Abstract
Submergence stress due to flash floods reduces rice yield drastically in sensitive varieties. Maudamani is a high yielding popular rice variety but is highly susceptible to submergence stress. The selection of progenies carrying Sub1 and GW5 (wide-grain) enhanced the submergence stress tolerance and grain yield of theMaudamani variety by following the marker-assisted backcross breeding method. Foreground screening detected 14 BC1F1, 17 BC2F1, and 12 BC3F1 backcross progenies that carried the target QTLs for submergence tolerance and grain width. Background screening was performed in the progenies carrying the target QTL and enhanced the recovery of a recipient parent’s genome by upto 96.875% in the BC3 pyramided line. The BC3F1 plant containing the highest recipient parent genome content and the target QTLs was self-pollinated. In BC3F2 generation, the target QTLs the Sub1 and GW5 (wide-grain) alleles and recipient parent’s yield component QTL OsSPL14 were tracked for homozygous states in the progenies. Seven pyramided lines showed tolerance to submergence for 14 days and higher grain yield than both the parents. The pyramided lines were similar to the recipient parent for the majority of the studied morphological and quality traits. The pyramided lines are useful as cultivars and can serve as potential donors for transfer of Sub1, OsSPL14, Gn1a, GW5 (wide-grain), and SCM2 QTLs.
1. Introduction
Rice, the golden cereal, sustains life for millions of people around the world. Rice is important not only as a staple food but also for its association with life in India, which is seen in its use as worship material for important ceremonies and rituals since ancient time. Rice provides important compounds, namely carbohydrates, quality proteins, vitamins, specific oils, many minerals, dietary fiber, and a few phyto-compounds thatprovide added health benefits [1]. The crop is very unique in its adaptation and cultivation from very high elevations to below sea level. The crop covers around 160 million hectares of land around the world. The crop is cultivated as a rainfed crop in about 45% of the total rice cultivated area [1]. Rice crop provides livelihood to nearly 4 billion people, which constitutes about 55% of the global population. The crop generates about $206 billion global annual earnings, which is 17% of the total crop value [2]. In recent years, the crop has become highly affected by the adverse effects of climate change. The higher production from rainfed rice cultivation is now challenged by the climate change-related yield-reducing factors in India [3]. About 22 mha of rainfed rice area is cultivated in India, of which 90% is confined to the eastern region of the country [4]. Submergence tolerance and high grain yield along with resistance to major diseases and insect pests should be transferred to a superior variety to ensure stable production.
Although there has been impressive growth in rice production and productivity during recent years, still, the demand for rice continues to be higher, projecting upwards in the future due to the population growth rate being higher than the production increase rate of rice. Again, in the coming years, the production increase must be harvested under the adverse effects of climate change. In addition, the future production needs to be obtained from the least available land with less use of inputs and lower chemical usage and in a more environmentally friendly manner. The breeding strategy in this investigation focuses on simultaneous improvement of stress tolerance and yield in rice so as to fulfill future requirements [4]. Yield potential in a rice variety can be enhanced by pyramiding the suitable yield component QTLs absent in that variety. Among the yield component traits, grain number enhancement is controlled by QTLs Gn1a [5], Ghd7 [6], Ghd8 [7,8], APO1 [9], DEP1 [10,11], DEP2 [12], and DEP3 [13]. An increase in yield is also regulatedby the grain weight and grain dimension QTLs GW2 [14], GS3 [15], GS5 [16], and GW5 [17,18]. Tiller is controlled by the QTLs MOC1 [19], LRK1 [20], EP3 [21], and IPA1 [22,23]. High yield through higher grain filling is controlled by the QTLs GIF1 [11] and FLO [24,25]. Transfer of these yield component QTLs into a variety lacking these QTLs will be a useful way of raising the yield potential further in the variety.
Unpredictable and flash floods during rainy season are now a common occurrence in India, particularly in the eastern region of the country. This is a major cause of yield reduction in susceptible varieties affected by submergence stress. The Maudamani variety produces 7 to 9 t/ha normally but harvests upto 11 t/ha grain yield under favorable conditions. However, total crop failure occurs if the crop is affected by flash flood causing submergence for more than a week. The submergence tolerance QTL, Sub1, confers tolerance to submergence for about two weeks [26,27]. Recently, gene-based markers are available for transfer of Sub1 QTL through marker-assisted breeding. Submergence tolerance has been improved in many high yielding varieties including Swarna using this QTL introgression [28,29,30,31,32]. The transfer of Sub1 QTL from Swarna-Sub1 to Maudamani may contribute negligible undesirable genetic effects from the donor variety as the donor parent is a popular variety [33].
The yield component QTL, GW5, is associated with reduced grain width and the effects of the QTL was consistent under multiple environmental conditions. The loss of a GW5 segment resulted in wide-grain genotypes in most japonica and indica rice [18]. The donor parent, Swarna-Sub1, developed through the marker-assisted backcross breeding approach, carries the Sub1 QTL for submergence tolerance [1]. Additionally, the presence of yield component QTLs Gn1a, GW5, and SCM2 were also confirmed in the parent by the parental line validation study. Additionally, the recipient parent, Maudamani, a high yielding super rice variety, showed the presence of yield component QTLs OsSPL14, Gn1a, and SCM2 via the parental line validation study in this investigation. We report here the successful development of pyramided lines in a Maudamani background carrying Sub1, OsSPL14, and GW5 (wide grain) QTLs in a homozygous state for submergence tolerance and high grain yield.
2. Materials and Methods
2.1. Plant Materials and Breeding Program
The rice variety Swarna-Sub1 carrying Sub1 QTL for submergence tolerance and yield component QTLs SCM2 and GW5 was used as the donor male parent in a hybridization program. The recipient parent, Maudamani, is a high yielding variety of eastern India inbuilt with the yield component QTLs OsSPL14, Gn1a, and SCM2 but shows susceptibility to submergence stress. The recipient parent was crossed with the donor variety, Swarna-Sub1, during a dry season in 2014 as per the scheme depicted for the marker-assisted breeding (Figure 1). The donor and recipient parents were obtained from the gene bank of National Rice Research Institute (NRRI), Cuttack, India. One true F1 plant was hybridized with the recipient parent during the rainy season in 2014 to generate BC1F1 generation seeds. True hybridity was checked using the direct Sub1 marker, Sub1-A203, and with a co-dominant marker, RM8300, as well. The BC1F1 seeds were grown, and the progenies were screened for the target genes, yield component, and submergence tolerance QTLs by using the established molecular markers (Table 1). The grain size QTL, GW5, is associated with narrow grained rice, and hence, negative selection was performed to obtain the Maudamani grain width. In the background selection, progenies of the BC1F1 generation carrying the two target QTLs were screened using the polymorphic markers. The foreground positive progenies containing the highest genome content of the recurrent parent was hybridized with the recipient parent Maudamani to obtain BC2F1 seeds. During the dry season in 2015, BC2F1 seeds were harvested. Among the derived progenies detected with the target QTLs in BC2F1 generation, those with highest recurrent genome content was again crossed with the recipient parent during the wet season in 2015 to produce BC3F1 seeds. The background analysis of BC3F1 progenies was performed during the dry season in 2016. The BC3F1 plant containing the highest recipient genome content along with two target QTLs wereselfed in the dry season in 2016. BC3F2 progenies were genotyped to search for the presence of homozygosity for the target QTLs and recipient parent’s yield QTLs during the wet season in 2016. The seed increase in the pyramided progenies detected with homozygous target genes were increased during the dry season in 2017. Evaluations for agronomic and other traits were performed during the wet seasons in 2017, 2018, and 2019.

Figure 1.
Breeding scheme used for transfer of Sub1 and GW5 (wide-grain) for submergence tolerance and yield component QTLs through marker-assisted backcross breeding into popular variety, ‘Maudamani’ (Inside the parentheses, number of hybrids/derived progenies were raised in the generation).

Table 1.
Molecular markers used for screening of yield component QTLs and submergence tolerance during foreground selection.
2.2. Genomic DNA Isolation, Polymerase Chain Reaction and Marker Analysis
Genomic DNA was isolated following the standard extraction protocol [36]. APCR reaction was performed following the procedure used in a previous publication [32]. The information regarding chromosome number, position, and sequence of the primers used in the polymerase chain reaction are presented in Table 1. Eight gene-specific and tightly linked markers for the two target QTLs and four recipient QTLs were used in foreground selection (Table 1). These markers information were taken from earlier publications reported for these target traits [3,5,14,18,23,33,35,36]. A total of 644 publicly available SSR markers were used for the study of polymorphism between the two parents. The polymorphic markers detected were used for background selection (Table 2). Agarose gel electrophoresis was used to separate the amplification products obtained from PCR reactions. The images were recorded in a gel documentation system (SynGene, Cambridge, UK). Data analysis and dendrogram construction were performed following the standard publications [37,38,39]. Graphical Geno Types (GGT) Version 2.0 software was used to construct the genome recovery graph of recipient parent in the pyramided lines based on the SSR marker data [40].

Table 2.
Polymorphic microsatellite markers obtained between the rice varieties Maudamani and Swarna-Sub1.
2.3. Screening for Submergence Tolerance
The BC3F4 generation pyramided lines and parents were transplanted in the screening tank of ICAR NRRI, Cuttack, at around 3 weeks’ seedling age during the wet seasons in 2018 and 2019. The screening trial was laid out in a randomized complete block design (RBD) with three replications/entries accommodating a population size of 66 plants/entry. The experiment materials were transplanted witha spacing of 15 × 20 cm2 by providing three rows/entry. Two weeks of complete submergence stress upto 1.5 water depth was maintained in the tank. De-submergence was performed just after completion of the 14-day stress period, and subsequently, regeneration was assessedone week after de-submergence. The data recording and scoring the genotypes were collected following the procedures of earlier publications [1,3].
2.4. Evaluation of the Pyramided Lines for Various Traits
The seedlings of 25-day-old pyramided lines carrying Sub1 and yield QTLs were transplanted along with the parents during the wet seasons in 2017, 2018, and 2019. A plot slot size of 12 m2 was provided to each entry, with 40 plants per row, at a spacing of 15 × 20 cm2, and planted in RBD with three replications in the research farm of NRRI, Cuttack. The data for ten plants for morpho-quality traits viz., plant height, panicles/plant, panicle weight (g), number of filled grains, total spikelets, number of primary branches, secondary branches and number of tertiary branches per panicle, grain length, grain breadth, 1000-grain weight, milling (%), head rice recovery (%), and amylose content (%) from each entry and replications were recorded. Plot yield and days to 50% flowering were recorded on a whole plot basis. The standard protocols published for head rice recovery [41] and gel consistency [42] were adopted. Amylose content in the grains of the pyramided and parental lines was estimated following the standard procedures described in an earlier publication [43]. An analysis of the various morpho-quality traits of the pyramided and parental lines were analyzed using SAS 2008, version 9.2 [44]. The Principal Component analysis (PCA) for the pyramided and parental lines was performed by using multivariate analysis (Past Software version 4.03) data of the 15 morphological traits. A scatter plot was generated by using two major components: Principal Component 1 (PC1) and Principal Component 2 (PC2). The Eigen value and percentage of variance were generated by the interaction of a variance–covariance matrix. The interaction between all morphological traits was depicted through biplot graph in the matrix. All of the plots and results of PCA were generated as per a standard procedure following previous publications [45,46,47].
3. Results
3.1. Development of Improved Lines
3.1.1. Validation of Donor and Recipient Parents for the Target Traits
The target QTLs controlling the traits were validated in the donor and recipient parents before starting hybridization and selection. The presence of submergence tolerance QTL Sub1 and the yield component QTLs Gn1a and GW5 was confirmed in the parent, Swarna-Sub1 (Figure 1). The recipient parent is a high yielding popular variety. The genetic basis of high yield was checked by validating the presence of QTLs contributing higher yield through Gn1a, OsSPL14, and SCM2 in the recipient parent Maudamani. The yield component QTLs common in both parents were observed to be Gn1a and SCM2 (Figure 2). The gene-based and tightly linked molecular markers for Sub1 QTL and the direct marker for the yield component QTL, GW5, were used for validation and tracking of the target genes in the parental and backcross-derived lines (Table 1). A parental polymorphism survey was performed by using 644 simple sequence repeats markers covering all of the chromosomes. A total of 57 polymorphic markers were detected between the two parents and deployed for background screening (Table 2).
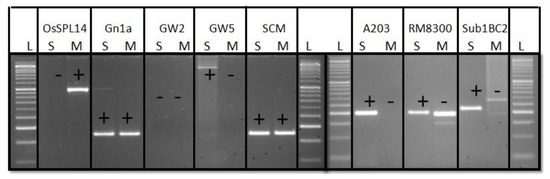
Figure 2.
PCR amplification of OsSPL14, Gn1a, GW5 and SCM2 gene specific markers for yield component QTLs and the markers controlling submergence tolerance, Sub1 by deploying Sub1-A203 and Sub1-BC2 in both the parents Maudamani and Swarna-Sub1. S: Swarna-Sub1; M: Maudamani; L—Molecular weight marker (50 bp plus ladder) are the gel lanes.
3.1.2. Marker-Assisted Selection in BC1F1 Generation
Maudamani was hybridized with ‘Swarna-Sub1′, and 870 F1 seeds were obtained. The hybridity in F1 plants was confirmed by genotyping the hybrid plants using the Sub1 specific marker. One true F1 plant was crossed with the recipient parent, Maudamani, and a total of 132 BC1F1 seeds were generated. The backcross generation was grown, and foreground screening was performed in all of the BC1F1 plants using the two markers for the QTLs, Sub1 and GW5 (Figure 3).
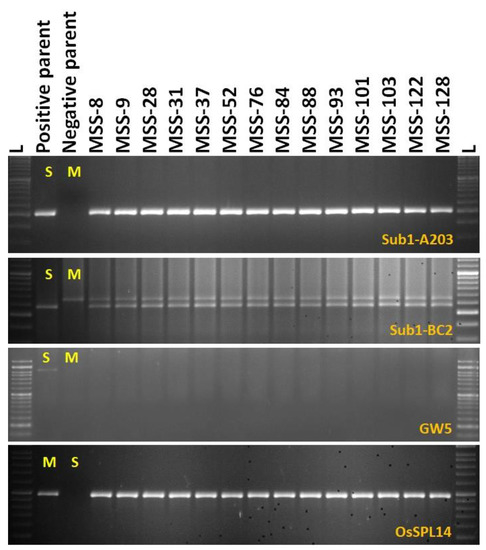
Figure 3.
PCR amplification of markers for submergence tolerance, Sub1 deploying Sub1-A203 and Sub1-BC2 along with yield QTLs GW5 and OsSPL14 in BC1F1 progenies. L: Molecular weight marker (50 bp plus ladder) and lanes on the top of the gel indicate BC1F1 progenies.
The screening results of the BC1F1 progenies of the cross revealed the presence of Sub1 QTL in 78 derivatives detected by the markers A203 and Sub1-BC2 (200 bp and 240 bp). Screening for the presence of GW5 (wide-grain) gene controlling the grain width and weight identified 14 progenies to carry both traits. Negative selection was performed for GW5 in order to obtain the Maudamani grain width and weight. The other yield component QTL desired, OsSPL14, and validated in the recipient parent, Maudamani, isalso inherited in these progenies. Additionally, the common yield component QTLs present in both parents namely, Gn1a and SCM2, detected from both parents, are expected to be in homozygous states in these progenies. Background screening was performed in the 14 BC1F1 foreground-positive progenies by using 57 SSR markers. Out of these 14 plants, the progeny carrying maximum recipient genome content was selected for the next backcross. The recipient parent’s genome content in those 14 progenies varied from 64.58 to 81.25% with an average value of 76.26% (Table 3). The backcross derivatives MSS128 and MSS84showed the highest recurrent genome content of 81.25%. The BC1F1 lines generated from MSS84 and MSS128 were backcrossed with the recipient parent, Maudamani, to obtain BC2F1 seeds.

Table 3.
Genotyping of backcross progenies for two QTLs and recovery of recipient parent’s genome in the foreground positive backcross progenies.
3.1.3. Marker-Assisted Selection in BC2F1 Generation
One hundred and sixty-nine BC2F1 plants were grown in the field for selection. The target QTLs were tracked by foreground selection using gene-specific and linked markers. The genotyping results of 169 BC2F1 progenies showed 93 positive progenies for Sub1 QTL. These 93 positive progenies were checked for the presence/absence of the GW5 (wide-grain) gene using gene-specific markers. Seventeen plants with the desired QTLs were identified for further background selection (Figure 4). The other yield QTLs OsSPL14 desired from Maudamani, Gn1a and SCM2, from both parents, is also present in those 17 progenies. Background screening for recovery of the recipient parent genome in those 17 identified plants containing the target QTLs ranged from 82.29 to 91.67% with an average of 87.74% (Table 3). The plant MSS 128-102 showing 91.67% of the Maudamani genome content was used for the next BC3 back crossing.
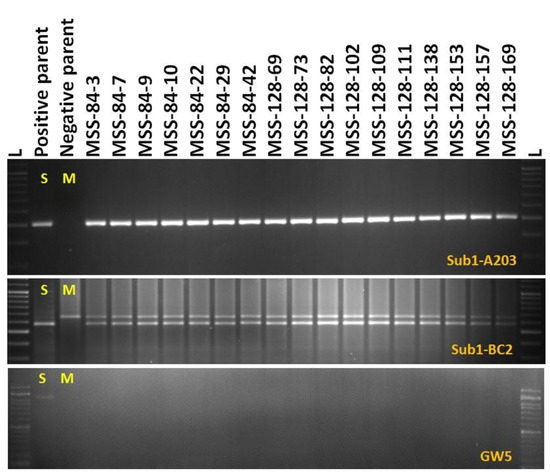
Figure 4.
PCR amplification of gene specific markers for submergence tolerance, Sub1 using Sub1-A203 and Sub1-BC2 along with yield component QTLs GW5 and OsSPL14 in BC2F1 progenies. L—Molecular weight marker (50 bp plus ladder) and Lanes on the top of the gel indicate BC2F1 progenies.
3.1.4. Marker-Assisted Selection in BC3F1 and BC3F2 Generations
The BC3F1 seeds were generated by crossing BC2F1 plant no. MSS 128-102 and the recurrent parent ‘Maudamani’. A total of 144 BC3F1 seeds were generated and raised for molecular screening by foreground and background selections. The genotyping results for the target QTL, Sub1, were positive in 87 progenies. Those 87 Sub1 carrier plants were genotyped for checking the presence/absence of the GW5 QTL. This analysis identified 12 plants positive for GW5 and further genotyped for background screening (Figure 5). The background analysis using 57 SSR markers in these 12 plants detected 92.7 to 96.875% recurrent parent’s genome recovery, with an average of 94.88% (Table 3). The highest recurrent genome containing plant MSS 128-102-97 was selfed, and 31.5g seeds were produced for further evaluation in BC3F2 generation. Around one third of the selfed seeds were raised, and 618 BC3F2 plants were subjected to foreground screening, of which seven plants were identified to be homozygous. Additionally, the yield QTL being inherited from the Maudamani parent was checked for its homozygous state. In addition, the inheritances of yield component QTLs Gn1a and SCM2 from both parents to BC3F2 progenies were also validated through foreground analysis using gene-specific markers. The genotyping results of 618 BC3F2 progenies detected seven plants containing three QTLs, namely Sub1, GW5 (wide-grain), and OsSPL14, along with the yield component QTLs Gn1a and SCM2 in homozygous condition (Figure 6). The seed increase in these seven pyramided lines was assessed for evaluation in BC3F4 generation for various morphological and quality traits. Cluster analysis with agro-morphologic and quality traits showed distinct clusters of Swarna-Sub1, the donor line forming one group (Figure 7A).Additionally, a dendrogram was generated by using the alleles detected using the SSR markers, which grouped the developed pyramided and parental lines into two main groups (Figure 7B). Eight genotypes were accommodated in cluster I along with the recipient parent ‘Maudamani’, while the donor parent for submergence tolerance and yield component QTL remained in cluster II. The backcross derived lines in the cluster I were found to form different subclusters based on the 15 agro-morphologic traits studied but were similarto the recipient parent ‘Maudamani’ for the majority of the studied morphological and quality traits. The pyramided lines MSS 128-102-97-117, MSS128-102-97-601, MSS128-102-97-613, and MSS128-102-97-617 were almost similar in terms of genome recovery among themselves and with recipient parent ‘Maudamani’ (Figure 7C).
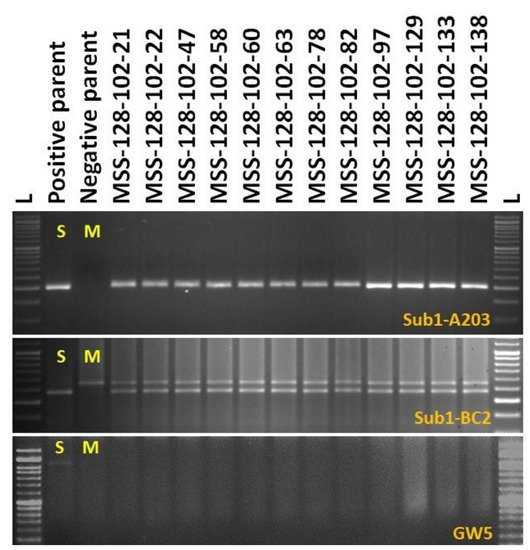
Figure 5.
PCR amplification of markers for submergence tolerance, Sub1 deploying Sub1-A203 and Sub1-BC2 along with yield component QTLs Gw5 and OsSPL14 in BC3F1 progenies. L: Molecular weight marker and lanes on the top of the gel indicate BC3F1 progenies.
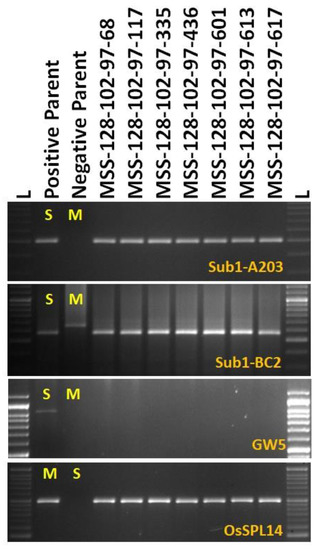
Figure 6.
PCR amplification of gene specific markers for submergence tolerance, Sub1 deploying Sub1-A203 and Sub1-BC2 along with yield component QTLs GW5 and OsSPL14 in BC3F2 progenies. L—Molecular weight marker (50 bp plus ladder) and lanes on the top of the gel indicate BC3F2 progenies.
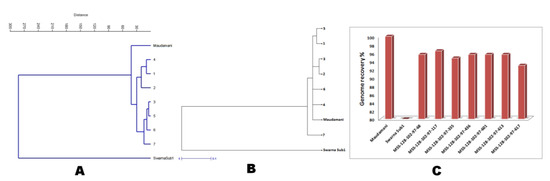
Figure 7.
Seven pyramided lines along with parents in (A) dendrogram showing relatedness based on 16 morphologic and quality traits; (B) Dendrogram showing the genetic relationship between lines based on 57 microsatellite markers and (C) % contribution of recurrent genome in the pyramided lines. The numbers indicate the pyramided lines, 1: MSS128-102-97-68; 2: MSS128-102-97-117; 3: MSS128-102-97-335; 4: MSS128-102-97-436; 5: MSS128-102-97-601; 6: MSS128-102-97-613; 7: MSS128-102-97-617.
3.2. Analysis of Recipient Genome Recovery on the Carrier Chromosomes in the Pyramided Lines
The background analysis for recipient genome recovery and genetic drag linked to the donor segments were assessed using 57 background and 8 foreground markers. The markers were carefully selected for all of the chromosomes to obtain maximum coverage in background screening. The foreground analysis detected seven BC3F2 pyramided lines for the presence of homozygous target QTLs in the progenies. The Sub1 carrier chromosome 9 showed linkage drag of the donor fragment on both sides of the marker A203 and Sub1-BC2 in all seven NILs (Figure 8). The GW5 (wide-grain) present on the chromosome 5 showed no drag of the donor segment in all pyramided lines except MSS128-102-97-613 and MSS128-102-97-335, where a drag was noticed in between RM7452 and RM440 (Figure 8).
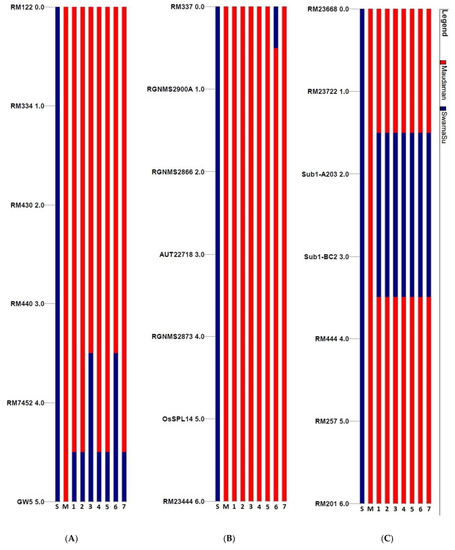
Figure 8.
Analyses of QTLs stacking in carrier chromosomes associated with submergence tolerance and yield component QTLs in 7 pyramided lines (A) GW5 (wide-grain) yield component QTL present on chromosome 5 (B) OsSPL14 yield component QTL present on chromosome 8 and (C) Sub1 QTL on chromosome 9 in the BC3F3 progenies of Maudamani/Swarna-Sub1. The numbers indicate the pyramided lines, 1: MSS128-102-97-68; 2: MSS128-102-97-117; 3: MSS128-102-97-335; 4: MSS128-102-97-436; 5: MSS128-102-97-601; 6: MSS128-102-97-613; 7: MSS128-102-97-617.
3.3. Evaluation of the Pyramided Lines for Submergence Tolerance
Nine genotypes including seven BC3F4 pyramided lines carrying target QTLs were evaluated under the controlled submergence screening tank for confirmation of the submergence tolerance trait in the pyramided lines. The test genotypes were exposed to two weeks of submergence stress. After one week of de-submergence, all seven pyramided lines showed regeneration ability from 85 to 95% while the donor parent ‘Swarna-Sub1′ showed regeneration of 95% (Figure 9). No regeneration was found in the sensitive parent ‘Maudamani’. The pyramided lines MSS128-102-97-117 and MSS128-102-97-436 had similar regeneration abilitiesto that of the Swarna-Sub1 parent. Pyramided lines viz., MSS128-102-97-68, MSS128-102-97-335, MSS128-102-97-601, MSS128-102-97-617, and MSS128-102-97-613 showed regeneration abilities of 90%.
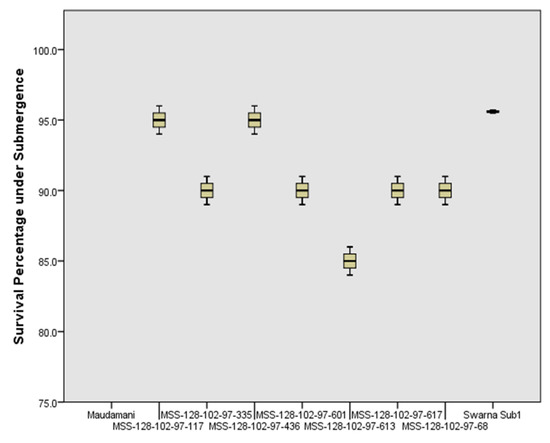
Figure 9.
Percent plant regenerated in the pyramided lines carrying Sub1 QTL along with the parents under control screening facility after one week of de-submergence from 14 days of submergence stress.
3.4. Evaluation of Pyramided Lines for Agro-Morphologic, Yield Components and Grain Quality Traits
The pyramided lines carrying submergence tolerance and yield component QTLs in the background of the Maudamani variety were evaluated for various traits during the wet seasons in 2017, 2018, and 2019. The pyramided lines were compared with both the popular rice varieties Maudamani and Swarna-Sub1. The recipient parent ‘Maudamani’ produced a pooled mean grain yield of 8.69 t/ha. The pyramided lines MSS128-102-97-436, MSS128-102-97-117, MSS128-102-97-617, MSS128-102-97-601, MSS128-102-97-68, and MSS128-102-97-335 produced more yield than the recipient parent Maudamani (Table 4). The pyramided lines MSS128-102-97-117 and MSS128-102-97-436 produced >9 t/ha grain yield, showing an advantage of >5% over the recipient popular variety Maudamani. However, the agro-morphologic traits of all of the pyramided lines were not similar tothat of the parent, Maudamani. The target morphologic traits controlled by the yield component QTLs viz., no. of primary branches, secondary branches and tertiary branches per panicle, and panicle weight finally influencing grain yield in the pyramided lines were observed to be almost similar within each subcluster (Table 4; Figure 10). Much of the grain quality and the cooking characters of the recipient parent such as milling (%), head rice recovery (%), kernel length (mm), kernel breadth (mm), kernel length after cooking, gel consistency, amylose content (%), and alkali spreading value were retained in a few pyramided lines (Table 4). The placement pattern of the parents and the pyramided lines in the quadrants of the genotype-by-trait biplot diagram constructed based on 15 agro-morphologic, yield, and component traits over three years showed similarity among the pyramided lines (Figure 11). The pyramided lines were found in the first and second quadrants along with the recipient parent ‘Maudamani’. The pyramided lines closer to each other were almost similar in grain yield, grain quality, and the other studied parameters (Figure 11). These genotypes closer to Maudamani are good candidates for further evaluation and release as cultivars in various parts of the country. The variation observed for the first principal component was61.7%, while 15.1% was explained for the second component.

Table 4.
Agro-morphologic and grain quality parameters of the pyramided lines along with parents pooled over 3 seasons under field evaluation.

Figure 10.
Panicle photographs of 2 parents and 7 pyramided lines evaluated in BC3F4 generation during wet season, 2018. M: Maudamani, S: Swarna-Sub1 and the numbers in the figure indicate the pyramided lines, 1: MSS128-102-97-68; 2: MSS128-102-97-117; 3: MSS128-102-97-335; 4: MSS128-102-97-436; 5: MSS128-102-97-601; 6: MSS128-102-97-613; 7: MSS128-102-97-617.
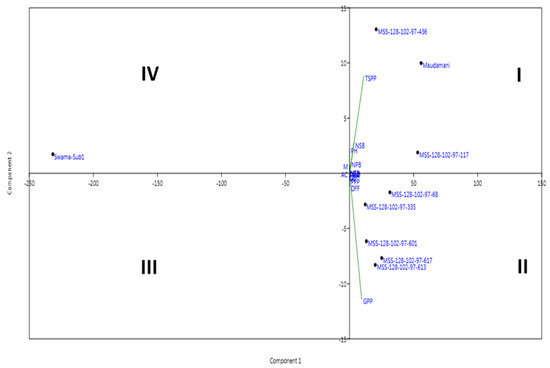
Figure 11.
Genotype-trait biplot diagram of 7 pyramided lines carrying Sub1 and GW5 (wide-grain) allele along with the parents for the first two principal components. DFF—Days to 50% flowering; PH—Plant height; PN—Panicles/plant; TW—1000-grain weight; Milling (%); HRR—Head rice recovery (%); AC—Amylose content (%); NPB—Number of primary branches/panicle; NSB—Number of secondary branches/Panicle; NTB—number of tertiary branches/Panicles; GL—Grain length; GB—Grain breadth, and YLD—plot yield.
4. Discussion
The inclusion of marker-aided selection increases the accuracy in transfer of the target genes/QTLs into a recipient variety in a backcross breeding program. In the present marker-assisted breeding program, we successfully developed pyramided lines showing submergence tolerance and yield component QTL in the high yielding background of Maudamani without altering the main features of the recipient variety. Here, the QTLs were transferred simultaneously into the popular variety. In addition, it was possible to reduce the breeding duration for developing a variety compared to the classical backcross breeding approach. In this breeding program, three backcrosses and one selfing generation were utilized to transfer the target QTLs into the popular variety. The desired traits that were lacking in the popular variety could be improved in less time and with more precision. Such examples of variety development by precise transfer of genes and with a shorterduration through marker-assisted breeding are available in rice crop [29,30,48,49,50,51].
Previous reports of many successful gene transfer and pyramiding cases have been published in rice crop [47,48,49,50,51,52,53,54,55,56,57,58,59,60]. This study of QTL pyramiding for submergence tolerance and yield component QTLs is clearly different from earlier gene pyramiding work. Earlier gene stacking publications on bacterial blight resistance with submergence tolerance transfer into rice varieties, namely improved Lalat and improved Tapaswini, have been published [58,59]. However, here, yield improvement and submergence tolerance through gene pyramiding is a typical example of gene stacking. Other publications using MAB breeding are mostly for the development of cultivars through pyramiding of resistance genes for insects and diseases in rice [48,49,52,53,54,55,56,57,58].
In this investigation, the presence of the OsSPL14, Gn1a, and SCM2 yield QTLs were detected in the popular variety Maudamani while Gn1a, SCM2, and GW5 were confirmed in the variety Swarna-Sub1. These two varieties are good sources of yield component QTLs in indica rice. The yield component QTL OsSPL14 showed negative regulation for tiller/panicle number per plant in rice. In our study, OsSPL14 is present in a homozygous state in Maudamani and produces low to moderate tiller numbers (average, 8.86/plant). However, Swarna-Sub1 lacks this QTL and produces high tiller numbers (average, 13.93/plant). As expected, the pyramided lines also showed similar trends in tiller no./plant to that of the recipient parent, Maudamani, but different from that of Swarna-Sub1 (i.e., not a higher tiller number). The current pyramided lines will serve as potential sources of QTLs containing Sub1+ OsSPL14+ Gn1a + gw5 + SCM2 and may be suitable as cultivars. The research results of a mapping study of Gn1a QTL indicated the source of the grain-number controlling trait from a japonica variety, Habataki [5]. The presence of SCM2 provides non-lodging to the rice culm, and the japonica variety Habataki was the source for the QTL [33]. Similarly, the donor line for high panicle branching was from the ST-12 variety. The yield QTL GW5 is responsible for grain width and weight [15,61,62]. A 1212 bp deletion is responsible for enhanced grain width and weight, whereas the presence of it reduces the grain width, thereby making it a narrow grain [18]. Now, the pyramided lines in a Maudamani background carrying five yield QTLs along with submergence tolerance genes are much better and potential sources for transfer to indica rice rather than from different japonica varieties.
Work on gene pyramiding for the transfer of various traits in rice has been published earlier [48,52,53,54,55,60]. By using this precision breeding for transfer of target traits, pyramids containing Sub1+ OsSPL14+ Gn1a + gw5 + SCM2 QTLs along with recipient parents’ genome of >95% in the pyramided lines was possible. The undesirable drag expected from the donor genome may come from theselection of additional unlinked loci in backcross generations [51]. In our investigation, such effects were detected in the elite pyramided lines while transferring the Sub1 QTL into the Maudamani background. The graphical representation of genotyping data as seen in the diagram constructed for the pyramided lines showed the linkage drag on the chromosome carrying the target QTLs (Figure 5). This region was previouslyreported to be a recombination hotspot [18]. However, no linkage drag was observed on the chromosome 8 carrying OsSPL14 as the QTL was inherited from the recipient parent and was not from the donor parent. Less linkage drag from donor parents was also reported by earlier researchers assessingmarker-assisted breeding in rice using more background markers [49,54,55]. Here, the donor parent was a popular variety, and hence, the drag may not show any undesirable effects in the developed pyramided lines (Figure 5). Similar findings in other publications suggest the use of an improved variety as the donor results in less or no undesirable drag compared to the wild and landraces source [49,50,51].
A few elite pyramided lines hadimportant features similar to the recipient parent though variation was seen among the pyramided lines. The dendrogram drawn based on the studied traits indicated grouping of the pyramided and parental lines into main three clusters with similarity within the clusters (Figure 7A). All of the pyramided lines and recipient parents were observed in quadrants I and II in the biplot diagram drawn based on the 15 morpho-quality traits, indicating minor variations among the lines (Figure 11). An evaluation of the pyramided lines for yield and quality traits showed higher yieldsin pyramided lines MSS128-102-97-436, MSS128-102-97-117, MSS128-102-97-617, MSS128-102-97-601, MSS128-102-97-68, and MSS128-102-97-335 than the recipient parent (Table 4). The transfer of traits and achieving similar or better yield in the pyramided lines were also reported earlier in a few gene-pyramiding publications [48,49,52,53,54,55,56,57].
The biplot diagram places the six pyramided lines closer to each other, while the donor parent is quite far away and placed in a separate quadrant. This shows resemblance among the pyramided and recipient lines and no undesirable drag from the donor parent during transfer of the target genes into the pyramided lines. The performance of a few pyramided lines was better than the recipient parent in yield, quality, and morphological traits (Table 4). The analysis of background genotyping results showed higher recovery of the recipient parent’s genome in a few pyramided lines than the expected value in various backcross generations. Again, it revealed that transfer of Sub1 and the yield component QTLs into one genetic background may not show antagonistic effects for yield and other traits [1,48,54,56].
5. Conclusions
The pyramided lines MSS128-102-97-436, MSS128-102-97-117, MSS128-102-97-617, MSS128-102-97-601, MSS128-102-97-68, and MSS128-102-97-335 showed higher yield and submergence tolerance than the recipient parent Maudamani. The higher yield obtained in the pyramided lines might be due to an accumulation of additional yield QTLs, and no yield penalty happened due to these QTLs. In addition, the elite pyramided lines in the background of the popular variety ‘Maudamani’ may serve as potential donors of QTLs possessing Sub1+ OsSPL14+ gw5 + SCM2 in future breeding programs. Moreover, a few promising pyramided lines may be released as cultivars for flood-prone target regions in the country. Much of the grain quality and thecooking characteristics of the recipient parents such as milling %, head rice recovery %, kernel length (mm), kernel breadth (mm), kernel length after cooking, gel consistency, amylose content (%), and alkali spreading value were retained in few pyramided lines. The quality features of the popular rice variety Maudamani remained unchanged along with high grain yield. This study established the application of marker-assisted selection for transferring abiotic stresses tolerance and enhancing yield in rice.
Author Contributions
S.K.P. conceptualized the work, provided the resources, procured the funding, and supervised the work. S.K.P. wrote the original draft, E.P. and S.K.P. reviewedand edited the manuscript. S.K.P. and J.M. planned, executed, and supervised the work. E.P., S.P., S.R.B. and S.P.M. performed the investigation. Formal analysis andsoftware work were performed by E.P. and S.R.B., E.P., S.P. and S.P.M. performed the data curation. S.P. and S.P.M. validated the work. E.P., S.P. and S.R.B. visualized the work. All authors have read and agreed to the published version of the manuscript.
Funding
No externally aided fund was availed for this research work. However, the institute’s internal funding was used for this investigation.
Institutional Review Board Statement
The authors declare that this study complies with the current laws of the countries in which the experiments were performed.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data generated or analyzed in this study are included in this article.
Acknowledgments
The authors are highly grateful to the Head, Crop Improvement Division and Director, ICAR-NRRI, Cuttack for encouraging the team and for providing all the necessary facilities.
Conflicts of Interest
The authors declare that there is no competing interest and that the article is submitted without any commercial or economic interest that could be generated as a potential conflict of interest.
Abbreviations
| BC1F1 | Backcross generation 1 |
| BC2F1 | Backcross generation 2 |
| BC3F1 | Backcross generation 3 |
| GC | Gel consistency |
| QTL | Quantitative trait loci |
| MSS | Maudamani Swarna-Sub1 |
| Sub1 | Submergence tolerance |
| RBD | Randomized block design |
References
- Pradhan, S.K.; Pandit, E.; Pawar, S.; Baksh, S.Y.; Mukherjee, A.K.; Mohanty, S.P. Development of flash-flood tolerant and durable bacterial blight resistant versions of mega rice variety ‘Swarna’ through marker-assisted backcross breeding. Sci. Rep. 2019, 9, 12810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Food and Agriculture Organization. Food and Agriculture Organization of the United Nations. Rice Mark. Monit. 2017, 20, 1–38. [Google Scholar]
- Pradhan, S.K.; Barik, S.R.; Sahoo, J.; Pandit, E.; Nayak, D.K.; Pani, D.R.; Anandana, A. Comparison of Sub1 markers and their combinations for submergence tolerance and analysis of adaptation strategies of rice in rainfed lowland ecology. Comptes Rendus Biol. 2015, 338, 650–659. [Google Scholar] [CrossRef]
- Pradhan, S.K.; Chakraborti, M.; Chakraborty, K.; Behera, L.; Meher, J.; Subudhi, H.N.; Mishra, S.K.; Pandit, E.; Reddy, J.N. Genetic Improvement of Rainfed Shallow-lowland Rice for Higher Yield and Climate Resilience. In Rice Research for Enhancing Productivity, Profitability and Climate Resilience; Pathak, H., Nayak, A.K., Jena, M., Singh, O.N., Samal, P., Sharma, S.G., Eds.; ICAR-National Rice Research Institute: Cuttack, India, 2018; pp. 107–121. Available online: https://icar-nrri.in/wp-content/uploads/2019/02/Rice_Research_book_nrri.pdf (accessed on 19 May 2021).
- Ashikari, M.; Sakakibara, H.; Lin, S.; Yamamoto, T.; Takashi, T.; Nishimura, A.; Angeles, E.R.; Qian, Q.; Kitano, H.; Matsuoka, M. Cytokinin oxidase regulates rice grain production. Science 2005, 309, 741–745. [Google Scholar] [CrossRef]
- Ikeda, K.; Ito, M.; Nagasawa, N.; Kyozuka, J.; Nagato, Y. Rice ABERRANT PANICLE ORGANIZATION 1, encoding an F-box protein, regulates meristem fate. Plant J. 2007, 51, 1030–1040. [Google Scholar] [CrossRef]
- Wang, E.; Wang, J.; Zhu, X.; Hao, W.; Wang, L.; Li, Q.; Zhang, L.; He, W.; Lu, B.; Lin, H.; et al. Controlofricegrain-fillingandyieldbyagenewithapotentialsignatureofdomestication. Nat. Genet. 2008, 40, 1370–1374. [Google Scholar] [CrossRef]
- Huang, X.; Qian, Q.; Liu, Z.; Sun, H.; He, S.; Luo, D.; Xia, G.; Chu, C.; Li, J.; Fu, X. Natural variation at the DEP1 locus enhances grain yield in rice. Nat. Genet. 2009, 41, 494–497. [Google Scholar] [CrossRef]
- Xue, W.; Xing, Y.; Weng, X.; Zhao, Y.; Tang, W.; Wang, L.; Zhou, H.; Yu, S.; Xu, C.; Li, X.; et al. Natural variation in Ghd7is an important regulator of heading date and yield potential in rice. Nat. Genet. 2008, 40, 761–767. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Xu, J.; Guo, H.; Jiang, L.; Chen, S.; Yu, C.; Zhou, Z.; Hu, P.; Zhai, H.; Wan, J. DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol. 2010, 153, 1747–1758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, W.H.; Wang, P.; Chen, H.X.; Zhou, H.J.; Li, Q.P.; Wang, C.R.; Ding, Z.H.; Zhang, Y.S.; Yu, S.B.; Xing, Y.Z.; et al. A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Mol. Plant 2011, 4, 319–330. [Google Scholar] [CrossRef]
- Yuki, A.; Keiko, M.; Tsuyu, A.; Izumi, K.; Masahiro, Y.; Hidemi, K.; Yukimoto, I. The SMALL AND ROUND SEED1 (SRS1/DEP2) gene is involved in the regulation of seed size in rice. Genes Genet. Syst. 2010, 85, 327–339. [Google Scholar]
- Qiao, Y.L.; Piao, R.H.; Shi, J.X.; Lee, S.I.; Jiang, W.Z.; Kim, B.K.; Lee, J.; Han, L.; Ma, W.; Koh, H.J. Fine mapping and candidate gene analysis of dense and erect panicle 3, DEP3, which confers high grain yield in rice (Oryza sativa L.). Theor. Appl. Genet. 2011, 122, 1439–1449. [Google Scholar] [CrossRef] [PubMed]
- Song, X.-J.; Huang, W.; Shi, M.; Zhu, M.-Z.; Lin, H.-X. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 2007, 39, 623–630. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.; Xing, Y.; Mao, H.; Lu, T.; Han, B.; Xu, C.; Li, X.; Zhang, Q. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 2006, 112, 1164–1171. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Fan, C.; Xing, Y.; Jiang, Y.; Luo, L.; Sun, L.; Shao, D.; Xu, C.; Li, X.; Xiao, J.; et al. Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 2011, 43, 1266–1269. [Google Scholar] [CrossRef] [PubMed]
- Shomura, A.; Izawa, T.; Ebana, K.; Ebitani, T.; Kanegae, H.; Konishi, S.; Yano, M. Deletion in a gene associated with grain size increased yields during rice domestication. Nat. Genet. 2008, 40, 1023–1028. [Google Scholar] [CrossRef]
- Weng, J.; Gu, S.; Wan, X.; Gao, H.; Guo, T.; Su, N.; Lei, C.; Zhang, X.; Cheng, Z.; Guo, X.; et al. Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res. 2008, 18, 1199–1209. [Google Scholar] [CrossRef]
- Li, X.; Qian, Q.; Fu, Z.; Wang, Y.; Xiong, G.; Zeng, D.; Wang, X.; Liu, X.; Teng, S.; Hiroshi, F.; et al. Control of tillering in rice. Nature 2003, 422, 618–621. [Google Scholar] [CrossRef]
- Zha, X.; Luo, X.; Qian, X.; He, G.; Yang, M.; Li, Y.; Yang, J. Over-expression of the rice LRK1 gene improves quantitative yield components. Plant Biotechnol. J. 2009, 7, 611–620. [Google Scholar] [CrossRef]
- Piao, R.; Jiang, W.; Ham, T.-H.; Choi, M.-S.; Qiao, Y.; Chu, S.-H.; Park, J.-H.; Woo, M.-O.; Jin, Z.; An, G.; et al. Map-based cloning of the ERECT PANICLE 3 gene in rice. Theor. Appl. Genet. 2009, 119, 1497–1506. [Google Scholar] [CrossRef]
- Jiao, Y.; Wang, Y.; Xue, D.; Wang, J.; Yan, M.; Liu, G.; Dong, G.; Zeng, D.; Lu, Z.; Zhu, X.; et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 2010, 42, 541–544. [Google Scholar] [CrossRef] [PubMed]
- Miura, K.; Ikeda, M.; Matsubara, A.; Song, X.-J.; Ito, M.; Asano, K.; Matsuoka, M.; Kitano, H.; Ashikari, M. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 2010, 42, 545–549. [Google Scholar] [CrossRef]
- Qiao, Y.; Lee, S.-I.; Piao, R.; Jiang, W.; Ham, T.-H.; Chin, J.-H.; Piao, Z.; Han, L.; Kang, S.-Y.; Koh, H.-J. Fine mapping and candidate gene analysis of the floury endosperm gene, FLO(a), in rice. Mol. Cells 2010, 29, 167–174. [Google Scholar] [CrossRef] [PubMed]
- She, K.-C.; Kusano, H.; Koizumi, K.; Yamakawa, H.; Hakata, M.; Imamura, T.; Fukuda, M.; Naito, N.; Tsurumaki, Y.; Yaeshima, M.; et al. A novel factor FLOURY ENDOSPERM2 is involved in regulation of rice grain size and starch quality. Plant Cell 2010, 22, 3280–3294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, K.; Mackill, D.J. A major locus for submergence tolerance mapped on rice chromosome 9. Mol. Breed. 1996, 2, 219–224. [Google Scholar] [CrossRef]
- Chen, M.; Presting, G.; Barbazuk, W.B.; Goicoechea, J.L.; Blackmon, B.; Fang, G.; Kim, H.; Frisch, D.; Yu, Y.; Sun, S.; et al. An integrated physical and genetic map of the rice genome. Plant Cell 2002, 14, 537–545. [Google Scholar] [CrossRef]
- Neeraja, C.N.; Maghirang-Rodriguez, R.; Pamplona, A.; Heuer, S.; Collard, B.C.; Septiningsih, E.M.; Vergara, G.; Sanchez, D.; Xu, K.; Ismail, A.M.; et al. A marker-assisted backcross approach for developing submergence-tolerant rice cultivars. Theor. Appl. Genet. 2007, 115, 767–776. [Google Scholar] [CrossRef]
- Iftekharuddaula, K.M.; Newaz, M.A.; Salam, M.A.; Ahmed, H.U.; Mahbub, M.A.A.; Septiningsih, E.M.; Collard, B.C.Y.; Sanchez, D.L.; Pamplona, A.M.; Mackill, D.J. Rapid and high-precision marker assisted backcrossing to introgress the SUB1 QTL into BR11, the rainfed lowland rice mega variety of Bangladesh. Euphytica 2011, 178, 83–97. [Google Scholar] [CrossRef]
- Manivong, P.; Korinsak, S.; Siangliw, J.L.; Vanavichit, A.; Toojinda, T. Marker-assisted selection to improve submergence tolerance, blast resistance and strong fragrance in glutinous rice. Thai J. Genet. 2014, 7, 110–122. [Google Scholar]
- Khush, G.S.; Mackill, D.J.; Sidhu, G.S. Breeding Rice for Resistance to Bacterial Leaf Blight; IRRI: Manila, Philippines, 1989; pp. 207–217. [Google Scholar]
- Pradhan, S.K.; Pandit, E.; Pawar, S.; Naveenkumar, R.; Barik, S.R.; Mohanty, S.P.; Nayak, D.K.; Ghritlahre, S.K.; Rao, D.S.; Reddy, J.N.; et al. Linkage disequilibrium mapping for grain Fe and Zn enhancing QTLs useful for nutrient dense rice breeding. BMC Plant Biol. 2020, 20, 57. [Google Scholar] [CrossRef] [Green Version]
- Ookawa, T.; Hobo, T.; Yano, M.; Murata, K.; Ando, T.; Miura, H.; Asano, K.; Ochiai, Y.; Ikeda, M.; Nishitani, R.; et al. New approach for rice improvement using a pleiotropic QTL gene for lodging resistance and yield. Nat. Commun. 2010, 1, 132. [Google Scholar] [CrossRef] [Green Version]
- Xu, K.; Xu, X.; Fukao, T.; Canlas, P.; Maghirang-Rodriguez, R.; Heuer, S.; Ismail, A.M.; Bailey-Serres, J.; Ronald, P.C.; Mackill, D.J. Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 2006, 442, 705–708. [Google Scholar] [CrossRef] [Green Version]
- Septiningsih, E.M.; Pamplona, A.M.; Sanchez, D.L.; Neeraja, C.N.; Vergara, G.V.; Heuer, S.; Ismail, A.M.; Mackill, D.J. Development of submergence tolerant rice cultivars: The Sub1 locus and beyond. Ann. Bot. 2009, 103, 151–160. [Google Scholar] [CrossRef] [Green Version]
- Dellaporta, S.L.; Wood, J.; Hicks, J.B. A plant DNA mini preparation: Version II. Plant Mol. Biol. Rep. 1983, 1, 19–21. [Google Scholar] [CrossRef]
- Pavalíce, A.; Hrda, S.; Flegr, J. Free Tree—freeware program for construction of phylogenetic trees on the basis of distance data and bootstrap/jackknife analysis of the tree robustness. Application in the RAPD analysis of genus Frenkelia. Folia Biol. (Pragua) 1999, 45, 97–99. [Google Scholar]
- Hampl, V.; Pavlicek, A.; Flegr, J. Construction and bootstrap analysis of DNA fingerprinting based phylogenetic trees with the freeware program FreeTree: Application to trichomonad parasites. Int. J. Syst. Evol. Microbiol. 2001, 51, 731–735. [Google Scholar] [CrossRef]
- Page, R.D. TreeView: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 1996, 12, 357–358. [Google Scholar] [PubMed] [Green Version]
- Van Berloo, R. GGT: Software for display of graphical genotypes. J. Hered. 1999, 90, 328–330. [Google Scholar] [CrossRef] [Green Version]
- Tan, Y.F.; Li, J.X.; Yu, S.B.; Xing, Y.Z.; Xu, C.G.; Zhang, Q. The three important traits for cooking and eating quality of rice grains are controlled by a single locus in an elite rice hybrid, Shanyou63. Theor. Appl. Genet. 1999, 99, 642–648. [Google Scholar] [CrossRef]
- Cagampang, G.B.; Perez, C.M.; Juliano, B.O. A gel consistency test for eating quality of rice. J. Sci. Food Agric. 1973, 24, 1589–1594. [Google Scholar] [CrossRef]
- Juliano, B.O. Rice quality screening with the Rapid ViscoAnalyser. In Applicntions of the Rapid ViscoAnalyser; Walker, C.E., Hazelton, J.L., Eds.; Newport Scientific: Sydney, Australia, 1996; p. 19. [Google Scholar]
- SAS Institute Inc. Statistical Analysis System, version 9.2.; SAS Institute Inc.: Cary, NC, USA, 2008. [Google Scholar]
- Pandit, E.; Tasleem, S.; Nayak, D.K.; Barik, S.R.; Mohanty, D.P.; Das, S.; Pradhan, S.K. Genome-wide association mapping reveals multiple QTLs governing tolerance response for seedling stage chilling stress in Indica rice. Front. Plant Sci. 2017, 8, 552. [Google Scholar] [CrossRef] [Green Version]
- Pandit, E.; Panda, R.K.; Sahoo, A.; Pani, D.R.; Pradhan, S.K. Genetic relationship and structure analyses of root growth angle for improvement of drought avoidance in early and mid-early maturing rice genotypes. Rice Sci. 2020, 27, 124–132. [Google Scholar] [CrossRef]
- Pradhan, S.K.; Pandit, E.; Pawar, S.; Bharati, B.; Chatopadhyay, K.; Singh, S.; Dash, P.; Reddy, J.N. Association mapping reveals multiple QTLs for grain protein content in rice useful for biofortification. Mol. Genet. Genom. 2019, 294, 963–983. [Google Scholar] [CrossRef]
- Pradhan, S.K.; Nayak, D.K.; Mohanty, S.; Behera, L.; Barik, S.R.; Pandit, E.; Lenka, S.; Anandan, A. Pyramiding of three bacterial blight resistance genes for broad-spectrum resistance in deepwater rice variety, Jalmagna. Rice 2015, 8, 19. [Google Scholar] [CrossRef]
- Sundaram, R.M.; Vishnupriya, M.R.; Biradar, S.K.; Laha, G.S.; Reddy, G.A.; Rani, N.S.; Sarma, N.P.; Sonti, R.V. Marker assisted introgression of bacterial blight resistance in Samba Mahsuri, an elite indica rice variety. Euphytica 2008, 160, 411–422. [Google Scholar] [CrossRef]
- Pradhan, S.K.; Nayak, D.K.; Pandit, E.; Behera, L.; Anandan, A.; Mukherjee, A.K.; Lenka, S.; Barik, D.P. Incorporation of bacterial blight resistance genes into lowland rice cultivar through marker assisted backcross breeding. Phytopathology 2016, 6, 710–718. [Google Scholar] [CrossRef] [Green Version]
- Nayak, D.K.; Pandit, E.; Mohanty, S.; Barik, D.P.; Pradhan, S.K. Marker assisted selection in back cross progenies for transfer of bacterial leaf blight resistance genes into a popular lowland rice cultivar. Oryza 2015, 52, 163–168. [Google Scholar]
- Sonti, R.V. Bacterial leaf blight of rice: New insights from molecular genetics. Curr. Sci. 1998, 74, 206–212. [Google Scholar]
- Sanchez, A.C.; Brar, D.S.; Huang, N.; Li, Z.; Khush, G.S. Sequence tagged site markers-assisted selection for three bacterial blight resistance genes in rice. Crop Sci. 2000, 40, 792–797. [Google Scholar] [CrossRef]
- Singh, S.; Sidhu, J.S.; Huang, N.; Vikal, Y.; Li, Z.; Brar, D.S.; Dhaliwal, H.S.; Khush, G.S. Pyramiding three bacterial blight resistance genes (xa-5, xa-13 and Xa-21) using marker-assisted selection into indica rice cultivar PR-106. Theor. Appl. Genet. 2001, 102, 1011–1015. [Google Scholar] [CrossRef]
- Perez, L.M.; Redona, E.D.; Mendioro, M.S.; Vera Cruz, C.M.; Leung, H. Introgression of Xa4, Xa7 and Xa21 for resistance to bacterial blight in thermo-sensitive genetic male sterile rice (Oryzasativa L.) for the development of two-line hybrids. Euphytica 2008, 164, 627–636. [Google Scholar] [CrossRef]
- Dokku, P.; Das, K.M.; Rao, G.J.N. Pyramiding of four resistance genes of bacterial blight in Tapaswini, an elite rice cultivar, through marker-assisted selection. Euphytica 2013, 192, 87–96. [Google Scholar] [CrossRef]
- Pradhan, S.K.; Nayak, D.K.; Pandit, E.; Barik, S.R.; Mohanty, S.P.; Anandan, A.; Reddy, J.N. Characterization of morpho-quality traits and validation of bacterial blight resistance in pyramided rice genotypes under various hotspots of India. Aust. J. Crop. Sci. 2015, 9, 127–134. [Google Scholar]
- Das, G.; Rao, G.J.N. Molecular marker assisted gene stacking for biotic and abiotic stress resistance genes in an elite rice cultivar. Front. Plant Sci. 2015, 6, 698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, G.; Rao, G.J.; Varier, M.; Prakash, A.; Prasad, D. Improved Tapaswini having four BB resistance genes pyramided with six genes/QTLs, resistance/tolerance to biotic and abiotic stresses in rice. Sci. Rep. 2018, 8, 2413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pradhan, S.K.; Barik, S.R.; Nayak, D.K.; Pradhan, A.; Pandit, E.; Nayak, P.; Das, S.R.; Pathak, H. Genetics, Molecular Mechanisms and Deployment of Bacterial Blight Resistance Genes in Rice. Crit. Rev. Plant Sci. 2020, 39, 360–385. [Google Scholar] [CrossRef]
- Mohanty, S.P.; Kumbhakar, S.; Pandit, E.; Barik, S.R.; Mohanty, D.P.; Nayak, D.K.; Singh, N.R.; Pradhan, S.K. Molecular screening of yield component QTLs for strong culm, grain number and grain width using gene specific markers in indica-tropical japonica derived rice lines. Oryza 2016, 53, 136–143. [Google Scholar]
- Mohapatra, S.; Pandit, E.; Mohanty, S.P.; Barik, S.R.; Pawar, S.; Nayak, D.K.; Subudhi, H.N.; Das, L.; Pradhan, S.K. Molecular and phenotypic analyses of yield components QTLs in IR64 backcross progenies and popular high yielding rice varieties of India. Oryza 2018, 55, 271–277. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).