolism by the Carbon-Nitrogen Status in Tobacco: Nitrate Inhibits Large Sectors of Phenylpropanoid Metabolism. Plant J. 2006, 46, 533–548. [Google Scholar] [CrossRef]
- Scheible, W.-R.; Lauerer, M.; Schulze, E.-D.; Caboche, M.; Stitt, M. Accumulation of Nitrate in the Shoot Acts As a Signal to Regulate Shoot-Root Allocation in Tobacco. Plant J. 1997, 11, 671–691. [Google Scholar] [CrossRef] [Green Version]
- Urbanczyk-Wochniak, E.; Fernie, A.R. Metabolic profiling reveals altered nitrogen nutrient regimes have diverse effects on the metabolism of hydroponically-grown tomato (Solanum lycopersicum) plants. J. Exp. Bot. 2005, 56, 309–321. [Google Scholar] [CrossRef] [Green Version]
- Gaufichon, L.; Rothstein, S.J.; Suzuki, A. Asparagine Metabolic Pathways in Arabidopsis. Plant Cell Physiol. 2015, 57, 675–689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Igamberdiev, A. Origins and Metabolism of Formate in Higher Plants. Plant Physiol. Biochem. 1999, 37, 503–513. [Google Scholar] [CrossRef]
- Vanlerberghe, G.C.; Martyn, G.D.; Dahal, K. Alternative Oxidase: A Respiratory Electron Transport Chain Pathway Essential for Maintaining Photosynthetic Performance During Drought Stress. Physiol. Plant. 2016, 157, 322–337. [Google Scholar] [CrossRef]
- Noguchi, K.; Terashima, I. Responses of Spinach Leaf Mitochondria to Low N Availability. Plant Cell Environ. 2005, 29, 710–719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saroussi, S.I.; Wittkopp, T.M.; Grossman, A.R. The Type II NADPH Dehydrogenase Facilitates Cyclic Electron Flow, Energy-Dependent Quenching, and Chlororespiratory Metabolism During Acclimation of Chlamydomonas Reinhardtii to Nitrogen Deprivation. Plant Physiol. 2016, 170, 1975–1988. [Google Scholar] [CrossRef] [Green Version]
- Sunil, B.; Saini, D.; Bapatla, R.B.; Aswani, V.; Raghavendra, A.S. Photorespiration Is Complemented by Cyclic Electron Flow and the Alternative Oxidase Pathway to Optimize Photosynthesis and Protect Against Abiotic Stress. Photosynth. Res. 2019, 139, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Wan, H.; Chang, S.; Hu, J.-P.; Tian, Y.-X.; Tian, X.-H. Molecular Dynamics Simulations of Ternary Complexes: Comparisons of LEAFY Protein Binding to Different DNA Motifs. J. Chem. Inf. Model. 2015, 55, 784–794. [Google Scholar] [CrossRef]
- Chen, S.; Wang, X.J.; Zhang, L.Y.; Lin, S.S.; Liu, D.C.; Wang, Q.Z.; Cai, S.Y.; El-Tanbouly, R.; Gan, L.J.; Wu, H.; et al. Identification and characterization of tomato gibberellin 2-oxidases (ga2oxs) and effects of fruit-specific slga2ox1 overexpression on fruit and seed growth and development. Hortic. Res. 2016, 3. [Google Scholar] [CrossRef] [Green Version]
- Fan, X.R.; Naz, M.; Fan, X.; Xuan, W.; Miller, A.J.; Xu, G.H. Plant Nitrate Transporters: From Gene Function to Application. J. Exp. Bot. 2017, 68, 2463–2475. [Google Scholar] [CrossRef] [PubMed]
- Castaings, L.; Camargo, A.; Pocholle, D.; Gaudon, V.; Texier, Y.; Boutet-Mercey, S.; Taconnat, L.; Renou, J.-P.; Daniel-Vedele, F.; Fernandez, E.; et al. The Nodule Inception-Like Protein 7 Modulates Nitrate Sensing and Metabolism in Arabidopsis. Plant J. 2009, 57, 426–435. [Google Scholar] [CrossRef] [PubMed]
- Konishi, M.; Yanagisawa, S. An NLP-Binding Site in the 3’ Flanking Region of the Nitrate Reductase Gene Confers Nitrate-Inducible Expression in Arabidopsis Thaliana (L.) Heynh. Soil Sci. Plant Nutr. 2013, 59, 612–620. [Google Scholar] [CrossRef] [Green Version]
- Zhao, M.; Ding, H.; Zhu, J.-K.; Zhang, F.; Li, W. Involvement of miR169 in the nitrogen-starvation Responses in Arabidopsis. New Phytol. 2011, 190, 906–915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alvarez, J.M.; Riveras, E.; Vidal, E.; Gras, D.E.; Contreras-López, O.; Tamayo, K.P.; Aceituno, F.; Gómez, I.; Ruffel, S.; Lejay, L.; et al. Systems Approach Identifies TGA1 and TGA4 Transcription Factors As Important Regulatory Components of the Nitrate Response Of Arabidopsis Thalianaroots. Plant J. 2014, 80, 1–13. [Google Scholar] [CrossRef]
- Para, A.; Li, Y.; Marshall-Colón, A.; Varala, K.; Francoeur, N.J.; Moran, T.M.; Edwards, M.B.; Hackley, C.; Bargmann, B.; Birnbaum, K.D.; et al. Hit-and-Run Transcriptional Control by BZIP1 Mediates Rapid Nutrient Signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 10371–10376. [Google Scholar] [CrossRef] [Green Version]
- Guan, P.; Wang, R.; Nacry, P.; Breton, G.; Kay, S.A.; Pruneda-Paz, J.L.; Davani, A.; Crawford, N.M. Nitrate Foraging by Arabidopsisroots Is Mediated by the Transcription Factor TCP20 through the Systemic Signaling Pathway. Proc. Natl. Acad. Sci. USA 2014, 111, 15267–15272. [Google Scholar] [CrossRef] [Green Version]
- Widiez, T.; El Kafafi, E.S.; Girin, T.; Berr, A.; Ruffel, S.; Krouk, G.; Vayssières, A.; Shen, W.-H.; Coruzzi, G.M.; Gojon, A.; et al. High nitrogen insensitive 9 (hni9)-mediated systemic repression of root no3- uptake is associated with changes in histone methylation. Proc. Natl. Acad. Sci. USA 2011, 108, 13329–13334. [Google Scholar] [CrossRef] [Green Version]
- Canales, J.; Contreras-López, O.; Álvarez, J.M.; Gutiérrez, R.A. Nitrate Induction of Root Hair Density Is Mediated by TGA1/TGA4 and CPC Transcription Factors in Arabidopsis Thaliana. Plant J. 2017, 92, 305–316. [Google Scholar] [CrossRef] [Green Version]
- Thum, K.E.; Shin, M.J.; Gutiérrez, R.A.; Mukherjee, I.; Katari, M.S.; Nero, D.; Shasha, D.; Coruzzi, G.M. An Integrated Genetic, Genomic and Systems Approach Defines Gene Networks Regulated by the Interaction of Light and Carbon Signaling Pathways in Arabidopsis. BMC Syst. Biol. 2008, 2, 31. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; Wu, D.; Kong, F.; Lin, K.; Zhang, H.; Li, G. The Arabidopsis Thaliana Nuclear Factor Y Transcription Factors. Front. Plant Sci. 2017, 7, 1425. [Google Scholar] [CrossRef] [Green Version]
- Sainju, U.M.; Dris, R.; Singh, B. Mineral nutrition of tomato. J. Food Agric. Environ. 2003, 1, 176–183. [Google Scholar]
- Ho, L. The Mechanism of Assimilate Partitioning and Carbohydrate Compartmentation in Fruit in Relation to the Quality and Yield of Tomato. J. Exp. Bot. 1996, 47, 1239–1243. [Google Scholar] [CrossRef]
- Kanai, S.; Adu-Gymfi, J.; Lei, K.; Ito, J.; Ohkura, K.; Moghaieb, R.E.; El-Shemy, H.; Mohapatra, R.; Mohapatra, P.K.; Saneoka, H.; et al. N-Deficiency Damps Out Circadian Rhythmic Changes of Stem Diameter Dynamics in Tomato Plant. Plant Sci. 2008, 174, 183–191. [Google Scholar] [CrossRef]
- Parisi, M.; Giordano, L.; Pentangelo, A.; D’Onofrio, B.; Villari, G. Effects of different levels of nitrogen fertilization on yield and fruit quality in processing tomato. In Proceedings of the International Symposium Towards Ecologically Sound Fertilisation Strategies for Field Vegetable Production, Perugia, Italy, 7–10 June 2004; p. 129. [Google Scholar] [CrossRef]
- Petro-Turza, M. Flavor of Tomato and Tomato Products. Food Rev. Int. 1986, 2, 309–351. [Google Scholar] [CrossRef]
- Løvdal, T.; Olsen, K.M.; Slimestad, R.; Verheul, M.; Lillo, C. Synergetic Effects of Nitrogen Depletion, Temperature, and Light on the Content of Phenolic Compounds and Gene Expression in Leaves of Tomato. Phytochemistry 2010, 71, 605–613. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Renau-Morata, B.; Molina, R.-V.; Minguet, E.G.; Cebolla-Cornejo, J.; Carrillo, L.; Martí, R.; García-Carpintero, V.; Jiménez-Benavente, E.; Yang, L.; Cañizares, J.; et al. Integrative Transcriptomic and Metabolomic Analysis at Organ Scale Reveals Gene Modules Involved in the Responses to Suboptimal Nitrogen Supply in Tomato. Agronomy 2021, 11, 1320. https://doi.org/10.3390/agronomy11071320
Renau-Morata B, Molina R-V, Minguet EG, Cebolla-Cornejo J, Carrillo L, Martí R, García-Carpintero V, Jiménez-Benavente E, Yang L, Cañizares J, et al. Integrative Transcriptomic and Metabolomic Analysis at Organ Scale Reveals Gene Modules Involved in the Responses to Suboptimal Nitrogen Supply in Tomato. Agronomy. 2021; 11(7):1320. https://doi.org/10.3390/agronomy11071320
Chicago/Turabian StyleRenau-Morata, Begoña, Rosa-Victoria Molina, Eugenio G. Minguet, Jaime Cebolla-Cornejo, Laura Carrillo, Raúl Martí, Víctor García-Carpintero, Eva Jiménez-Benavente, Lu Yang, Joaquín Cañizares, and et al. 2021. "Integrative Transcriptomic and Metabolomic Analysis at Organ Scale Reveals Gene Modules Involved in the Responses to Suboptimal Nitrogen Supply in Tomato" Agronomy 11, no. 7: 1320. https://doi.org/10.3390/agronomy11071320
APA StyleRenau-Morata, B., Molina, R.-V., Minguet, E. G., Cebolla-Cornejo, J., Carrillo, L., Martí, R., García-Carpintero, V., Jiménez-Benavente, E., Yang, L., Cañizares, J., Canales, J., Medina, J., & Nebauer, S. G. (2021). Integrative Transcriptomic and Metabolomic Analysis at Organ Scale Reveals Gene Modules Involved in the Responses to Suboptimal Nitrogen Supply in Tomato. Agronomy, 11(7), 1320. https://doi.org/10.3390/agronomy11071320







