Characterization of Interactions between the Soybean Salt-Stress Responsive Membrane-Intrinsic Proteins GmPIP1 and GmPIP2
Abstract
:1. Introduction
2. Materials and Methods
2.1. Soybean Plant Materials and Treatments
2.2. RNA Extraction and cDNA Synthesis
2.3. Gene Expression Analysis
2.4. GmPIPs Gene Cloning and Constructs Preparation
2.5. Yeast Two-Hybrid Assay
2.6. Bioinformatics Analysis of Soybean PIPs
2.7. Statistical Analysis
3. Results
3.1. Sequence Analysis of Soybean GmPIPs
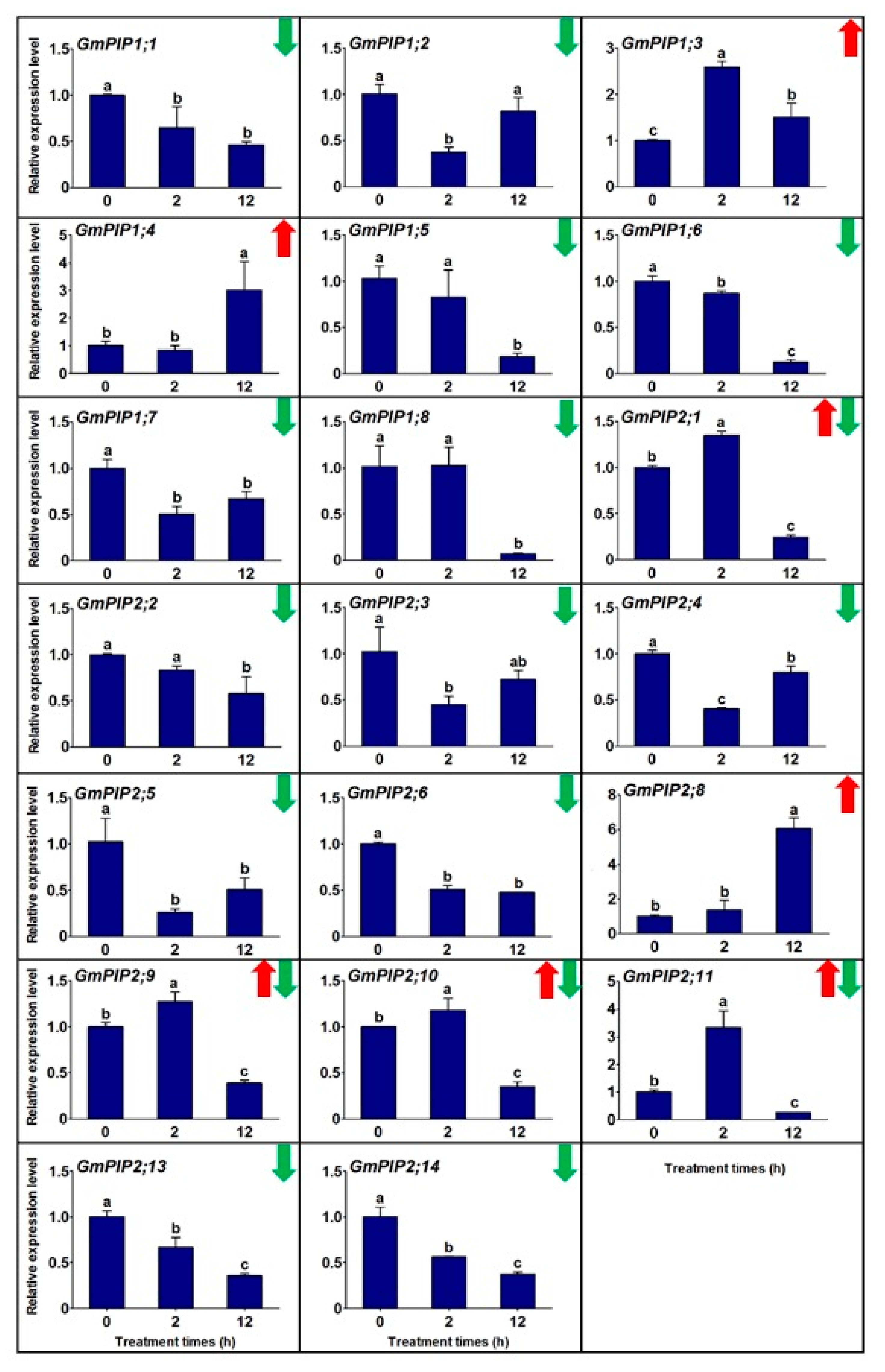
3.2. The Transcriptional Profiles of Gmpips under Salt Stress
3.3. Hetero- and Homotetramerization in GmPIP1s and GmPIP2s
3.4. Salt-Stress Treatment Enhanced the Interactions of PIPs
3.5. GmPIP2;9 Itself Could Form a Homodimer in Yeast Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yaneff, A.; Vitali, V.; Amodeo, G. PIP1 aquaporins: Intrinsic water channels or PIP2 aquaporin modulators? FEBS Lett. 2015, 589, 3508–3515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quigley, F.; Rosenberg, J.M.; Shachar-Hill, Y.; Bohnert, H.J. From genome to function: The Arabidopsis aquaporins. Genome Biol. 2001, 3, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Ali, Z.; Wang, C.; Xu, L.; Yi, J.; Xu, Z.; Liu, X.; He, X.; Huang, Y.; Khan, I. Genome-wide sequence characterization and expression analysis of major intrinsic proteins in soybean (Glycine max L.). PLoS ONE 2013, 8, e56312. [Google Scholar] [CrossRef] [PubMed]
- Kayum, M.A.; Park, J.-I.; Nath, U.K.; Biswas, M.K.; Kim, H.-T.; Nou, I.-S. Genome-wide expression profiling of aquaporin genes confer responses to abiotic and biotic stresses in Brassica Rapa. BMC Plant Biol. 2017, 17, 23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deokar, A.A.; Tar'an, B. Genome-wide analysis of the aquaporin gene family in chickpea (Cicer arietinum L.). Front. Plant Sci. 2016, 7, 1802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beese-Sims, S.E.; Lee, J.; Levin, D.E. Yeast Fps1 glycerol facilitator functions as a homotetramer. Yeast 2011, 28, 815–819. [Google Scholar] [CrossRef] [PubMed]
- Gonen, T.; Sliz, P.; Kistler, J.; Cheng, Y.; Walz, T. Aquaporin-0 membrane junctions reveal the structure of a closed water pore. Nature 2004, 429, 193–197. [Google Scholar] [CrossRef] [PubMed]
- Harvengt, P.; Vlerick, A.; Fuks, B.; Wattiez, R.; Ruysschaert, J.M.; Homble, F. Lentil seed aquaporins form a hetero-oligomer which is phosphorylated by a Mg2+-dependent and Ca2+-regulated kinase. Biochem. J. 2000, 352, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Fetter, K.; Wilder, V.V.; Menachem, M.; Chaumont, F. Interactions between plasma membrane aquaporins modulate their water channel activity. Plant Cell 2004, 16, 215–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bienert, M.D.; Diehn, T.A.; Richet, N.; Chaumont, F.; Bienert, G.P. Heterotetramerization of plant PIP1 and PIP2 aquaporins is an evolutionary ancient feature to guide PIP1 plasma membrane localization and function. Front. Plant Sci. 2018, 9, 382. [Google Scholar] [CrossRef]
- Hachez, C.; Laloux, T.; Reinhardt, H.; Cavez, D.; Degand, H.; Grefen, C.; De Rycke, R.; Inzé, D.; Blatt, M.R.; Russinova, E. Arabidopsis SNAREs SYP61 and SYP121 coordinate the trafficking of plasma membrane aquaporin PIP2; 7 to modulate the cell membrane water permeability. Plant Cell 2014, 26, 3132–3147. [Google Scholar] [CrossRef] [Green Version]
- Qiang, X.; Yu, G.; Jiang, L.; Sun, L.; Zhang, S.; Wei, L.; Cheng, X. Thellungiella halophila ThPIP1 gene enhances the tolerance of the transgenic rice to salt stress. J. Integr. Agric. 2015, 14, 1911–1922. [Google Scholar] [CrossRef]
- Liu, S.; Fukumoto, T.; Gena, P.; Feng, P.; Sun, Q.; Li, Q.; Matsumoto, T.; Kaneko, T.; Zhang, H.; Zhang, Y.; et al. Ectopic expression of a rice plasma membrane intrinsic protein (OsPIP1;3) promotes plant growth and water uptake. Plant J. 2020, 102, 779–796. [Google Scholar] [CrossRef]
- Mahdieh, M.; Mostajeran, A.; Horie, T.; Katsuhara, M. Drought stress alters water relations and expression of PIP-type aquaporin genes in Nicotiana tabacum plants. Plant Cell Physiol. 2008, 49, 801–813. [Google Scholar] [CrossRef]
- Vandeleur, R.K.; Mayo, G.; Shelden, M.C.; Gilliham, M.; Kaiser, B.N.; Tyerman, S.D. The role of plasma membrane intrinsic protein aquaporins in water transport through roots: Diurnal and drought stress responses reveal different strategies between isohydric and anisohydric cultivars of grapevine. Plant Physiol. 2009, 149, 445–460. [Google Scholar] [CrossRef] [Green Version]
- Bellati, J.; Alleva, K.; Soto, G.; Vitali, V.; Jozefkowicz, C.; Amodeo, G. Intracellular pH sensing is altered by plasma membrane PIP aquaporin co-expression. Plant Mol. Biol. 2010, 74, 105–118. [Google Scholar] [CrossRef]
- Jozefkowicz, C.; Rosi, P.; Sigaut, L.; Soto, G.; Pietrasanta, L.I.; Amodeo, G.; Alleva, K. Loop A is critical for the functional interaction of two Beta vulgaris PIP aquaporins. PLoS ONE 2013, 8, e57993. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.; Tong, J.; He, X.; Xu, Z.; Xu, L.; Wei, P.; Huang, Y.; Brestic, M.; Ma, H.; Shao, H. A novel soybean intrinsic protein gene, GmTIP2;3, involved in responding to osmotic stress. Front. Plant Sci. 2016, 6, 1237. [Google Scholar] [CrossRef] [Green Version]
- Lu, L.; Dong, C.; Liu, R.; Zhou, B.; Wang, C.; Shou, H. Roles of soybean plasma membrane intrinsic protein GmPIP2; 9 in drought tolerance and seed development. Front. Plant Sci. 2018, 9, 530. [Google Scholar] [CrossRef] [Green Version]
- Zhou, L.; Wang, C.; Liu, R.; Han, Q.; Vandeleur, R.K.; Du, J.; Tyerman, S.; Shou, H. Constitutive overexpression of soybean plasma membrane intrinsic protein GmPIP1; 6 confers salt tolerance. BMC Plant Biol. 2014, 14, 181. [Google Scholar] [CrossRef] [Green Version]
- Dajic, Z. Salt stress. In Physiology and Molecular Biology of Stress Tolerance in Plants; Springer: Dordrecht, The Netherlands, 2006; pp. 41–99. [Google Scholar]
- Lv, X.; Jin, Y.; Wang, Y. De novo transcriptome assembly and identification of salt-responsive genes in sugar beet M14. Comput. Biol. Chem. 2018, 75, 1–10. [Google Scholar] [CrossRef]
- Wu, J.; Jiang, Y.; Liang, Y.; Chen, L.; Chen, W.; Cheng, B. Expression of the maize MYB transcription factor ZmMYB3R enhances drought and salt stress tolerance in transgenic plants. Plant Physiol. Biochem. 2019, 137, 179–188. [Google Scholar] [CrossRef]
- Shoemaker, R.C.; Schlueter, J.; Doyle, J.J. Paleopolyploidy and gene duplication in soybean and other legumes. Curr. Opin. Plant Biol. 2006, 9, 104–109. [Google Scholar] [CrossRef]
- Plant Genome Duplication Database. Available online: http://chibba.agtec.uga.edu/duplication/ (accessed on 8 August 2019).
- RiceFREND. Available online: http://ricefrend.dna.affrc.go.jp (accessed on 8 August 2019).
- SMART. Available online: http://smart.embl-heidelberg.de (accessed on 1 September 2019).
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739, MEGA 5. Available online: https://www.megasoftware.net/ (accessed on 23 August 2019). [CrossRef] [Green Version]
- Brito, C.; Dinis, L.-T.; Ferreira, H.; Rocha, L.; Pavia, I.; Moutinho-Pereira, J.; Correia, C.M. Kaolin particle film modulates morphological, physiological and biochemical olive tree responses to drought and rewatering. Plant Physiol. Biochem. 2018, 133, 29–39. [Google Scholar] [CrossRef]
- Pavia, I.; Roque, J.; Rocha, L.; Ferreira, H.; Castro, C.; Carvalho, A.; Silva, E.; Brito, C.; Gonçalves, A.; Lima-Brito, J. Zinc priming and foliar application enhances photoprotection mechanisms in drought-stressed wheat plants during anthesis. Plant Physiol. Biochem. 2019, 140, 27–42. [Google Scholar] [CrossRef]
- Sun, K.; Wang, H.; Xia, Z. The maize bHLH transcription factor bHLH105 confers manganese tolerance in transgenic tobacco. Plant Sci. 2019, 280, 97–109. [Google Scholar] [CrossRef]
- Afzal, Z.; Howton, T.; Sun, Y.; Mukhtar, M.S. The roles of aquaporins in plant stress responses. J. Dev. Biol. 2016, 4, 9. [Google Scholar] [CrossRef] [Green Version]
- Ispolatov, I.; Yuryev, A.; Mazo, I.; Maslov, S. Binding properties and evolution of homodimers in protein–protein interaction networks. Nucleic Acids Res. 2005, 33, 3629–3635. [Google Scholar] [CrossRef] [Green Version]
- Zargar, S.M.; Nagar, P.; Deshmukh, R.; Nazir, M.; Wani, A.A.; Masoodi, K.Z.; Agrawal, G.K.; Rakwal, R. Aquaporins as potential drought tolerance inducing proteins: Towards instigating stress tolerance. J. Proteom. 2017, 169, 233–238. [Google Scholar] [CrossRef]
- Tournaire-Roux, C.; Sutka, M.; Javot, H.; Gout, E.; Gerbeau, P.; Luu, D.-T.; Bligny, R.; Maurel, C. Cytosolic pH regulates root water transport during anoxic stress through gating of aquaporins. Nature 2003, 425, 393–397. [Google Scholar] [CrossRef] [PubMed]

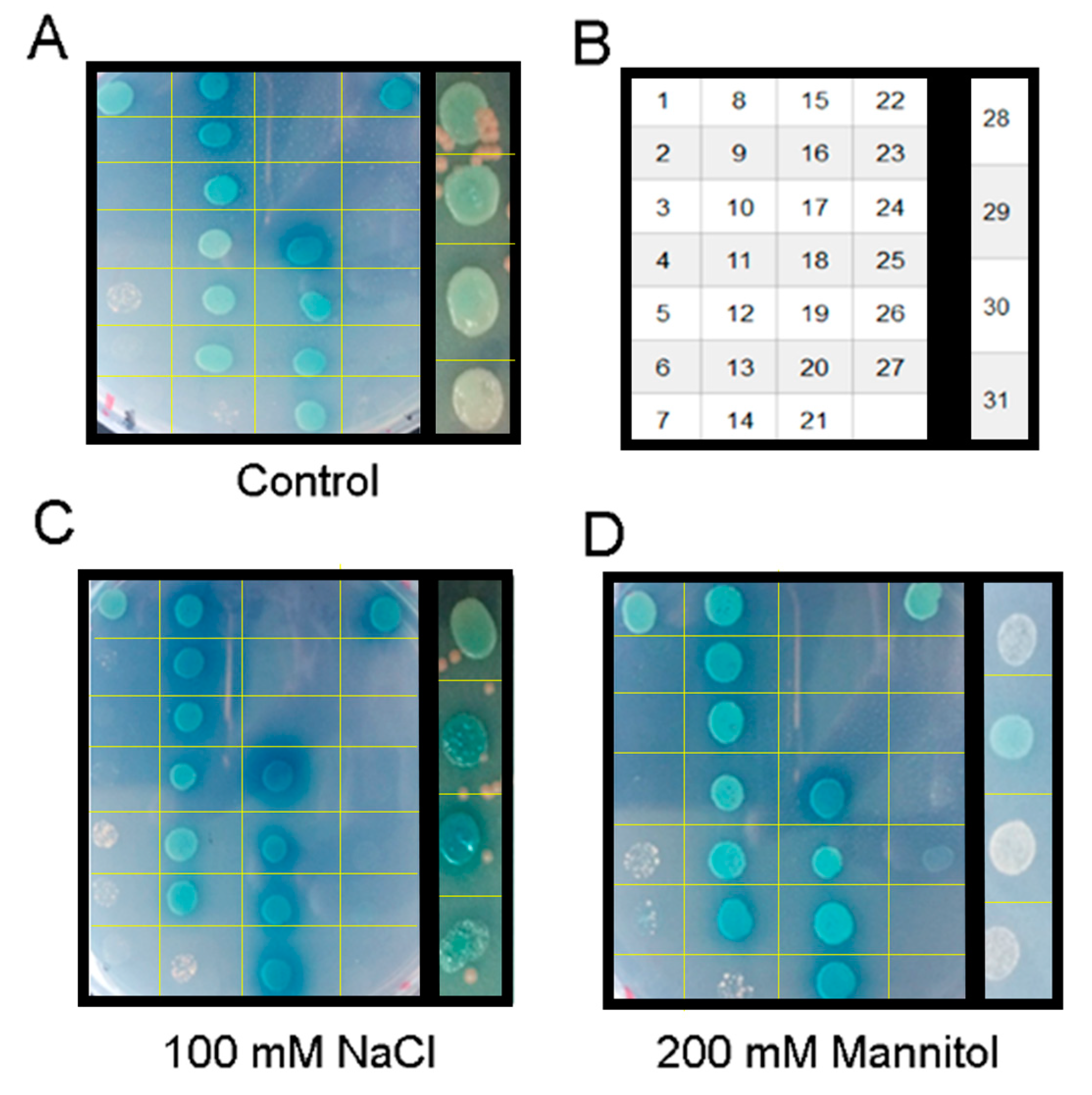

| Strengthened by Salt Stress | |||
|---|---|---|---|
| Hetero-interaction between GmPIP1s and 2s | GmPIP1;5 | GmPIP2;2 | |
| GmPIP1;5 | GmPIP2;4 | ||
| GmPIP1;5 | GmPIP2;6 | ||
| GmPIP1;5 | GmPIP2;8 | ||
| GmPIP1;5 | GmPIP2;9 | ||
| GmPIP1;5 | GmPIP2;11 | Yes | |
| GmPIP1;5 | GmPIP2;13 | Yes | |
| GmPIP1;6 | GmPIP2;2 | ||
| GmPIP1;6 | GmPIP2;4 | ||
| GmPIP1;6 | GmPIP2;6 | ||
| GmPIP1;6 | GmPIP2;8 | ||
| GmPIP1;6 | GmPIP2;9 | Yes | |
| GmPIP1;6 | GmPIP2;11 | Yes | |
| GmPIP1;6 | GmPIP2;13 | Yes | |
| Hetero-interaction between GmPIP1s | GmPIP1;6 | GmPIP1;7 | Yes |
| Hetero-interaction between GmPIP2s | Not detected | ||
| Homo-interaction between GmPIP | GmPIP2;9 | Yes | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Qi, W.; Lu, H.; Shao, H.; Zhang, D. Characterization of Interactions between the Soybean Salt-Stress Responsive Membrane-Intrinsic Proteins GmPIP1 and GmPIP2. Agronomy 2021, 11, 1312. https://doi.org/10.3390/agronomy11071312
Liu J, Qi W, Lu H, Shao H, Zhang D. Characterization of Interactions between the Soybean Salt-Stress Responsive Membrane-Intrinsic Proteins GmPIP1 and GmPIP2. Agronomy. 2021; 11(7):1312. https://doi.org/10.3390/agronomy11071312
Chicago/Turabian StyleLiu, Jia, Weicong Qi, Haiying Lu, Hongbo Shao, and Dayong Zhang. 2021. "Characterization of Interactions between the Soybean Salt-Stress Responsive Membrane-Intrinsic Proteins GmPIP1 and GmPIP2" Agronomy 11, no. 7: 1312. https://doi.org/10.3390/agronomy11071312
APA StyleLiu, J., Qi, W., Lu, H., Shao, H., & Zhang, D. (2021). Characterization of Interactions between the Soybean Salt-Stress Responsive Membrane-Intrinsic Proteins GmPIP1 and GmPIP2. Agronomy, 11(7), 1312. https://doi.org/10.3390/agronomy11071312







