Implementation of a Generalized Additive Model (GAM) for Soybean Maturity Prediction in African Environments
Abstract
:1. Introduction
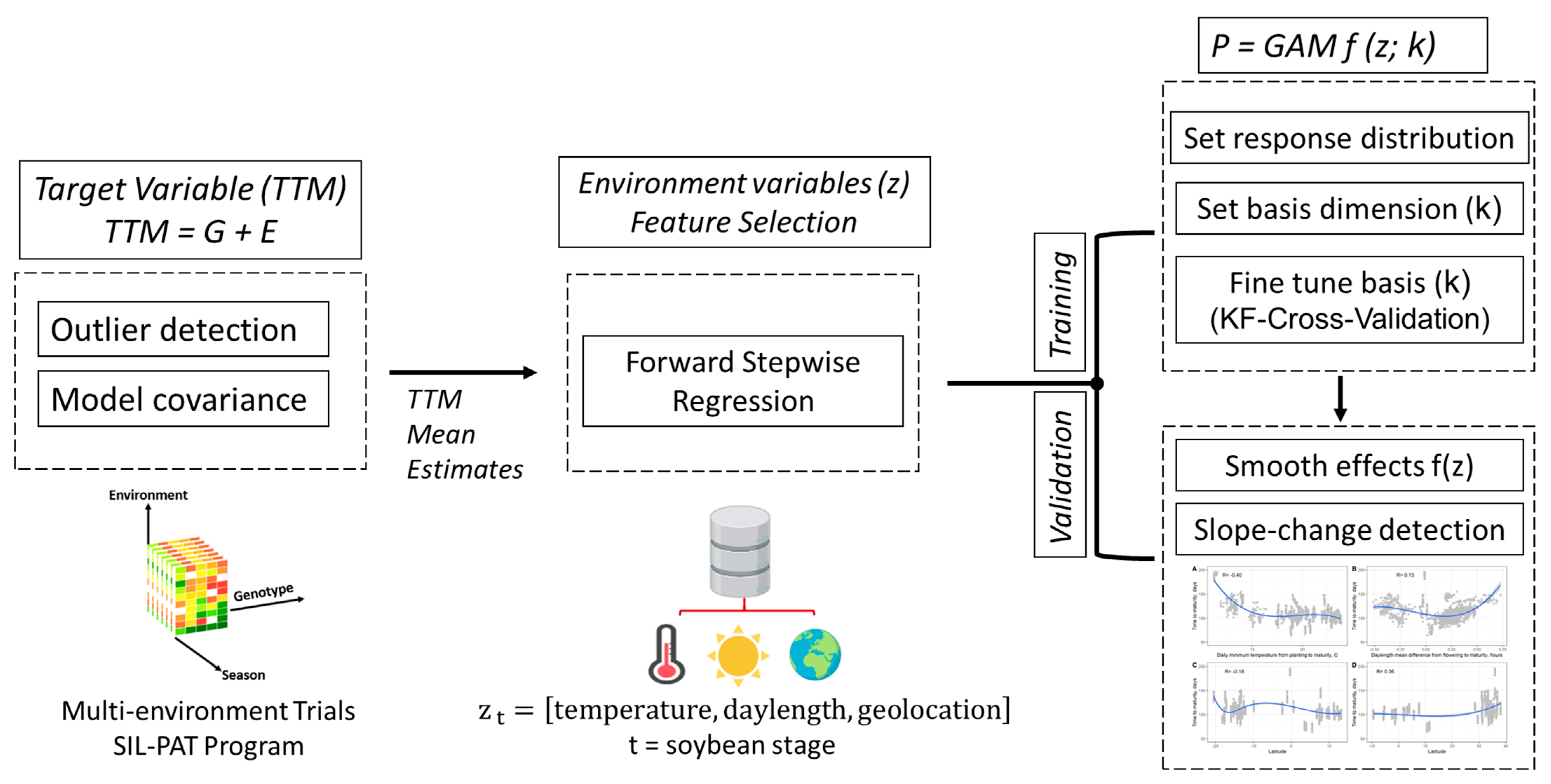
2. Materials and Methods
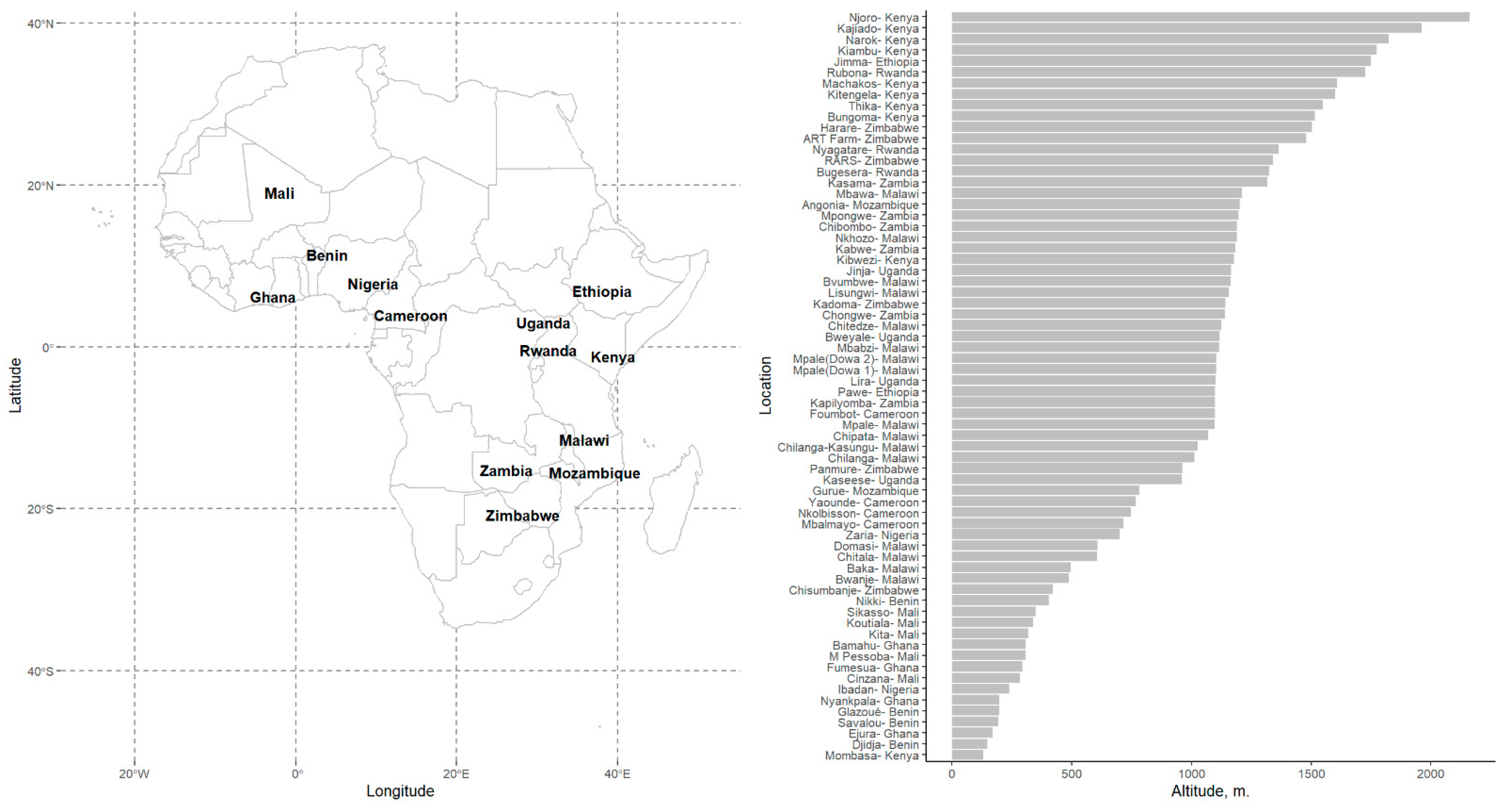
2.1. Pre-Modeling Exploratory Analysis: Soybean Maturity Time
2.2. Modeling Soybean Time to Maturity as a Function of Environment
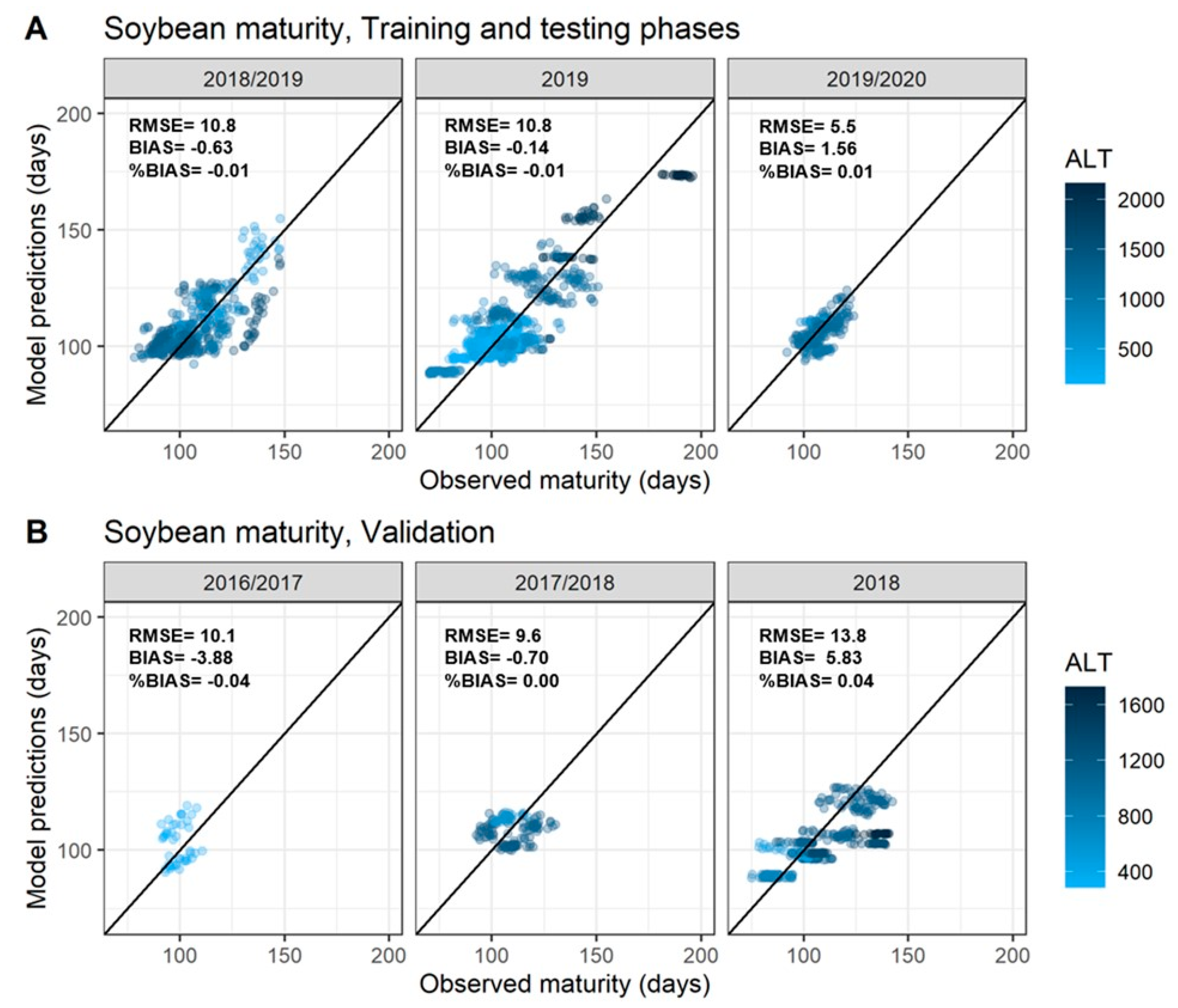
3. Results
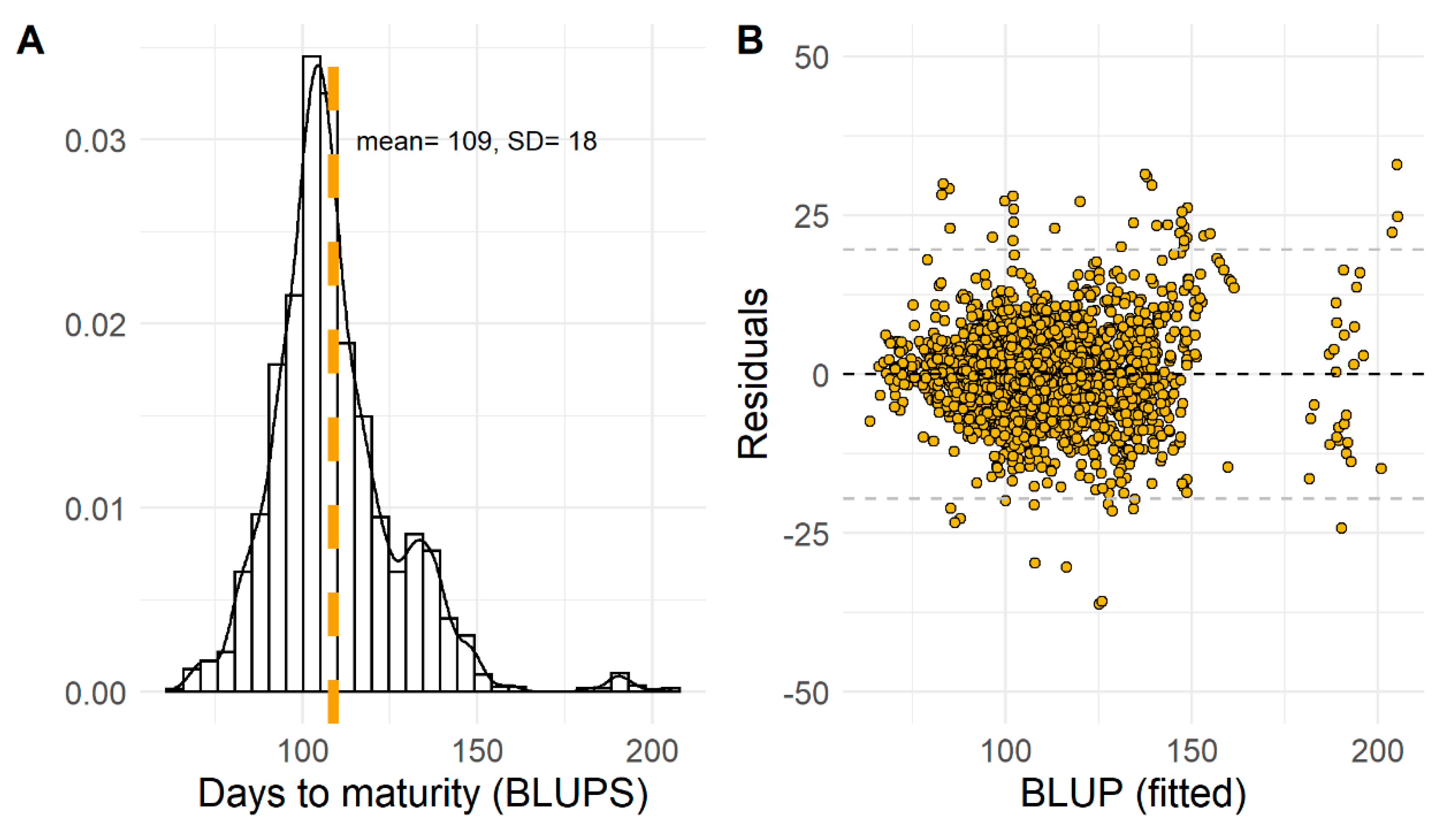
3.1. Exploratory Analysis of Soybean Maturity Timing
3.2. Best Features to Characterize Soybean Time to Maturity (TTM)
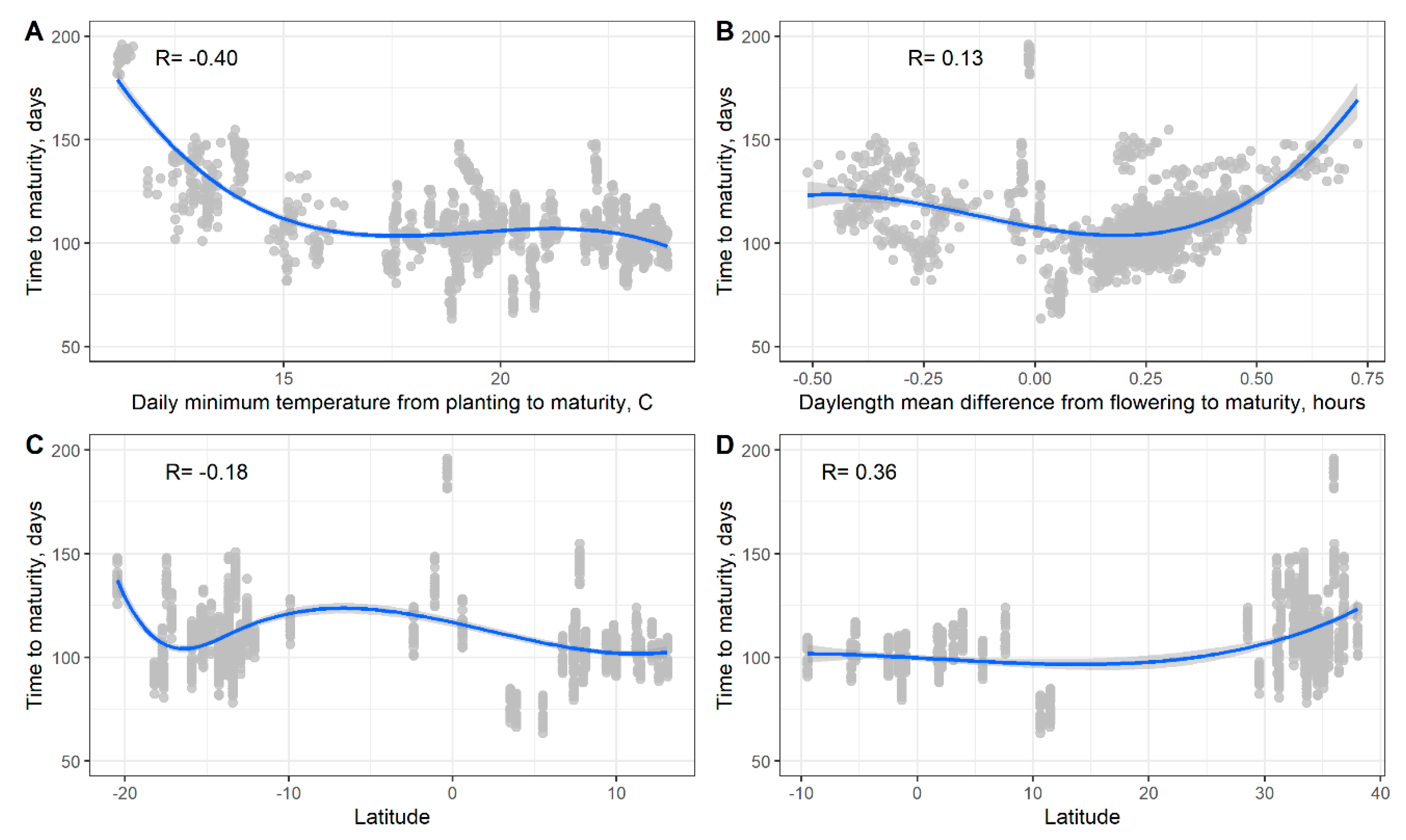
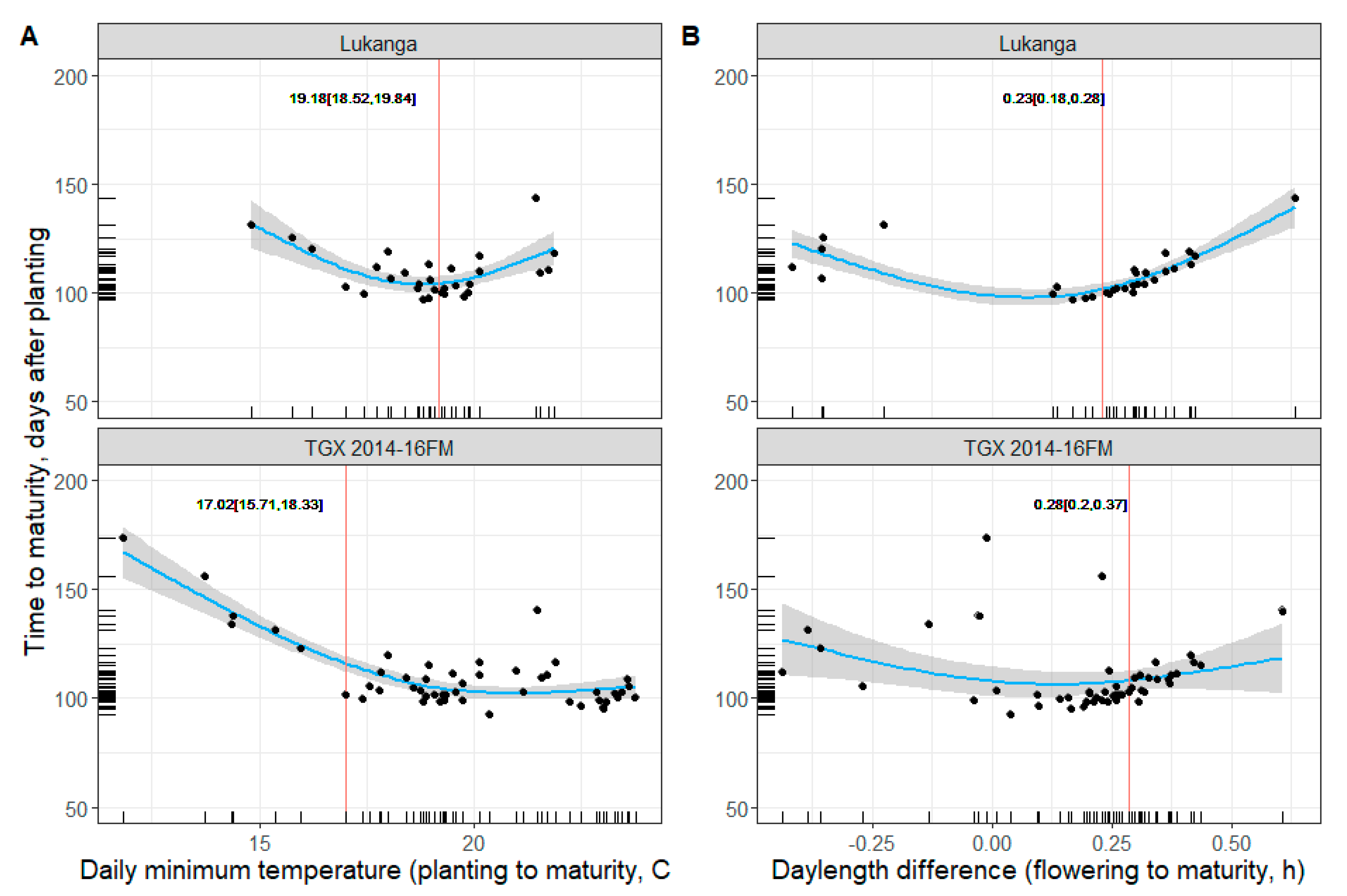
3.3. Soybean Maturity Response to Temperature and Daylength
4. Discussion and Implications
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Carsky, R.J.; Berner, D.K.; Oyewole, B.D.; Dashiell, K.; Schulz, S. Reduction of Striga hermonthica parasitism on maize using soybean rotation. Int. J. Pest Manag. 2000, 46, 115–120. [Google Scholar] [CrossRef]
- Sinclair, T.R.; Marrou, H.; Soltani, A.; Vadez, V.; Chandolu, K.C. Soybean production potential in Africa. Glob. Food Secur. 2014, 3, 31–40. [Google Scholar] [CrossRef] [Green Version]
- Khojely, D.M.; Ibrahim, S.E.; Sapey, E.; Han, T. History, current status, and prospects of soybean production and research in sub-Saharan Africa. Crop. J. 2018, 6, 226–235. [Google Scholar] [CrossRef]
- Foyer, C.H.; Siddique, K.H.; Tai, A.P.; Anders, S.; Fodor, N.; Wong, F.-L.; Ludidi, N.; Chapman, M.A.; Ferguson, B.J.; Considine, M.J.; et al. Modelling predicts that soybean is poised to dominate crop production across Africa. Plant Cell Environ. 2018, 42, 373–385. [Google Scholar] [CrossRef]
- Keyser, J.C.; Van Gent, R.V. Zambia Competitiveness Report; The World Bank, Environmental, Rural, and Social Development Unit: Washington, DC, USA, 2007. [Google Scholar]
- Soybean Innovation Lab. Soybean Innovation Lab 2020. Available online: https://www.soybeaninnovationlab.illinois.edu (accessed on 25 February 2021).
- Tropical Soybean Information Portal. Tropicalsoybean. 2020. Available online: https://www.tropicalsoybean.com/databases (accessed on 25 February 2021).
- Santos, M.D.F. University of Illinois at Urbana-Champaign Soybean Varieties in Sub-Saharan Africa. Afr. J. Food Agric. Nutr. Dev. 2020, 19, 15136–15139. [Google Scholar] [CrossRef]
- Leles, E. Pan-African Soybean Variety Trials Database Supports Decision-Making Across Africa. Agrilinks 2021. Available online: https://www.agrilinks.org/post/pan-african-soybean-variety-trials-database-supports-decision-making-across-africa (accessed on 24 February 2021).
- Ersoz, E.S.; Martin, N.F.; Stapleton, A.E. On to the next chapter for crop breeding: Convergence with data science. Crop. Sci. 2020, 60, 639–655. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.X.; Kyei-Boahen, S.; Zhang, J.; Zhang, M.H.; Freeland, T.B.; Watson, C.E.; Liu, X. Modifications of Optimum Adaptation Zones for Soybean Maturity Groups in the USA. Crop. Manag. 2007, 6, 1–11. [Google Scholar] [CrossRef]
- Mourtzinis, S.; Conley, S. Delineating Soybean Maturity Groups across the United States. Agron. J. 2017, 109, 1397–1403. [Google Scholar] [CrossRef] [Green Version]
- Cooper, R.L. A delayed flowering barrier to higher soybean yields. Field Crop. Res. 2003, 82, 27–35. [Google Scholar] [CrossRef]
- Cober, E.R.; Stewart, D.W.; Voldeng, H.D. Photoperiod and Temperature Responses in Early-Maturing, Near-Isogenic Soybean Lines. Crop. Sci. 2001, 41, 721–727. [Google Scholar] [CrossRef]
- Scott, W.O.; Aldrich, S.R. Modern Soybean Production; S & A Publications: Champaign, IL, USA, 1983. [Google Scholar]
- Bernardo, R. Reinventing quantitative genetics for plant breeding: Something old, something new, something borrowed, something BLUE. Heredity 2020, 125, 375–385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piepho, H.-P.; Möhring, J.; Schulz-Streeck, T.; Ogutu, J.O. A stage-wise approach for the analysis of multi-environment trials. Biom. J. 2012, 54, 844–860. [Google Scholar] [CrossRef]
- Buntaran, H.; Piepho, H.; Schmidt, P.; Rydén, J.; Halling, M.; Forkman, J. Cross-validation of stagewise mixed-model analysis of Swedish variety trials with winter wheat and spring barley. Crop. Sci. 2020, 60, 2221–2240. [Google Scholar] [CrossRef]
- Major, D.J.; Johnson, D.R.; Tanner, J.W.; Anderson, I.C. Effects of Daylength and Temperature on Soybean Development 1. Crop. Sci. 1975, 15, 174–179. [Google Scholar] [CrossRef]
- aWhere | Climate Smart Weather Insights Backed by AI. 2021. Available online: https://www.awhere.com/ (accessed on 25 February 2021).
- Campbell, G.S.; Norman, J.M. An Introduction to Environmental Biophysics; Springer: New York, NY, USA, 2000. [Google Scholar]
- Teh, C.B.S. Introduction to Mathematical Modeling of Crop Growth: How the Equations Are Derived and Assembled into a Computer Model; Brown Walker Press: Boca Raton, FL, USA, 2006. [Google Scholar]
- Amemiya, T. Selection of Regressors. Int. Econ. Rev. 1980, 21, 331. [Google Scholar] [CrossRef]
- Dawson, J.C.; Endelman, J.B.; Heslot, N.; Crossa, J.; Poland, J.; Dreisigacker, S.; Manès, Y.; Sorrells, M.E.; Jannink, J.-L. The use of unbalanced historical data for genomic selection in an international wheat breeding program. Field Crop. Res. 2013, 154, 12–22. [Google Scholar] [CrossRef] [Green Version]
- James, G.; Witten, D.; Hastie, T.; Tibshirani, R. (Eds.) Moving Beyond Linearity. In An Introduction to Statistical Learning: With Applications in R; Springer: New York, NY, USA, 2013; pp. 265–301. [Google Scholar]
- Roberts, M.J.; Key, N. Agricultural Payments and Land Concentration: A Semiparametric Spatial Regression Analysis. Am. J. Agric. Econ. 2008, 90, 627–643. [Google Scholar] [CrossRef]
- Stauffer, R.; Mayr, G.J.; Messner, J.W.; Umlauf, N.; Zeileis, A. Spatio-temporal precipitation climatology over complex terrain using a censored additive regression model. Int. J. Clim. 2017, 37, 3264–3275. [Google Scholar] [CrossRef] [Green Version]
- Lawler, J.J.; White, D.; Neilsonjand, R.P.; Blaustein, A.R. Predicting climate-induced range shifts: Model differences and model reliability. Glob. Chang. Biol. 2006, 12, 1568–1584. [Google Scholar] [CrossRef] [Green Version]
- Chen, K.; O’Leary, R.A.; Evans, F.H. A simple and parsimonious generalised additive model for predicting wheat yield in a decision support tool. Agric. Syst. 2019, 173, 140–150. [Google Scholar] [CrossRef]
- De Rosa, D.; Basso, B.; Fasiolo, M.; Friedl, J.; Fulkerson, B.; Grace, P.R.; Rowlings, D.W. Predicting pasture biomass using a statistical model and machine learning algorithm implemented with remotely sensed imagery. Comput. Electron. Agric. 2021, 180, 105880. [Google Scholar] [CrossRef]
- Rosenheim, J.A.; Cass, B.N.; Kahl, H.; Steinmann, K.P. Variation in pesticide use across crops in California agriculture: Economic and ecological drivers. Sci. Total. Environ. 2020, 733, 138683. [Google Scholar] [CrossRef]
- Muggeo, V.M.R. Segmented: An R Package to Fit Regression Models with Broken-Line Relationships. R News 2008, 8, 20–25. [Google Scholar]
- Küchenhoff, H.; Carroll, R.J. Segmented Regression with Errors in Predictors: Semi-Parametric and Parametric Methods. Stat. Med. 1997, 16, 169–188. [Google Scholar] [CrossRef]
- Alliprandini, L.F.; Abatti, C.; Bertagnolli, P.F.; Cavassim, J.E.; Gabe, H.L.; Kurek, A.; Matsumoto, M.N.; De Oliveira, M.A.R.; Pitol, C.; Prado, L.C.; et al. Understanding Soybean Maturity Groups in Brazil: Environment, Cultivar Classification, and Stability. Crop. Sci. 2009, 49, 801–808. [Google Scholar] [CrossRef]
- Malosetti, M.; Ribaut, J.-M.; Van Eeuwijk, F.A. The statistical analysis of multi-environment data: Modeling genotype-by-environment interaction and its genetic basis. Front. Physiol. 2013, 4, 44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- George, T.; Bartholomew, D.; Singleton, P. Effect of temperature and maturity group on phenology of field grown nodulating and nonnodulating soybean isolines. Biotronics 1990, 19, 49–59. [Google Scholar]
- Lawn, R.; Byth, D. Response of soya beans to planting date in south-eastern Queensland. II.* Vegetative and reproductive development. Aust. J. Agric. Res. 1974, 25, 723–737. [Google Scholar] [CrossRef]
- Egli, D. Cultivar maturity and potential yield of soybean. Field Crop. Res. 1993, 32, 147–158. [Google Scholar] [CrossRef]
- Sinclair, T.R.; Hinson, K. Soybean Flowering in Response to the Long-Juvenile Trait. Crop. Sci. 1992, 32, 1242–1248. [Google Scholar] [CrossRef]
- Muggeo, V.M.R. Estimating regression models with unknown break-points. Stat. Med. 2003, 22, 3055–3071. [Google Scholar] [CrossRef] [PubMed]

| Temperature (Celsius, °C) | |||
|---|---|---|---|
| Label | Description | Mean (1 SD) | Range (2 IQR) |
| TMAXF | Daily maximum temperature to flowering | 28.1(2.0) | 22.4–32.5 (2.4) |
| TMINMF | Daily minimum temperature to flowering | 19.7 (3.2) | 11.2–23.9 (3.8) |
| TMEANF | Daily mean temperature to flowering | 23.9 (2.5) | 16.8–28.2 (3.3) |
| TMAXM | Daily maximum temperature to maturity | 28.5 (2.0) | 22.5–32.9 (2.8) |
| TMINMM | Daily minimum temperature to maturity | 19.8 (2.8) | 11.8–23.9 (3.8) |
| TMEANM | Daily mean temperature to maturity | 24.1 (2.3) | 17.2–28.4 (3.4) |
| Daylength (h, hours) | |||
| DLMEANF | Mean daylength to flowering | 12.5 (0.5) | 11.4–13.3 (0.5) |
| DLMEANM | Mean daylength to maturity | 12.4 (0.3) | 11.7–13.1 (0.4) |
| Difference-based variables (Flowering to Maturity) | |||
| TMINMDIFF | Minimum temperature difference (°C) | −0.1 (0.9) | −4, −1.12 (0.5) |
| TMEANDIFF | Mean temperature difference (°C) | −0.3 (0.7) | −3.5, 1.0 (0.4) |
| TMAXDIFF | Maximum temperature difference (°C) | −0.4 (0.7) | −3.0, 1.0 (0.6) |
| DLMEANDIFF | Daylength difference (h) | 0.2 (0.2) | −0.5, 0.7 (0.2) |
| Location-based variables | |||
| Long | Longitude, (Degrees, deg) | 23.1 (15.6) | −9.5, 38.0 (28.6) |
| Lat | Latitude, (Degrees, deg) | −5.1 (11.4) | −20.5, 13.1 (21.7) |
| ALT | Altitude, (Meters, m) | 858 (470) | 148–2160 (752) |
| Factor | 2 DF Model | 2 DF Residuals | 1 RSS | Adj-R 2 | 3 RSE (Days) |
|---|---|---|---|---|---|
| Genotype (G) | 174 | 2648 | 882,623 | 0.12 | 18.2 |
| Location (L) | 67 | 2755 | 330,979 | 0.68 | 10.9 |
| Season (S) | 8 | 2814 | 1,017,289 | 0.05 | 19.0 |
| G + L | 241 | 2581 | 225,891 | 0.76 | 9.3 |
| G + S | 182 | 2640 | 864,933 | 0.14 | 18.10 |
| E = L + S | 75 | 2747 | 280,997 | 0.73 | 10.1 |
| G + E | 249 | 2557 | 121,887 | 0.87 | 6.9 |
| Random Effects | Group | σ2 | σ (days) | σ [95% CI] | n |
| G | 65.69 | 8.10 | [7.20, 9.16] | 175 | |
| E | 357.35 | 18.90 | [16.36, 21.92] | 73 | |
| Error | 47.80 | 6.91 | [6.72, 7.10] | ||
| Fixed Effects | Intercept [95% CI] | 108.9 [95% CI: 104.87, 113.05] | |||
| Goodness of fit | AIC = 19,984 | ||||
| BIC = 19,918 | |||||
| R2 = 0.89 | |||||
| Feature Subset | AIC | Adj-R 2 | 1 Cp | 2 HSP | 3 APC |
|---|---|---|---|---|---|
| TMINM + DLMEANDIFF + lat + long | 13,817 | 0.36 | 5.04 | 0.13 | 0.63 |
| TMEANM + DLMEANDIFF + lat + long | 14,014 | 0.29 | 7.09 | 0.14 | 0.71 |
| TMINDIFF + DLMEANDIFF + lat + long | 13,962 | 0.31 | 5.0 | 0.14 | 0.68 |
| ALT + DLMEANDIFF + lat + long | 14,134 | 0.24 | 5.0 | 0.15 | 0.76 |
| Model | 5-Fold Cross Validation | AIC | BIC | |||||
|---|---|---|---|---|---|---|---|---|
| 1 RMSE (Days) | R2-Adjusted | |||||||
| Training | Testing | Training | Testing | |||||
| lm ~ TMINM | 15.34 | 15.79 | 0.29 | 0.30 | 14,000 | 14,016 | ||
| gam ~ f(TMINM, k = 3) | 13.55 | 13.67 | 0.46 | 0.46 | 13,542 | 13,564 | ||
| lm ~ (TMINM + DLMEANDIFF) | 14.96 | 15.48 | 0.32 | 0.33 | 13,924 | 13,945 | ||
| gam ~ f(TMINM, k = 3) + DLMEANDIFF | 12.50 | 12.70 | 0.54 | 0.53 | 13,280 | 13,307 | ||
| gam ~ f(TMINM, k = 3) + f(DLMEANDIFF, k = 3) | 10.78 | 11.02 | 0.66 | 0.65 | 12,793 | 12,825 | ||
| lm ~ (TMINM + DLMEANDIFF + lat + long) | 14.90 | 15.48 | 0.34 | 0.32 | 13,817 | 13,850 | ||
| gam ~ f(TMINM, k = 3) + f(DLMEANDIFF, k = 3) + lat + long | 10.05 | 10.3 | 0.70 | 0.69 | 12,576 | 12,620 | ||
| gam ~ f(TMINM, k = 3) + f(DLMEANDIFF, k = 3) + f(lat, long, k = 4) | 9.99 | 10.35 | 0.70 | 0.69 | 12,564 | 12,613 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marcillo, G.S.; Martin, N.F.; Diers, B.W.; Da Fonseca Santos, M.; Leles, E.P.; Chigeza, G.; Francischini, J.H. Implementation of a Generalized Additive Model (GAM) for Soybean Maturity Prediction in African Environments. Agronomy 2021, 11, 1043. https://doi.org/10.3390/agronomy11061043
Marcillo GS, Martin NF, Diers BW, Da Fonseca Santos M, Leles EP, Chigeza G, Francischini JH. Implementation of a Generalized Additive Model (GAM) for Soybean Maturity Prediction in African Environments. Agronomy. 2021; 11(6):1043. https://doi.org/10.3390/agronomy11061043
Chicago/Turabian StyleMarcillo, Guillermo S., Nicolas F. Martin, Brian W. Diers, Michelle Da Fonseca Santos, Erica Pontes Leles, Godfree Chigeza, and Josy H. Francischini. 2021. "Implementation of a Generalized Additive Model (GAM) for Soybean Maturity Prediction in African Environments" Agronomy 11, no. 6: 1043. https://doi.org/10.3390/agronomy11061043
APA StyleMarcillo, G. S., Martin, N. F., Diers, B. W., Da Fonseca Santos, M., Leles, E. P., Chigeza, G., & Francischini, J. H. (2021). Implementation of a Generalized Additive Model (GAM) for Soybean Maturity Prediction in African Environments. Agronomy, 11(6), 1043. https://doi.org/10.3390/agronomy11061043






