Agrobacterium-Mediated Transformation of pMDC140 Plasmid Containing the Wheatwin2 Gene into the Tadong Rice Genome
Abstract
1. Introduction
2. Materials and Methods
2.1. Synthesis of wwin2 Gene and Integration into a pMDC140 Vector
2.2. Transformation of pMDC140-wwin2 into Agrobacterium tumefaciens
2.3. Agrobacterium-Mediated Rice Calli Transformation
2.4. Histochemical GUS Assay of the Transformed Calli
2.5. Validation of Transformation Using Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR)
2.6. Confirmation of Gene Expression Using Quantitative Real-Time Polymerase Chain Reaction (qPCR)
3. Results
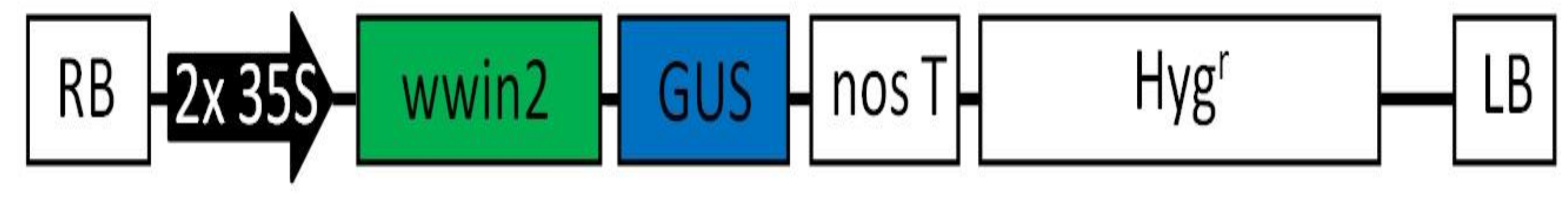
3.1. Mapping of the Newly Constructed pMDC140 T-DNA Region
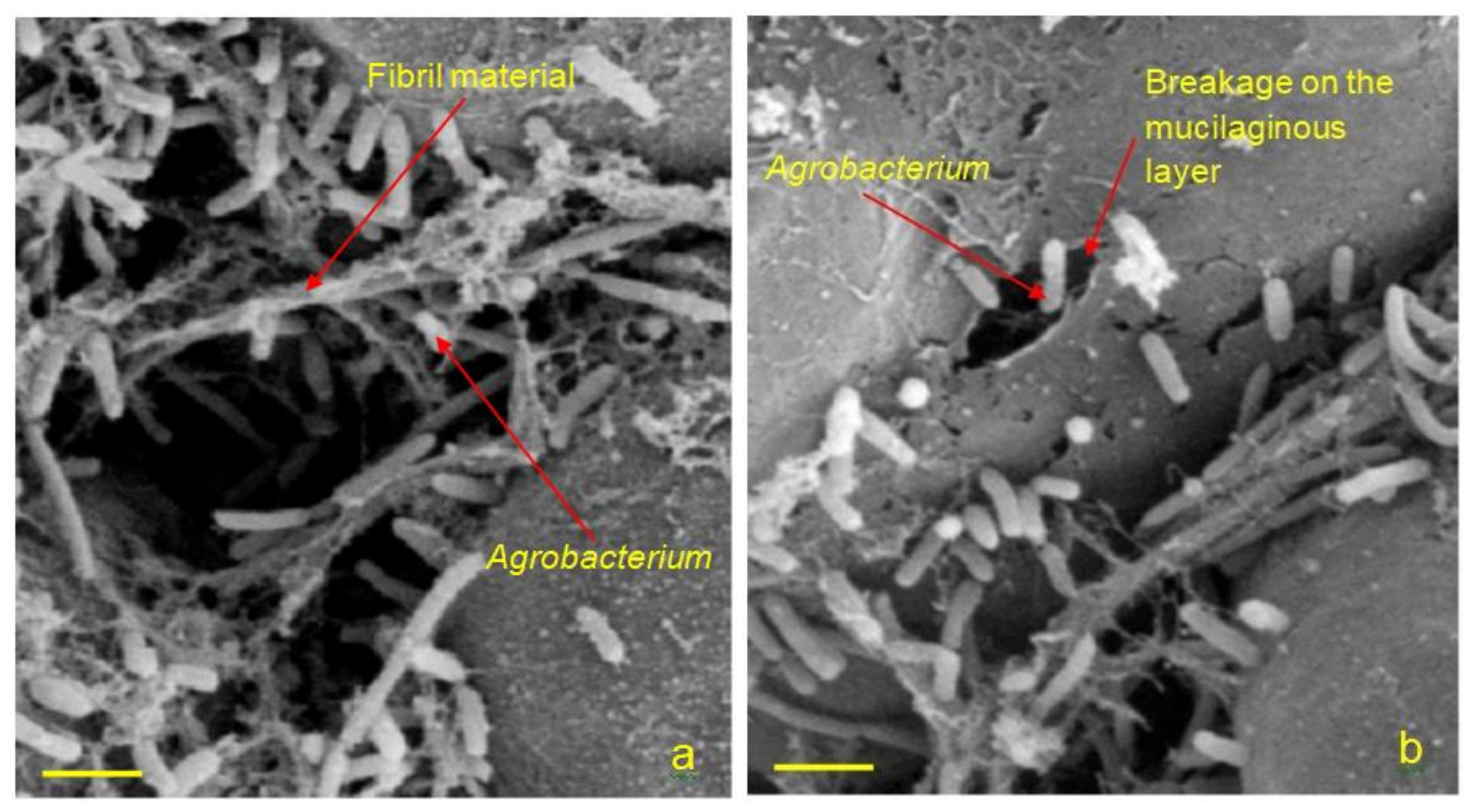
3.2. SEM Imaging on Agrobacterium-Transformed Rice Callus Surface
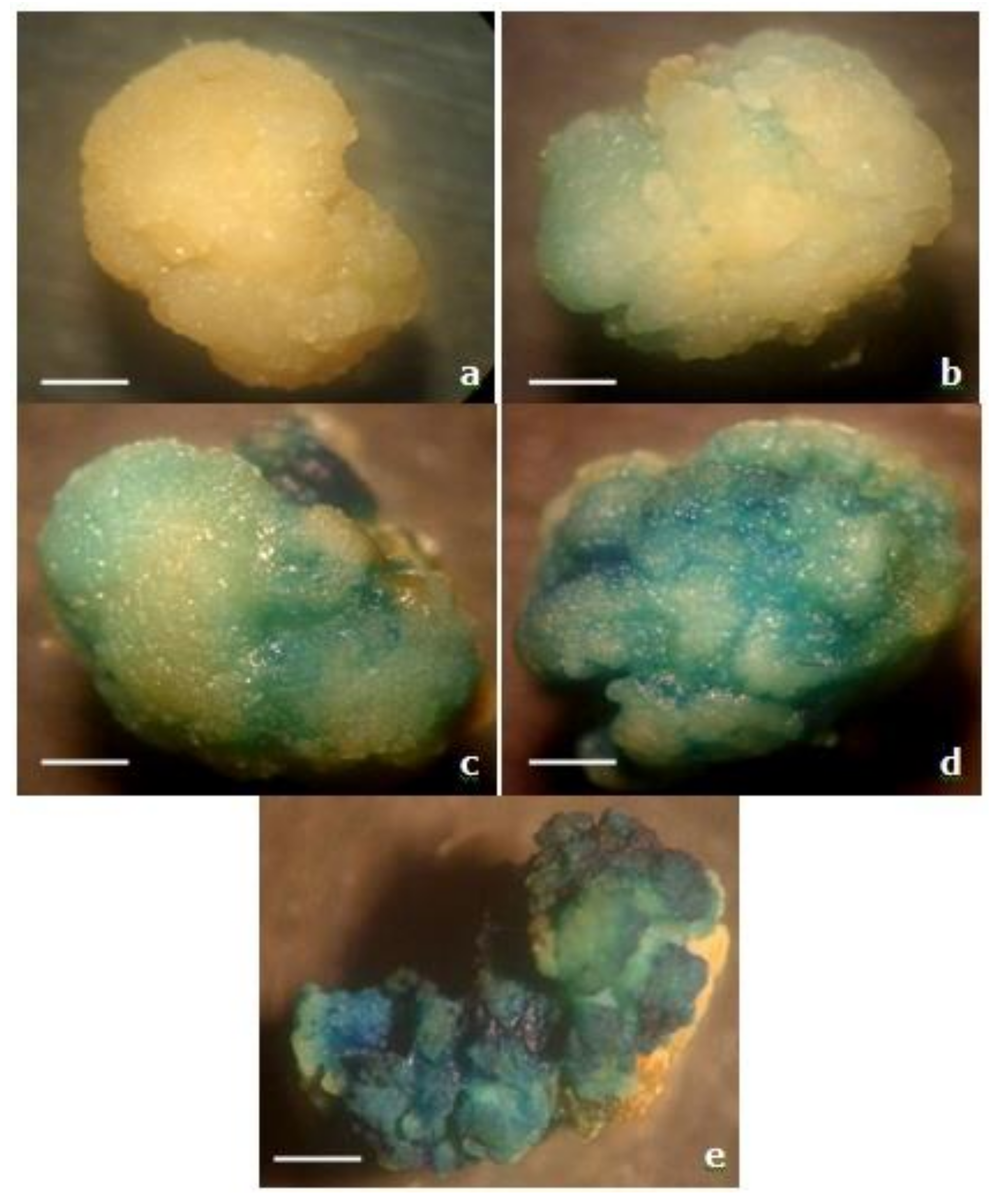
3.3. Microscopic Observation of GUS Expression in the Transformed Calli
3.4. RT-PCR to Confirm the Transformation in Rice Calli
3.5. Expressions of wwin2 and hptII Genes in the Transformed Calli with sps Gene as an Endogenous Reference
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhao, K.; Wright, M.; Kimball, J.; Eizenga, G.; McClung, A.; Kovach, M.; Tyagi, W.; Ali, M.L.; Tung, C.W.; Reynolds, A.; et al. Genomic diversity and introgression in O. sativa reveal the impact of domestication and breeding on the rice genome. PLoS ONE 2010, 5, e10780. [Google Scholar] [CrossRef] [PubMed]
- Chong, E.T.J.; Goh, L.P.W.; Wong, J.J.; Abdul Aziz, Z.; Surugau, N.; Latip, M.A.; Lee, P.-C. Genetic diversity and relationship of Sabah traditional rice varieties as revealed by RAPD markers. Pertanika J. Trop. Agric. Sci. 2018, 41, 177–190. [Google Scholar]
- Kim, S.R.; Torollo, G.; Yoon, M.R.; Kwak, J.; Lee, C.K.; Prahalada, G.D.; Choi, I.R.; Yeo, U.S.; Jeong, O.Y.; Jena, K.K.; et al. Loss-of-function alleles of heading date 1 (HD1) are associated with adaptation of temperate japonica rice plants to the tropical region. Front. Plant Sci. 2018, 9, 1827. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Torollo, G.; Ndayiragijie, A.; Bizimana, J.B.; Choi, I.R.; Gulles, A.; Yeo, U.S.; Jeong, O.Y.; Venkatanagappa, S.; Kim, B.K. Genetic relationship of tropical region-bred temperate japonica rice (Oryza sativa) plants and their grain yield variations in three different tropical environments. Plant Breed. 2018, 137, 857–864. [Google Scholar] [CrossRef]
- Chong, E.T.J.; Goh, L.P.W.; Latip, M.A.; Abdul Aziz, Z.; Surugau, N.; Lee, P.-C. Genetic diversity of upland traditional rice varieties in Malaysian Borneo based on mitochondrial cytochrome c oxidase 3 gene analysis. AIMS Agric. Food 2021, 6, 235–246. [Google Scholar] [CrossRef]
- Food and Agriculture Organization. Available online: http://www.fao.org/faostat/en/#data/QC/visualize (accessed on 5 February 2021).
- Food and Agricultural Organization. FAOSTAT Database; Food and Agricultural Organization: Rome, Italy, 2013. [Google Scholar]
- Rao, K.M. Rice Blast Disease; Daya Publishing House: Delhi, India, 1994; p. 1. [Google Scholar]
- Oh, Y.; Donofrio, N.; Pan, H.; Coughlan, S.; Brown, D.E.; Meng, S.; Mitchell, T.; Dean, R.A. Transcriptome analysis reveals new insight into appressorium formation and function in the rice blast fungus Magnaporthe oryzae. Genome Biol. 2008, 9, R85. [Google Scholar] [CrossRef]
- Nalley, L.; Tsiboe, F.; Durand-Morat, A.; Shew, A.; Thoma, G. Economic and environmental impact of rice blast pathogen (Magnaporthe oryzae) alleviation in the United States. PLoS ONE 2016, 11, e0167295. [Google Scholar] [CrossRef] [PubMed]
- Goh, L.P.W.; Chong, E.T.J.; Wong, J.J.; Abdul Aziz, Z.; Surugau, N.; Latip, M.A.; Lee, P.-C. Molecular identification of blast resistance and pathogenesis-related genes in various traditional paddy varieties from different divisions of Sabah, East Malaysia. Int. Food Res. J. 2018, 25, 626–631. [Google Scholar]
- Miah, G.; Rafii, M.Y.; Ismail, M.R.; Sahebi, M.; Hashemi, F.S.G.; Yusuff, O.; Usman, M.G. Blast disease intimidation towards rice cultivation: A review of pathogen and strategies to control. J. Anim. Plant Sci. 2017, 27, 1058–1066. [Google Scholar]
- Caporale, C.; Berardino, I.D.; Leonardi, L.; Bertini, L.; Cascone, A.; Buonocore, V.; Caruso, C. Wheat pathogenesis-related proteins of class 4 have ribonuclease activity. FEBS Lett. 2004, 575, 71–76. [Google Scholar] [CrossRef]
- Caruso, C.; Caporale, C.; Chilosi, G.; Vacca, F.; Bertini, L.; Magro, P.; Poerio, E.; Buonocore, V. Structural and antifungal properties of a pathogenesis-related protein from wheat kernel. J. Protein Chem. 1996, 15, 35–44. [Google Scholar] [CrossRef]
- Jain, D.; Khurana, J.P. Role of pathogenesis-related (PR) proteins in plant defense mechanism. In Molecular Aspects of Plant-Pathogen Interaction; Singh, A., Singh, I.K., Eds.; Springer: Singapore, 2018; pp. 265–281. [Google Scholar]
- Fiocchetti, F.; D’Amore, R.; De Palma, M.; Bertini, L.; Caruso, C.; Caporale, C.; Testa, A.; Cristinzio, G.; Saccardo, F.; Tucci, M. Constitutive over-expression of two wheat pathogenesis-related genes enhances resistance of tobacco plants to Phytophthora nicotianae. Plant Cell Tiss. Organ Cult. 2008, 92, 73–84. [Google Scholar] [CrossRef]
- Xu, R.Q.; Li, Q.Q.S. Protocol: Streamline cloning of genes into binary vectors in Agrobacterium via the Gateway TOPO vector system. Plant Methods 2008, 4, 4. [Google Scholar] [CrossRef] [PubMed]
- Popielarska, M.; Ślesak, H.; Góralski, G. Histological and SEM studies on organogenesis in endosperm-derived callus of kiwifruit (Actinidia deliciosa cv. Hayward). Acta Biol. Crac. Ser. Bot. 2006, 48, 97–104. [Google Scholar]
- Priya, A.M.; Pandian, S.K.; Manikandan, R. The effect of different antibiotics on elimination of Agrobacterium and high frequency Agrobacterium-mediated transformation of indica rice (Oryza sativa L.). Czech J. Genet. Plant Breed. 2012, 48, 120–130. [Google Scholar] [CrossRef]
- Ding, Y.; Jia, J.; Yang, L.; Wen, H.; Zhang, C.; Liu, W.; Zhang, D. Validation of a rice specific gene, sucrose phosphate synthase, used as the endogenous reference gene for qualitative and real-time quantitative PCR detection of transgenes. J. Agric. Food Chem. 2004, 52, 3372–3377. [Google Scholar] [CrossRef]
- Yang, L.; Ding, J.; Zhang, C.; Jia, J.; Weng, H.; Liu, W.; Zhang, D. Estimating the copy number of transgenes in transformed rice by real-time quantitative PCR. Plant Cell Rep. 2005, 23, 759–763. [Google Scholar] [CrossRef] [PubMed]
- Pajerowska-Mukhtar, K.M.; Wang, W.; Tada, Y.; Oka, N.; Tucker, C.L.; Fonseca, J.P.; Dong, X. The HSF-like transcription factor TBF1 is a major molecular switch for plant growth-to-defense transition. Curr. Biol. 2012, 22, 103–112. [Google Scholar] [CrossRef]
- Pal, A.; Borthkur, D. Transgenic overexpression of Leucaena β-carbonic anhydrases in tobacco does not affect carbon assimilation and overall biomass. Plant Biosyst. 2016, 150, 932–941. [Google Scholar] [CrossRef]
- Yordanov, Y.S.; Ma, C.; Yordanova, E.; Meilan, R.; Strauss, S.H.; Busov, V.B. BIG LEAF is a regulator of organ size and adventitious root formation in poplar. PLoS ONE 2017, 12, e0180527. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Yang, J.; Wang, Q.; Zhu, H.; Chen, Z.; Dao, Y.; Wang, K. Overexpression of the trehalose-6-phosphate phosphatase family gene AtTPPF improves the drought tolerance of Arabidopsis thaliana. BMC Plant Biol. 2019, 19, 381. [Google Scholar] [CrossRef] [PubMed]
- Bakhsh, A.; Anayol, E.; Ozcan, S.F. Comparison of transformation efficiency of five Agrobacterium tumefaciens strains in Nicotiana tabacum L. Emirates J. Food Agric. 2014, 26, 259–264. [Google Scholar] [CrossRef]
- Yadav, S.; Sharma, P.; Srivastava, A.; Desai, P.; Shrivastava, N. Strain specific Agrobacterium-mediated genetic transformation of Bacopa monnieri. J. Genet. Eng. Biotechnol. 2014, 12, 89–94. [Google Scholar] [CrossRef]
- Matthysse, A.G. Role of bacterial cellulose fibrils in Agrobacterium tumefaciens infection. J. Bacteriol. 1983, 154, 906–915. [Google Scholar] [CrossRef]
- Xu, Y.; Li, B.; Jia, F. A novel system for Agrobacterium-mediated transformation of wheat (Triticum aestivum L.) cells. Cell Res. 1993, 3, 49–60. [Google Scholar] [CrossRef]
- Abarca-Grau, A.M.; Penyalver, R.; López, M.M.; Marco-Noales, E. Pathogenic and non-pathogenic Agrobacterium tumefaciens, A. rhizogenes and A. vitis strains form biofilms on abiotic as well as on root surfaces. Plant Pathol. 2011, 60, 416–425. [Google Scholar] [CrossRef]
- Wakasa, Y.; Ozawa, K.; Takaiwa, F. Agrobacterium-mediated co-transformation of rice using two selectable marker genes derived from rice genome components. Plant Cell Rep. 2012, 31, 2075–2084. [Google Scholar] [CrossRef]
- Chakraborty, M.; Reddy, P.S.; Narasu, M.L.; Krishna, G.; Rana, D. Agrobacterium-mediated genetic transformation of commercially elite rice restorer line using nptII gene as a plant selection marker. Physiol. Mol. Biol. Plant 2016, 22, 51–60. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ali, S.; Ganai, B.A.; Kamili, A.N.; Bhat, A.A.; Mir, Z.A.; Bhat, J.A.; Tyagi, A.; Islam, S.T.; Mushtaq, M.; Yadav, P.; et al. Pathogenesis-related proteins and peptides as promising tools for engineering plants with multiple stress tolerance. Microbiol. Res. 2018, 212–213, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Wu, J.; Fan, S.; Li, W.; Dong, L.; Cheng, Q.; Xu, P.; Zhang, S. Isolation and characterization of a novel pathogenesis-related protein gene (GmPRP) with induced expression in soybean (Glycine max) during infection with Phytophthora sojae. PLoS ONE 2015, 10, e0129932. [Google Scholar] [CrossRef]
- Dai, L.; Wang, D.; Xie, X.; Zhang, C.; Wang, X.; Xu, Y.; Wang, Y.; Zhang, J. The novel gene VpPR4-1 from Vitis pseudoreticulata increases powdery mildew resistance in transgenic Vitis vinifera L. Front. Plant Sci. 2016, 7, 695. [Google Scholar] [CrossRef] [PubMed]
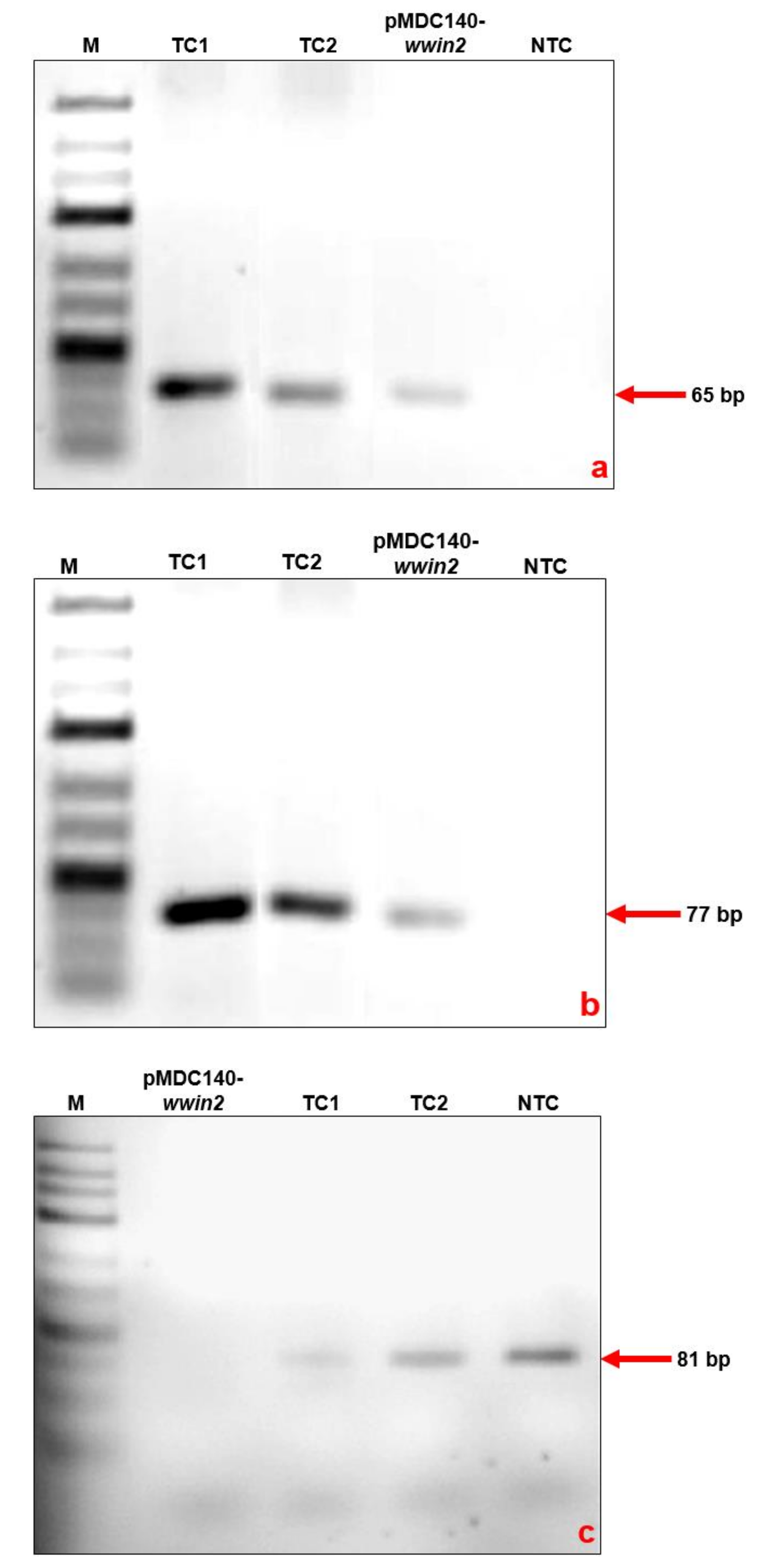
| Targeted Gene | Primer Sequence | Amplicon Size | Reference |
|---|---|---|---|
| Sucrose phosphate synthase (sps) | Forward: 5′-TTGCGCCTGAACGGATAT-3′ Reverse: 5′-CGGTTGATCTTTTCGGGATG-3′ | 81 bp | [20] |
| Hygromycin phosphotransferase II (hptII) | Forward: 5′-CTATTTCTTTGCCCTCGGACGA-3′ Reverse: 5′-GGACCGATGGCTGTGTAGAAG-3′ | 77 bp | [21] |
| Wheatwin2 (wwin2) | Forward: 5′-TGGACTGGGACACCGTCTTC-3′ Reverse: 5′-GAGGTGGCCCTGCTGGTA-3′ | 65 bp | This study |
| Gene | Mean CT Value ± SD | ||
|---|---|---|---|
| Transformed Callus 1 | Transformed Callus 2 | Non-Transformed Callus (Control) | |
| wwin2 | 29.79 ± 0.36 | 27.94 ± 0.29 | Undetermined |
| hptII | 27.64 ± 0.26 | 27.15 ± 0.96 | Undetermined |
| sps | 30.18 ± 1.59 | 22.20 ± 0.95 | 20.73 ± 0.75 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chong, E.T.J.; Wong, J.J.; Aziz, Z.A.; Tan, C.L.; Subramaniam, S.; Latip, M.A.; Lee, P.-C. Agrobacterium-Mediated Transformation of pMDC140 Plasmid Containing the Wheatwin2 Gene into the Tadong Rice Genome. Agronomy 2021, 11, 1072. https://doi.org/10.3390/agronomy11061072
Chong ETJ, Wong JJ, Aziz ZA, Tan CL, Subramaniam S, Latip MA, Lee P-C. Agrobacterium-Mediated Transformation of pMDC140 Plasmid Containing the Wheatwin2 Gene into the Tadong Rice Genome. Agronomy. 2021; 11(6):1072. https://doi.org/10.3390/agronomy11061072
Chicago/Turabian StyleChong, Eric Tzyy Jiann, Jovita Jun Wong, Zaleha Abdul Aziz, Chia Lock Tan, Sreeramanan Subramaniam, Mariam Abd. Latip, and Ping-Chin Lee. 2021. "Agrobacterium-Mediated Transformation of pMDC140 Plasmid Containing the Wheatwin2 Gene into the Tadong Rice Genome" Agronomy 11, no. 6: 1072. https://doi.org/10.3390/agronomy11061072
APA StyleChong, E. T. J., Wong, J. J., Aziz, Z. A., Tan, C. L., Subramaniam, S., Latip, M. A., & Lee, P.-C. (2021). Agrobacterium-Mediated Transformation of pMDC140 Plasmid Containing the Wheatwin2 Gene into the Tadong Rice Genome. Agronomy, 11(6), 1072. https://doi.org/10.3390/agronomy11061072






