Single Seed-Based High-Throughput Genotyping and Rapid Generation Advancement for Accelerated Groundnut Genetics and Breeding Research
Abstract
1. Introduction
2. Materials and Methods
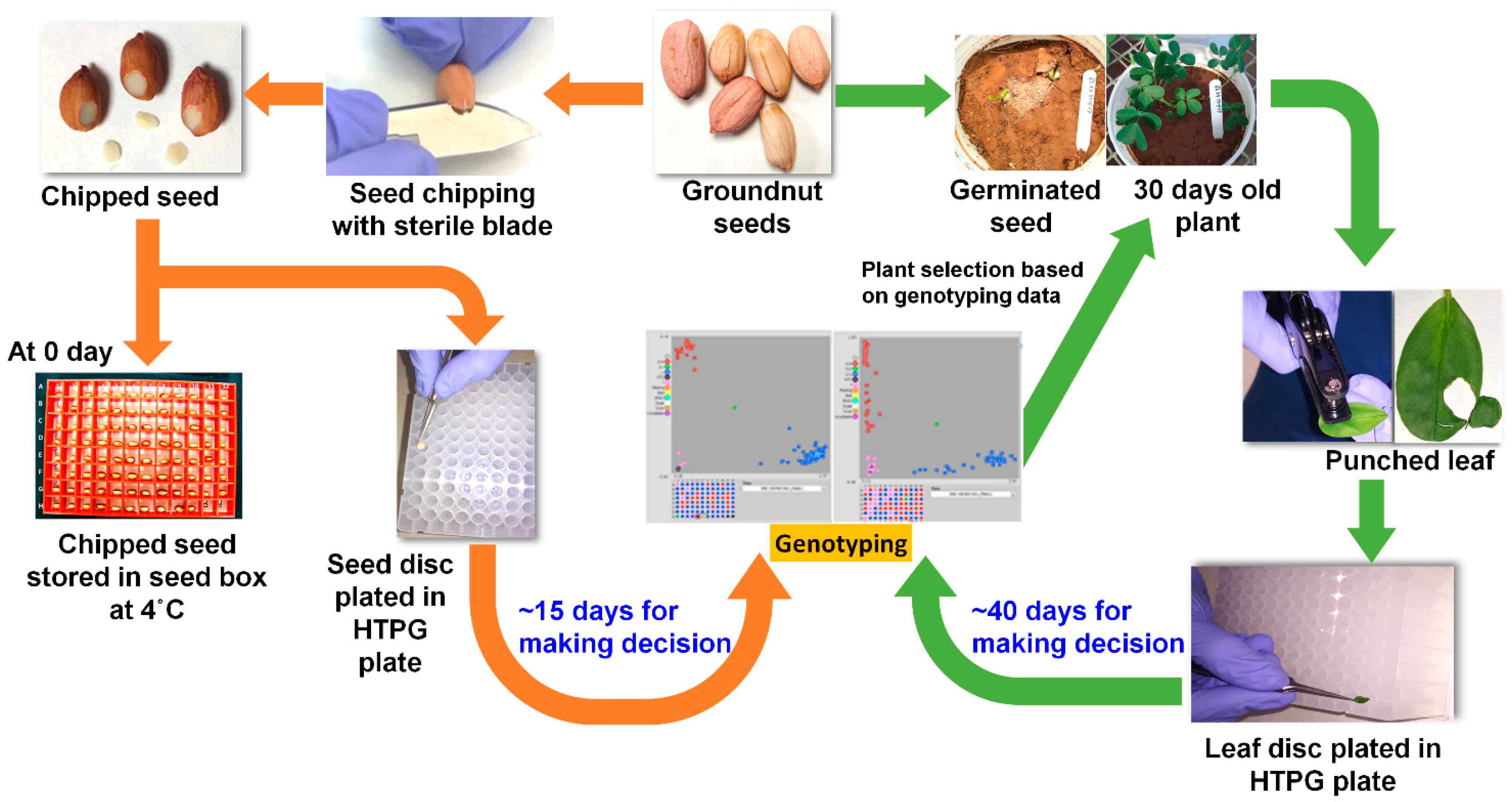
2.1. Plant Material and Generation Advancement
2.2. Sample Collection and DNA Extraction for Genotyping
2.3. Germination Assay for the Chipped and Control Seed Material
2.4. Comparison of DNA Quality and Quantity between Leaf Punching and Seed Chipping Followed by Genotyping Using KASP Markers
2.5. Rapid Generation Advancement House and Crop Growing Facility
2.6. Phenotype-Based Late Generation Selection vs. Marker-Based Early Generation Selection
2.7. Cost and Time Comparison among DNA, Leaf-Punching and Seed-Chipping Based Genotyping
3. Results
3.1. Comparison of Germination Percentage between Chipped and Non-Chipped (Control) Seeds
3.2. Comparison of DNA Quality and Quantity between Seed-Chip and Leaf-Disc Samples
3.3. Comparison of Genotyping Results between Seed-Chip and Leaf-Disc Samples
3.4. Optimization and Deployment of Seed-Chip Based Genotyping for Early Generation Selection in Groundnut Breeding
3.5. Phenotype Based Selection in Late Generation vs. Marker-Based Selection in Early Generations
3.6. Cost and Time Comparison for Genotyping among Different Sample Types (CTAB, Leaf-Punching, and Seed-Chipping)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pandey, M.K.; Roorkiwal, M.; Singh, V.K.; Ramalingam, A.; Kudapa, H.; Thudi, M.; Chitikineni, A.; Rathore, A.; Varshney, R.K. Emerging genomic tools for legume breeding: Current status and future prospects. Front. Plant Sci. 2016, 7, 455. [Google Scholar] [CrossRef]
- Varshney, R.K.; Pandey, M.K.; Bohra, A.; Singh, V.K.; Thudi, M.; Saxena, R.K. Toward the sequence-based breeding in legumes in the post-genome sequencing era. Theor. Appl. Genet. 2016, 132, 797–816. [Google Scholar] [CrossRef]
- Desmae, H.; Janila, P.; Okori, P.; Pandey, M.K.; Motagi, B.N.; Monyo, E.; Mponda, O.; Okello, D.; Sako, D.; Echeckwu, C.; et al. Genetics, genomics and breeding of groundnut (Arachis hypogaea L.). Plant Breed. 2019, 138, 425–444. [Google Scholar] [CrossRef]
- Pandey, M.K.; Pandey, A.K.; Kumar, R.; Nwosu, V.; Guo, B.; Wright, G.; Bhat, R.S.; Chen, X.; Bera, S.K.; Yuan, M.; et al. Translational genomics for achieving higher genetic gains in groundnut. Theor. Appl. Genet. 2020, 133, 1679–1702. [Google Scholar] [CrossRef]
- King, Z.; Serrano, J.; Boerma, H.R.; Li, Z. Non-toxic and efficient DNA extractions for soybean leaf and seed chips for high-throughput and large-scale genotyping. Biotechnol. Lett. 2014, 36, 1875–1879. [Google Scholar] [CrossRef]
- Arbelaez, J.D.; Tandayu, E.; Reveche, M.Y.; Jarana, A.; Van Rogen, P.; Sandager, L.; Stolt, P.; Ng, E.; Varshney, R.K.; Kretzschmar, T.; et al. Methodology: Ssb-MASS: A single seed-based sampling strategy for marker-assisted selection in rice. Plant Methods 2019, 15, 78. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Martinez, C.; Skinner, D.J.; Krivanek, A.F.; Crouch, J.H.; Xu, Y. Development of a seed DNA-based genotyping system for marker-assisted selection in maize. Mol. Breed. 2008, 22, 477. [Google Scholar] [CrossRef]
- Deshmukh, D.B.; Chaudhari, S.; Balram, M.; Durga Rani, C.; Sudini, H.K.; Murali, T.V.; Manohar, S.M.; Janila, P. A non-destructive seed sampling method for high throughput genotyping in groundnut. J. Res. PJTSAU 2018, 46, 20–27. [Google Scholar]
- Sujay, V.; Gowda, M.V.; Pandey, M.K.; Bhat, R.S.; Khedikar, Y.P.; Nadaf, H.L.; Gautami, B.; Sarvamangala, C.; Lingaraju, S.; Radhakrishan, T.; et al. Quantitative trait locus analysis and construction of consensus genetic map for foliar disease resistance based on two recombinant inbred line populations in cultivated groundnut (Arachis hypogaea L.). Mol. Breed. 2012, 30, 773–788. [Google Scholar] [CrossRef]
- Pandey, M.K.; Khan, A.W.; Singh, V.K.; Vishwakarma, M.K.; Shasidhar, Y.; Kumar, V.; Garg, V.; Bhat, R.S.; Chitikineni, A.; Janila, P.; et al. QTL-seq approach identified genomic regions and diagnostic markers for rust and late leaf spot resistance in groundnut (Arachis hypogaea L.). Plant Biotechnol. J. 2017, 15, 927–941. [Google Scholar] [CrossRef]
- Janila, P.; Variath, M.T.; Pandey, M.K.; Desmae, H.; Motagi, B.N.; Okori, P.; Manohar, S.S.; Rathnakumar, A.L.; Radhakrishnan, T.; Liao, B.; et al. Genomic tools in groundnut breeding program: Status and perspectives. Front. Plant Sci. 2016, 7, 289. [Google Scholar] [CrossRef]
- Chen, Z.; Wang, M.L.; Barkley, N.A.; Pittman, R.N. A simple allele-specific PCR assay for detecting FAD2 alleles in both A and B genomes of the cultivated peanut for high-oleate trait selection. Plant Mol. Biol. Rep. 2010, 28, 542–548. [Google Scholar] [CrossRef]
- Deshmukh, D.; Marathi, B.; Sudini, H.K.; Variath, M.T.; Chaudhari, S.; Manohar, S.S.; Rani, C.V.D.; Pandey, M.K.; Pasupuleti, J. Combining high oleic acid trait and resistance to late leaf spot and rust diseases in groundnut (Arachis hypogaea L.). Front. Genet. 2020, 11, 514. [Google Scholar] [CrossRef] [PubMed]
- Bohar, R.; Chitikineni, A.; Varshney, R.K. Genetic molecular markers to accelerate genetic gains in crops. Biotechniques 2020, 69. [Google Scholar] [CrossRef]
- Chu, Y.; Wu, C.L.; Holbrook, C.C.; Tillman, B.L.; Person, G.; Ozias-Akins, P. Marker-assisted selection to pyramid nematode resistance and the high oleic trait in peanut. Plant Genome 2011, 4, 110–117. [Google Scholar] [CrossRef]
- Varshney, R.K.; Pandey, M.K.; Janila, P.; Nigam, S.N.; Sudini, H.; Gowda, M.; Sriswathi, M.; Radhakrishnan, T.; Manohar, S.S.; Nagesh, P. Marker-assisted introgression of a QTL region to improve rust resistance in three elite and popular varieties of peanut (Arachis hypogaea L.). Theor. Appl. Genet. 2014, 127, 1771–1781. [Google Scholar] [CrossRef] [PubMed]
- Yeri, S.B.; Bhat, R.S. Development of late leaf spot and rust resistant backcross lines in JL 24 variety of groundnut (Arachis hypogaea L.). Electron. J. Plant Breed. 2016, 7, 37–41. [Google Scholar] [CrossRef]
- Kolekar, R.M.; Sukruth, M.; Shirasawa, K.; Nadaf, H.L.; Motagi, B.N.; Lingaraju, S.; Patil, P.V.; Bhat, R.S. Marker-assisted backcrossing to develop foliar disease-resistant genotypes in TMV 2 variety of peanut (Arachis hypogaea L.). Plant Breed. 2017, 136, 948–953. [Google Scholar] [CrossRef]
- Bera, S.K.; Kamdar, J.H.; Kasundra, S.V.; Dash, P.; Maurya, A.K.; Jasani, M.D.; Chandrashekar, A.B.; Manivannan, N.; Vasanthi, R.P.; Dobariya, K.L.; et al. Improving oil quality by altering levels of fatty acids through marker-assisted selection of ahfad2 alleles in peanut (Arachis hypogaea L.). Euphytica 2018, 214, 162. [Google Scholar] [CrossRef]
- Nawade, B.; Mishra, G.P.; Radhakrishnan, T.; Sangh, C.; Dobariya, J.R.; Kundu, R. Development of high oleic peanut lines through marker-assisted introgression of mutant ahFAD2 alleles and its fatty acid profiles under open-field and controlled conditions. 3 Biotech. 2019, 9, 243. [Google Scholar] [CrossRef] [PubMed]
- Bera, S.K.; Kamdar, J.H.; Kasundra, S.V.; Patel, S.V.; Jasani, M.D.; Maurya, A.K.; Dash, P.; Chandrashekar, A.B.; Rani, K.; Manivannan, N.; et al. Steady expression of high oleic acid in peanut bred by marker-assisted backcrossing for fatty acid desaturase mutant alleles and its effect on seed germination along with other seedling traits. PLoS ONE 2019, 14, e0226252. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Qi, F.; Sun, Z.; Miao, L.; Zhang, Z.; Liu, H.; Fang, Y.; Dong, W.; Tang, F.; Zheng, Z.; et al. Marker-assisted backcrossing to improve seed oleic acid content in four elite and popular peanut (Arachis hypogaea L.) cultivars with high oil content. Breed. Sci. 2019, 18107. [Google Scholar] [CrossRef] [PubMed]
- Shasidhar, Y.; Variath, M.T.; Vishwakarma, M.K.; Manohar, S.S.; Gangurde, S.S.; Sriswathi, M.; Sudini, H.K.; Dobariya, K.L.; Bera, S.K.; Radhakrishnan, T.; et al. Improvement of three popular Indian groundnut varieties for foliar disease resistance and high oleic acid using SSR markers and SNP array in marker-assisted backcrossing. Crop J. 2020, 8, 1–15. [Google Scholar] [CrossRef]
- Janila, P.; Nigam, S.N.; Pandey, M.K.; Nagesh, P.; Varshney, R.K. Groundnut improvement: Use of genetic and genomic tools. Front. Plant Sci. 2013, 4, 23. [Google Scholar] [CrossRef] [PubMed]
- Mace, E.S.; Buhariwalla, K.K.; Buhariwalla, H.K.; Crouch, J.H. A high-throughput DNA extraction protocol for tropical molecular breeding programs. Plant Mol. Biol. Rep. 2003, 21, 459–460. [Google Scholar] [CrossRef]
- Pandey, M.K.; Agarwal, G.; Kale, S.M.; Clevenger, J.; Nayak, S.N.; Sriswathi, M.; Chitikineni, A.; Chavarro, C.; Chen, X.; Upadhyaya, H.D.; et al. Development and evaluation of a high density genotyping ‘Axiom_Arachis’ array with 58 k SNPs for accelerating genetics and breeding in groundnut. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef]
- Bertioli, D.J.; Jenkins, J.; Clevenger, J.; Dudchenko, O.; Gao, D.; Seijo, G.; Leal Bertioli, S.C.; Ren, L.; Farmer, A.D.; Pandey, M.K.; et al. The genome sequence of peanut (Arachis hypogaea), a segmental allotetraploid. Nat. Genet. 2019, 51, 877–884. [Google Scholar] [CrossRef]
- Chen, X.; Lu, Q.; Liu, H.; Zhang, J.; Hong, Y.; Lan, H.; Li, H.; Wang, J.; Liu, H.; Li, S.; et al. Sequencing of cultivated peanut, Arachis hypogaea, yields insights into genome evolution and oil improvement. Mol. Plant. 2019, 12, 920–934. [Google Scholar] [CrossRef]
- Zhuang, W.; Chen, H.; Yang, M.; Wang, J.; Pandey, M.K.; Zhang, C.; Chang, W.C.; Zhang, L.; Zhang, X.; Tang, R.; et al. The genome of cultivated peanut provides insight into legume karyotypes, polyploid evolution and crop domestication. Nat. Genet. 2019, 51, 865–876. [Google Scholar] [CrossRef] [PubMed]
- Pandey, M.K.; Monyo, E.; Ozias-Akins, P.; Liang, X.; Guimarães, P.; Nigam, S.N.; Upadhyaya, H.D.; Janila, P.; Zhang, X.; Guo, B.; et al. Advances in Arachis genomics for peanut improvement. Biotechnol. Adv. 2012, 30, 639–651. [Google Scholar] [CrossRef]
- Varshney, R.; Thudi, M.; Pandey, M.K.; Tardieu, F.; Ojiewo, C.; Vadez, V.; Whitbread, A.M.; Siddique, K.H.; Nguyen, H.T.; Carberry, P.S.; et al. Accelerating genetic gains in legumes for the development of prosperous smallholder agriculture: Integrating genomics, phenotyping, systems modelling and agronomy. J. Exp. Bot. 2018, 69, 3293–3312. [Google Scholar] [CrossRef]
- Simpson, C.E.; Baring, M.R.; Schubert, A.M.; Melouk, H.A.; Lopez, Y.; Kirby, J.S. Registration of ‘OLin’peanut. Crop Sci. 2003, 43, 1880–1881. [Google Scholar] [CrossRef]
- Mishra, M.K.; Rani, N.S.; Ram, A.S.; Sreenath, H.L.; Jayarama, N. A simple method of DNA extraction from coffee seeds suitable for PCR analysis. Afr. J. Biotechnol. 2008, 7, 409–413. [Google Scholar]
- Lu, J.; Hou, J.; Ouyang, Y.; Luo, H.; Zhao, J.; Mao, C.; Han, M.; Wang, L.; Xiao, J.; Yang, Y.; et al. A direct PCR-based SNP marker–assisted selection system (D-MAS) for different crops. Mol. Breed. 2020, 40, 9. [Google Scholar] [CrossRef]
- Gupta, R.; Chandrashekar, U.S.; Chakrabarty, S.K.; Dadlani, M. A simple modified method of DNA extraction from seeds for PCR amplifications. Indian J. Agr. Sci. 2012, 82, 75. [Google Scholar]
- Biswas, C.; Dey, P.; Satpathy, S.; Sarkar, S.K.; Bera, A.; Mahapatra, B.S. A simple method of DNA isolation from jute (Corchorus olitorius) seed suitable for PCR-based detection of the pathogen Macrophominaphaseolina (Tassi) Goid. Lett. Appl. Microbiol. 2013, 56, 105–110. [Google Scholar] [CrossRef]
- Xia, Y.; Chen, F.; Du, Y.; Liu, C.; Bu, G.; Xin, Y.; Liu, B. A modified SDS-based DNA extraction method from raw soybean. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef]
- Kumar, R.P.; Sujatha, K.; Rao, K.S.; Natarajkumar, P.; Viraktamath, B.C.; Balachandran, S.M.; Biswal, A.K.; Sundaram, R.M. A protocol for isolation of DNA suitable for rapid seed and grain purity assessments in rice. Rice Genet. Newsl. 2007, 23, 92–95. [Google Scholar]
- Lopez, Y.; Smith, O.D.; Senseman, S.A.; Rooney, W.L. Genetic factors influencing high oleic acid content in Spanish market-type peanut cultivars. Crop Sci. 2001, 41, 51–56. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parmar, S.; Deshmukh, D.B.; Kumar, R.; Manohar, S.S.; Joshi, P.; Sharma, V.; Chaudhari, S.; Variath, M.T.; Gangurde, S.S.; Bohar, R.; et al. Single Seed-Based High-Throughput Genotyping and Rapid Generation Advancement for Accelerated Groundnut Genetics and Breeding Research. Agronomy 2021, 11, 1226. https://doi.org/10.3390/agronomy11061226
Parmar S, Deshmukh DB, Kumar R, Manohar SS, Joshi P, Sharma V, Chaudhari S, Variath MT, Gangurde SS, Bohar R, et al. Single Seed-Based High-Throughput Genotyping and Rapid Generation Advancement for Accelerated Groundnut Genetics and Breeding Research. Agronomy. 2021; 11(6):1226. https://doi.org/10.3390/agronomy11061226
Chicago/Turabian StyleParmar, Sejal, Dnyaneshwar B. Deshmukh, Rakesh Kumar, Surendra S. Manohar, Pushpesh Joshi, Vinay Sharma, Sunil Chaudhari, Murali T. Variath, Sunil S. Gangurde, Rajaguru Bohar, and et al. 2021. "Single Seed-Based High-Throughput Genotyping and Rapid Generation Advancement for Accelerated Groundnut Genetics and Breeding Research" Agronomy 11, no. 6: 1226. https://doi.org/10.3390/agronomy11061226
APA StyleParmar, S., Deshmukh, D. B., Kumar, R., Manohar, S. S., Joshi, P., Sharma, V., Chaudhari, S., Variath, M. T., Gangurde, S. S., Bohar, R., Singam, P., Varshney, R. K., Janila, P., & Pandey, M. K. (2021). Single Seed-Based High-Throughput Genotyping and Rapid Generation Advancement for Accelerated Groundnut Genetics and Breeding Research. Agronomy, 11(6), 1226. https://doi.org/10.3390/agronomy11061226










