Early Detection of Broad-Leaved and Grass Weeds in Wide Row Crops Using Artificial Neural Networks and UAV Imagery
Abstract
:1. Introduction
2. Materials and Methods
2.1. Description of Study Fields and UAV Flights
2.2. Digital Surface Model (DSM) and Orthomosaic Generation
2.3. Ground Truth Data
2.4. Image Analysis
2.4.1. Labeling of the Image Objects
2.4.2. Crop Detection
2.4.3. Artificial Neural Network Creation
2.5. Validation
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Variable | Importance | Normalized Importance (%) |
|---|---|---|
| Brightness | 0.040 | 100.0 |
| COMB1 | 0.040 | 100.0 |
| GLDV Entropy (all dir.) | 0.033 | 82.9 |
| VEG | 0.033 | 81.5 |
| NRGDI | 0.033 | 81.1 |
| Hue | 0.031 | 77.1 |
| r | 0.030 | 75.1 |
| CHM | 0.029 | 73.4 |
| ExGR | 0.029 | 71.7 |
| g | 0.029 | 71.5 |
| R/B | 0.029 | 71.4 |
| CIVE | 0.028 | 71.0 |
| ExR | 0.028 | 70.5 |
| GLCM Ang. 2nd moment (all dir.) | 0.028 | 68.8 |
| VARI | 0.028 | 68.7 |
| GLDV Ang. 2nd moment (all dir.) | 0.026 | 64.9 |
| ExG | 0.025 | 62.2 |
| GLCM Mean (all dir.) | 0.025 | 61.7 |
| GLCM Dissimilarity (all dir.) | 0.024 | 60.4 |
| R/G | 0.024 | 58.7 |
| NPCI | 0.023 | 57.7 |
| WI | 0.022 | 56.0 |
| Skewness | 0.022 | 54.3 |
| Compactness | 0.022 | 53.7 |
| COMB2 | 0.022 | 53.7 |
| GLCM Entropy (all dir.) | 0.021 | 52.4 |
| b | 0.020 | 50.8 |
| GLDV Mean (all dir.) | 0.020 | 50.5 |
| GLCM SD (all dir.) | 0.019 | 48.3 |
| Perimeter (polygon) (Pxl) | 0.019 | 47.7 |
| CLGM Contrast (all dir.) | 0.017 | 42.5 |
| Rectangular fit | 0.017 | 42.2 |
| Density | 0.016 | 38.8 |
| GLCM Homogeneity (all dir.) | 0.015 | 38.4 |
| GLCM Correlation (all dir.) | 0.014 | 34.6 |
| Compactness (polygon) | 0.013 | 33.3 |
| GLDV Contrast (all dir.) | 0.013 | 31.9 |
| SD of length of edges (polygon) (Pxl) | 0.012 | 30.6 |
| ExB | 0.012 | 29.5 |
| Shape index | 0.012 | 29.2 |
| Length/Width | 0.011 | 26.9 |
| Roundness | 0.010 | 25.8 |
| Number of segments | 0.008 | 20.1 |
| Average area represented by segments (Pxl) | 0.007 | 17.6 |
| Number of edges (polygon) | 0.006 | 15.9 |
| Radius of smallest enclosing ellipse | 0.006 | 15.2 |
| Radius of largest enclosed ellipse | 0.006 | 14.8 |
| Asymmetry | 0.004 | 10.7 |
| Variable | Importance | Normalized Importance (%) |
|---|---|---|
| Brightness | 0.058 | 100.0 |
| GLCM Homogeneity (all dir.) | 0.038 | 65.8 |
| GLCM ang. 2nd moment (all dir.) | 0.036 | 61.6 |
| ExGR | 0.034 | 58.3 |
| g | 0.033 | 56.9 |
| COMB2 | 0.032 | 55.0 |
| GLCM Entropy (all dir.) | 0.032 | 54.7 |
| GLDV Contrast (all dir.) | 0.030 | 50.9 |
| GLCM SD (all dir.) | 0.030 | 50.6 |
| NRGDI | 0.029 | 49.1 |
| R/G | 0.028 | 48.6 |
| VEG | 0.028 | 47.3 |
| GLDV Entropy (all dir.) | 0.028 | 47.2 |
| GLCM Mean (all dir.) | 0.025 | 43.5 |
| ExR | 0.025 | 42.0 |
| CIVE | 0.025 | 42.0 |
| GLCM Dissimilarity (all dir.) | 0.024 | 41.5 |
| GLDV ang. 2nd moment (all dir.) | 0.023 | 39.4 |
| WI | 0.021 | 36.5 |
| CLGM Contrast (all dir.) | 0.021 | 35.6 |
| GLDV Mean (all dir.) | 0.020 | 35.0 |
| Compactness | 0.020 | 34.6 |
| R/B | 0.020 | 33.6 |
| r | 0.020 | 33.5 |
| VARI | 0.019 | 32.7 |
| NPCI | 0.019 | 32.4 |
| ExG | 0.019 | 31.6 |
| GLCM Correlation (all dir.) | 0.018 | 31.2 |
| Rectangular fit | 0.017 | 29.2 |
| ExB | 0.016 | 27.4 |
| COMB1 | 0.015 | 25.6 |
| Shape index | 0.015 | 25.3 |
| Skewness | 0.014 | 24.1 |
| SD of length of edges (polygon) (Pxl) | 0.014 | 23.7 |
| Hue | 0.014 | 23.5 |
| b | 0.014 | 23.4 |
| Density | 0.013 | 22.6 |
| Average Area represented by segments (Pxl) | 0.013 | 21.9 |
| Length/Width | 0.013 | 21.7 |
| Radius of smallest enclosing ellipse | 0.013 | 21.6 |
| CHM | 0.011 | 19.6 |
| Roundness | 0.010 | 16.6 |
| Asymmetry | 0.010 | 16.3 |
| Compactness (polygon) | 0.009 | 16.0 |
| Radius of largest enclosed ellipse | 0.009 | 15.5 |
| Number of edges (polygon) | 0.009 | 15.3 |
| Number of segments | 0.009 | 14.7 |
| Perimeter (polygon) (Pxl) | 0.008 | 13.7 |
References
- Oerke, E.-C. Crop Losses to Pests. J. Agric. Sci. 2006, 144, 31–43. [Google Scholar] [CrossRef]
- Castillejo-González, I.L.; Peña-Barragán, J.M.; Jurado-Expósito, M.; Mesas-Carrascosa, F.J.; López-Granados, F. Evaluation of Pixel- and Object-Based Approaches for Mapping Wild Oat (Avena Sterilis) Weed Patches in Wheat Fields Using QuickBird Imagery for Site-Specific Management. Eur. J. Agron. 2014, 59, 57–66. [Google Scholar] [CrossRef]
- Castillejo-González, I.L.; de Castro, A.I.; Jurado-Expósito, M.; Peña, J.-M.; García-Ferrer, A.; López-Granados, F. Assessment of the Persistence of Avena Sterilis L. Patches in Wheat Fields for Site-Specific Sustainable Management. Agronomy 2019, 9, 30. [Google Scholar] [CrossRef] [Green Version]
- Jurado-Exposito, M.; Lopez-Granados, F.; Gonzalez-Andujar, J.L.; Garcia-Torres, L. Characterizing Population Growth Rate of Convolvulus Arvensis in Wheat-Sunflower No-Tillage Systems. Crop. Sci. 2005, 45, 2106–2112. [Google Scholar] [CrossRef] [Green Version]
- Jurado-Expósito, M.; López-Granados, F.; García-Torres, L.; García-Ferrer, A.; Sánchez de la Orden, M.; Atenciano, S. Multi-Species Weed Spatial Variability and Site-Specific Management Maps in Cultivated Sunflower. Weed Sci. 2003, 51, 319–328. [Google Scholar] [CrossRef]
- Directive 2009/128/EC of the European Parliament and of the Council of 21 October 2009 Establishing a Framework for Community Action to Achieve the Sustainable Use of Pesticides Text with EEA Relevance; 2009; p. 16.
- Regulation (EU) No 652/2014 of the European Parliament and of the Council of 15 May 2014 Laying down Provisions for the Management of Expenditure Relating to the Food Chain, Animal Health and Animal Welfare, and Relating to Plant. Health and Plant. Reproductive Material, Amending Council Directives 98/56/EC, 2000/29/EC and 2008/90/EC, Regulations (EC) No 178/2002, (EC) No 882/2004 and (EC) No 396/2005 of the European Parliament and of the Council, Directive 2009/128/EC of the European Parliament and of the Council and Regulation (EC) No 1107/2009 of the European Parliament and of the Council and Repealing Council Decisions 66/399/EEC, 76/894/EEC and 2009/470/EC; 2014; p. 32.
- Fernández-Quintanilla, C.; Peña, J.M.; Andújar, D.; Dorado, J.; Ribeiro, A.; López-Granados, F. Is the Current State of the Art of Weed Monitoring Suitable for Site-Specific Weed Management in Arable Crops? Weed Res. 2018, 58, 259–272. [Google Scholar] [CrossRef]
- Singh, V.; Rana, A.; Bishop, M.; Filippi, A.M.; Cope, D.; Rajan, N.; Bagavathiannan, M. Unmanned aircraft systems for precision weed detection and management: Prospects and challenges. In Advances in Agronomy; Elsevier: Amsterdam, The Netherlands, 2020; Volume 159, pp. 93–134. ISBN 978-0-12-820459-7. [Google Scholar]
- Borra-Serrano, I.; Peña, J.M.; Torres-Sánchez, J.; Mesas-Carrascosa, F.J.; López-Granados, F. Spatial Quality Evaluation of Resampled Unmanned Aerial Vehicle-Imagery for Weed Mapping. Sensors 2015, 15, 19688–19708. [Google Scholar] [CrossRef]
- Rasmussen, J.; Nielsen, J.; Garcia-Ruiz, F.; Christensen, S.; Streibig, J.C. Potential Uses of Small Unmanned Aircraft Systems (UAS) in Weed Research. Weed Res. 2013, 53, 242–248. [Google Scholar] [CrossRef]
- Torres-Sánchez, J.; López-Granados, F.; De Castro, A.I.; Peña-Barragán, J.M. Configuration and Specifications of an Unmanned Aerial Vehicle (UAV) for Early Site Specific Weed Management. PLoS ONE 2013, 8, e58210. [Google Scholar] [CrossRef] [Green Version]
- Gómez-Candón, D.; Castro, A.I.D.; López-Granados, F. Assessing the Accuracy of Mosaics from Unmanned Aerial Vehicle (UAV) Imagery for Precision Agriculture Purposes in Wheat. Precis. Agric. 2014, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Andújar, D.; Moreno, H.; Bengochea-Guevara, J.M.; de Castro, A.; Ribeiro, A. Aerial Imagery or On-Ground Detection? An Economic Analysis for Vineyard Crops. Comput. Electron. Agric. 2019, 157, 351–358. [Google Scholar] [CrossRef]
- Peña, J.M.; Torres-Sánchez, J.; Serrano-Pérez, A.; de Castro, A.I.; López-Granados, F. Quantifying Efficacy and Limits of Unmanned Aerial Vehicle (UAV) Technology for Weed Seedling Detection as Affected by Sensor Resolution. Sensors 2015, 15, 5609–5626. [Google Scholar] [CrossRef] [Green Version]
- López-Granados, F.; Torres-Sánchez, J.; Serrano-Pérez, A.; Castro AI, d.e.; Mesas-Carrascosa, F.-J.; Peña, J.-M. Early Season Weed Mapping in Sunflower Using UAV Technology: Variability of Herbicide Treatment Maps against Weed Thresholds. Precis. Agric. 2016, 17, 183–199. [Google Scholar] [CrossRef]
- López-Granados, F.; Torres-Sánchez, J.; Castro, A.-I.D.; Serrano-Pérez, A.; Mesas-Carrascosa, F.-J.; Peña, J.-M. Object-Based Early Monitoring of a Grass Weed in a Grass Crop Using High Resolution UAV Imagery. Agron. Sustain. Dev. 2016, 36, 67. [Google Scholar] [CrossRef]
- Peña, J.M.; Torres-Sánchez, J.; de Castro, A.I.; Kelly, M.; López-Granados, F. Weed Mapping in Early-Season Maize Fields Using Object-Based Analysis of Unmanned Aerial Vehicle (UAV) Images. PLoS ONE 2013, 8, e77151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, J.; Nuyttens, D.; Lootens, P.; He, Y.; Pieters, J.G. Recognising Weeds in a Maize Crop Using a Random Forest Machine-Learning Algorithm and near-Infrared Snapshot Mosaic Hyperspectral Imagery. Biosyst. Eng. 2018, 170, 39–50. [Google Scholar] [CrossRef]
- Huang, H.; Deng, J.; Lan, Y.; Yang, A.; Deng, X.; Zhang, L. A Fully Convolutional Network for Weed Mapping of Unmanned Aerial Vehicle (UAV) Imagery. PLoS ONE 2018, 13, e0196302. [Google Scholar] [CrossRef] [Green Version]
- Bah, M.D.; Hafiane, A.; Canals, R. Deep Learning with Unsupervised Data Labeling for Weed Detection in Line Crops in UAV Images. Remote Sens. 2018, 10, 1690. [Google Scholar] [CrossRef] [Green Version]
- de Castro, A.I.; Torres-Sánchez, J.; Peña, J.M.; Jiménez-Brenes, F.M.; Csillik, O.; López-Granados, F. An Automatic Random Forest-OBIA Algorithm for Early Weed Mapping between and within Crop Rows Using UAV Imagery. Remote Sens. 2018, 10, 285. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Liao, W.; Nuyttens, D.; Lootens, P.; Vangeyte, J.; Pižurica, A.; He, Y.; Pieters, J.G. Fusion of Pixel and Object-Based Features for Weed Mapping Using Unmanned Aerial Vehicle Imagery. Int. J. Appl. Earth Obs. Geoinf. 2018, 67, 43–53. [Google Scholar] [CrossRef]
- Blaschke, T.; Hay, G.J.; Kelly, M.; Lang, S.; Hofmann, P.; Addink, E.; Queiroz Feitosa, R.; van der Meer, F.; van der Werff, H.; van Coillie, F.; et al. Geographic Object-Based Image Analysis—Towards a New Paradigm. ISPRS J. Photogramm. Remote Sens. 2014, 87, 180–191. [Google Scholar] [CrossRef] [Green Version]
- Gutiérrez, P.A.; López-Granados, F.; Peña-Barragán, J.M.; Jurado-Expósito, M.; Hervás-Martínez, C. Logistic Regression Product-Unit Neural Networks for Mapping Ridolfia Segetum Infestations in Sunflower Crop Using Multitemporal Remote Sensed Data. Comput. Electron. Agric. 2008, 64, 293–306. [Google Scholar] [CrossRef]
- Zhu, X.; Tuia, D.; Mou, L.; Xia, G.-S.; Zhang, L.; Xu, F.; Fraundorfer, F. Deep learning in remote sensing: A comprehensive review and list of resources. IEEE Geosci. Remote Sens. Mag. 2017, 5, 8–36. [Google Scholar] [CrossRef] [Green Version]
- Schmidhuber, J. Deep Learning in Neural Networks: An Overview. Neural Netw. 2015, 61, 85–117. [Google Scholar] [CrossRef] [Green Version]
- Atkinson, P.M.; Tatnall, A.R.L. Introduction Neural Networks in Remote Sensing. Int. J. Remote Sens. 1997, 18, 699–709. [Google Scholar] [CrossRef]
- de Castro, A.-I.; Jurado-Expósito, M.; Gómez-Casero, M.-T.; López-Granados, F. Applying Neural Networks to Hyperspectral and Multispectral Field Data for Discrimination of Cruciferous Weeds in Winter Crops. Sci. World J. 2012. [Google Scholar] [CrossRef] [Green Version]
- Tang, L.; Tian, L.; Steward, B.L. Classification of Broadleaf and Grass Weeds Using Gabor Wavelets and an Artificial Neural Network. Trans. ASAE 2003, 46. [Google Scholar] [CrossRef] [Green Version]
- Andújar, D.; Escolà, A.; Dorado, J.; Fernández-Quintanilla, C. Weed Discrimination Using Ultrasonic Sensors. Weed Res. 2011, 51, 543–547. [Google Scholar] [CrossRef] [Green Version]
- Mesas-Carrascosa, F.J.; Rumbao, I.C.; Torres-Sánchez, J.; García-Ferrer, A.; Peña, J.M.; Granados, F.L. Accurate Ortho-Mosaicked Six-Band Multispectral UAV Images as Affected by Mission Planning for Precision Agriculture Proposes. Int. J. Remote Sens. 2017, 38, 2161–2176. [Google Scholar] [CrossRef]
- Mesas-Carrascosa, F.-J.; Torres-Sánchez, J.; Clavero-Rumbao, I.; García-Ferrer, A.; Peña, J.-M.; Borra-Serrano, I.; López-Granados, F. Assessing Optimal Flight Parameters for Generating Accurate Multispectral Orthomosaicks by UAV to Support Site-Specific Crop Management. Remote Sens. 2015, 7, 12793–12814. [Google Scholar] [CrossRef] [Green Version]
- Dandois, J.P.; Ellis, E.C. High Spatial Resolution Three-Dimensional Mapping of Vegetation Spectral Dynamics Using Computer Vision. Remote Sens. Environ. 2013, 136, 259–276. [Google Scholar] [CrossRef] [Green Version]
- McCoy, R.M. Field Methods in Remote Sensing; Guilford Press: New York, NY, USA, 2005; ISBN 978-1-59385-079-1. [Google Scholar]
- Baatz, M.; Schaepe, A. Multiresolution Segmentation: An Optimization Approach for High Quality Multi-Scale Image Segmentation (ECognition). Available online: http://www.ecognition.cc/download/baatz_schaepe.pdf (accessed on 18 April 2014).
- Torres-Sánchez, J.; López-Granados, F.; Peña, J.M. An Automatic Object-Based Method for Optimal Thresholding in UAV Images: Application for Vegetation Detection in Herbaceous Crops. Comput. Electron. Agric. 2015, 114, 43–52. [Google Scholar] [CrossRef]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I. Textural Features for Image Classification. IEEE Trans. Syst. Man Cybern. 1973, SMC-3, 610–621. [Google Scholar] [CrossRef] [Green Version]
- Weszka, J.S.; Dyer, C.R.; Rosenfeld, A. A Comparative Study of Texture Measures for Terrain Classification. IEEE Trans. Syst. Man Cybern. 1976, SMC-6, 269–285. [Google Scholar] [CrossRef]
- Everitt, J.H.; Villarreal, R. Detecting Huisache (Acacia farnesiana) and Mexican Palo-Verde (Parkinsonia aculeata) by Aerial Photography. Weed Sci. 1987, 35, 427–432. [Google Scholar] [CrossRef]
- Jiménez-Brenes, F.M.; López-Granados, F.; Torres-Sánchez, J.; Peña, J.M.; Ramírez, P.; Castillejo-González, I.L.; Castro, A.I. de Automatic UAV-Based Detection of Cynodon Dactylon for Site-Specific Vineyard Management. PLoS ONE 2019, 14, e0218132. [Google Scholar] [CrossRef] [PubMed]
- Gitelson, A.A.; Kaufman, Y.J.; Stark, R.; Rundquist, D. Novel Algorithms for Remote Estimation of Vegetation Fraction. Remote Sens. Environ. 2002, 80, 76–87. [Google Scholar] [CrossRef] [Green Version]
- Peñuelas, J.; Gamon, J.A.; Fredeen, A.L.; Merino, J.; Field, C.B. Reflectance Indices Associated with Physiological Changes in Nitrogen- and Water-Limited Sunflower Leaves. Remote Sens. Environ. 1994, 48, 135–146. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Stark, R.; Grits, U.; Rundquist, D.; Kaufman, Y.; Derry, D. Vegetation and Soil Lines in Visible Spectral Space: A Concept and Technique for Remote Estimation of Vegetation Fraction. Int. J. Remote Sens. 2002, 23, 2537–2562. [Google Scholar] [CrossRef]
- Woebbecke, D.M.; Meyer, G.E.; Von Bargen, K.; Mortensen, D.A. Color Indices for Weed Identification under Various Soil, Residue, and Lighting Conditions. Trans. ASAE 1995, 38, 259–269. [Google Scholar] [CrossRef]
- Guijarro, M.; Pajares, G.; Riomoros, I.; Herrera, P.J.; Burgos-Artizzu, X.P.; Ribeiro, A. Automatic Segmentation of Relevant Textures in Agricultural Images. Comput. Electron. Agric. 2011, 75, 75–83. [Google Scholar] [CrossRef] [Green Version]
- Meyer, G.E.; Hindman, T.W.; Laksmi, K. Machine Vision Detection Parameters for Plant Species Identification. In Proceedings of the Precision Agriculture and Biological Quality; International Society for Optics and Photonics, Boston, MA, USA, 14 January 1999; Volume 3543, pp. 327–335. [Google Scholar]
- Camargo Neto, J. A Combined Statistical-Soft Computing Approach for Classification and Mapping Weed Species in Minimum-Tillage Systems. Ph.D. Thesis, ETD Collection for University of Nebraska, Lincoln, NE, USA, 2004; pp. 1–170. [Google Scholar]
- Kataoka, T.; Kaneko, T.; Okamoto, H.; Hata, S. Crop Growth Estimation System Using Machine Vision. In Proceedings of the 2003 IEEE/ASME International Conference on Advanced Intelligent Mechatronics, Taipei, Taiwan, 14–19 September 2003; Volume 2, pp. b1079–b1083. [Google Scholar]
- Hague, T.; Tillett, N.D.; Wheeler, H. Automated Crop and Weed Monitoring in Widely Spaced Cereals. Precis. Agric. 2006, 7, 21–32. [Google Scholar] [CrossRef]
- Guerrero, J.M.; Pajares, G.; Montalvo, M.; Romeo, J.; Guijarro, M. Support Vector Machines for Crop/Weeds Identification in Maize Fields. Expert Syst. Appl. 2012, 39, 11149–11155. [Google Scholar] [CrossRef]
- IBM SPSS Statistics 26 Documentation. Available online: https://www.ibm.com/support/pages/ibm-spss-statistics-26-documentation (accessed on 11 January 2021).
- Møller, M.F. A Scaled Conjugate Gradient Algorithm for Fast Supervised Learning. Neural Netw. 1993, 6, 525–533. [Google Scholar] [CrossRef]
- Congalton, R.G. A Review of Assessing the Accuracy of Classifications of Remotely Sensed Data. Remote Sens. Environ. 1991, 37, 35–46. [Google Scholar] [CrossRef]
- Gibson, K.D.; Dirks, R.; Medlin, C.R.; Johnston, L. Detection of Weed Species in Soybean Using Multispectral Digital Images. Weed Technol. 2004, 18, 742–749. [Google Scholar] [CrossRef]
- Lottes, P.; Khanna, R.; Pfeifer, J.; Siegwart, R.; Stachniss, C. In Proceedings of the UAV-Based Crop and Weed Classification for Smart Farming, Singapore, 29 May–3 June 2017.
- Zisi, T.; Alexandridis, T.K.; Kaplanis, S.; Navrozidis, I.; Tamouridou, A.-A.; Lagopodi, A.; Moshou, D.; Polychronos, V. Incorporating Surface Elevation Information in UAV Multispectral Images for Mapping Weed Patches. J. Imaging 2018, 4, 132. [Google Scholar] [CrossRef] [Green Version]
- Veeranampalayam Sivakumar, A.N.; Li, J.; Scott, S.; Psota, E.; Jhala, A.J.; Luck, J.D.; Shi, Y. Comparison of Object Detection and Patch-Based Classification Deep Learning Models on Mid- to Late-Season Weed Detection in UAV Imagery. Remote Sens. 2020, 12, 2136. [Google Scholar] [CrossRef]
- Louargant, M.; Jones, G.; Faroux, R.; Paoli, J.-N.; Maillot, T.; Gée, C.; Villette, S. Unsupervised Classification Algorithm for Early Weed Detection in Row-Crops by Combining Spatial and Spectral Information. Remote Sens. 2018, 10, 761. [Google Scholar] [CrossRef] [Green Version]
- Hassler, S.C.; Baysal-Gurel, F. Unmanned Aircraft System (UAS) Technology and Applications in Agriculture. Agronomy 2019, 9, 618. [Google Scholar] [CrossRef] [Green Version]
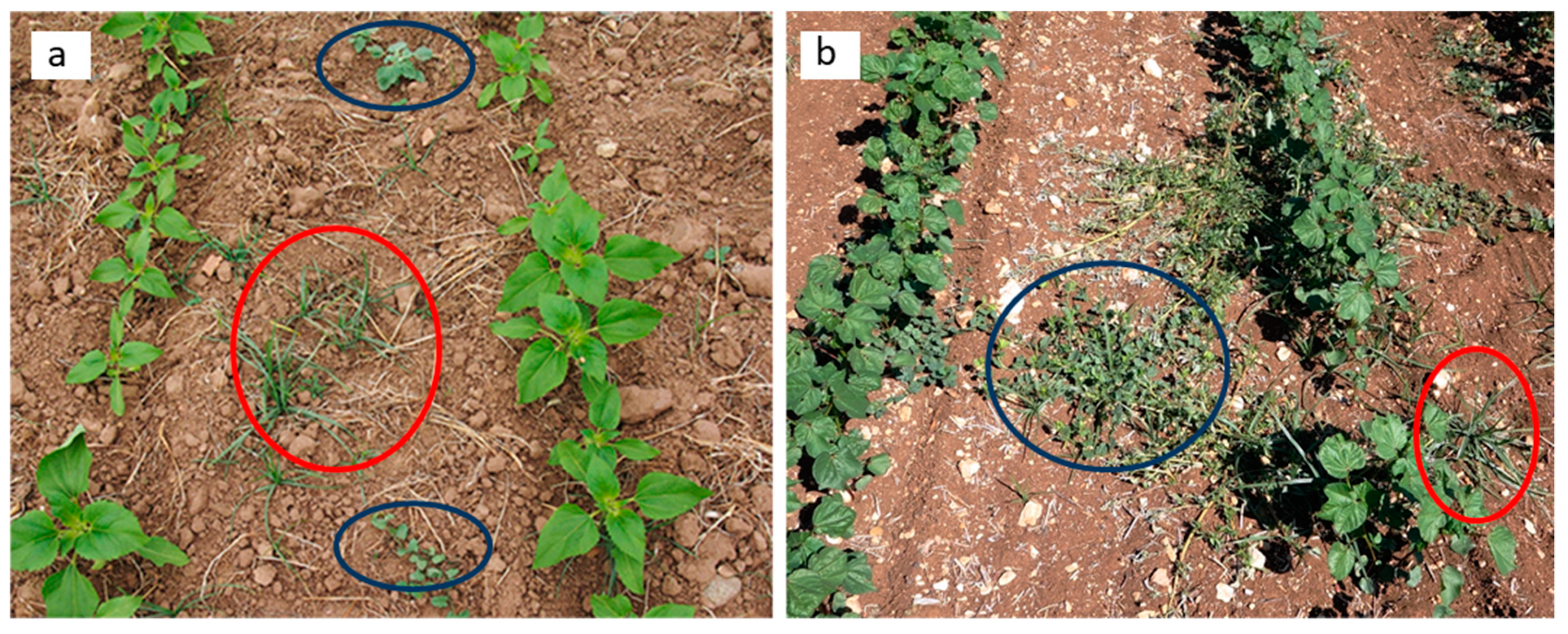
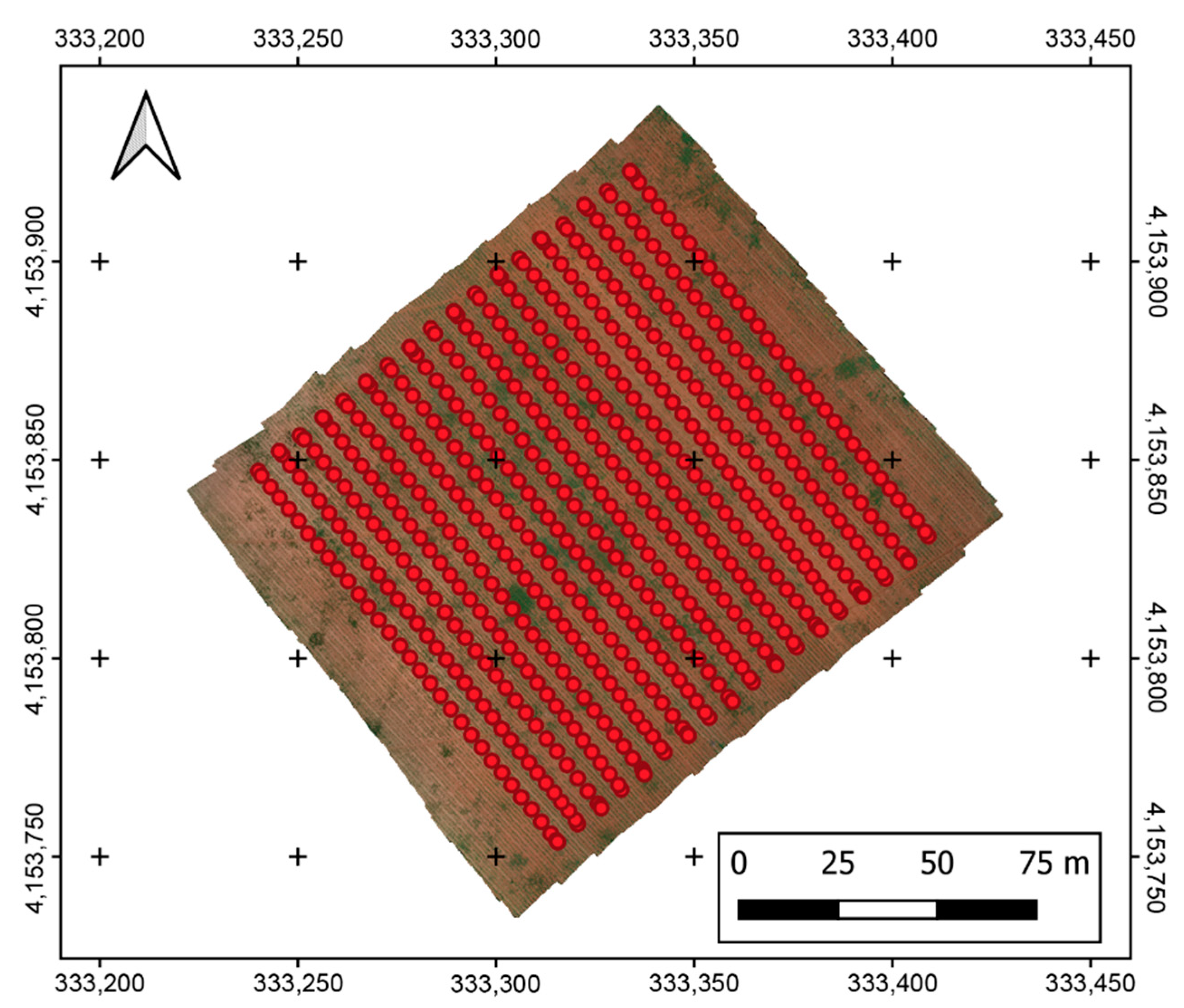
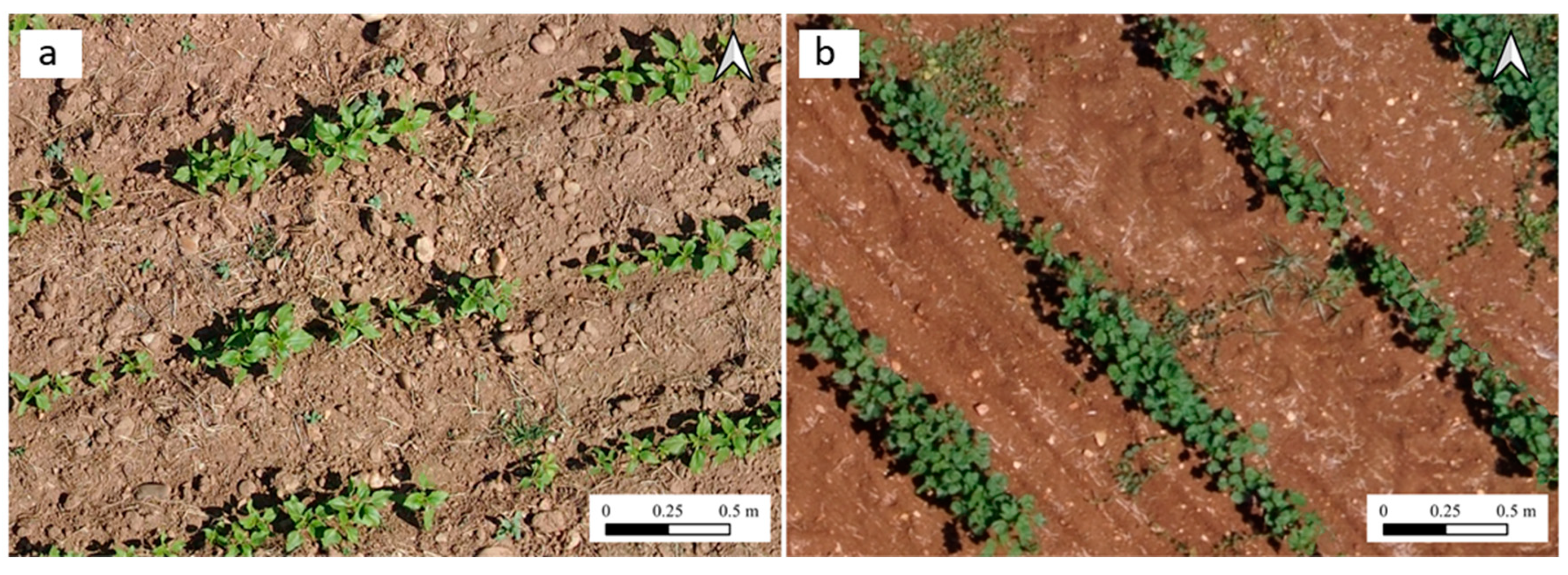

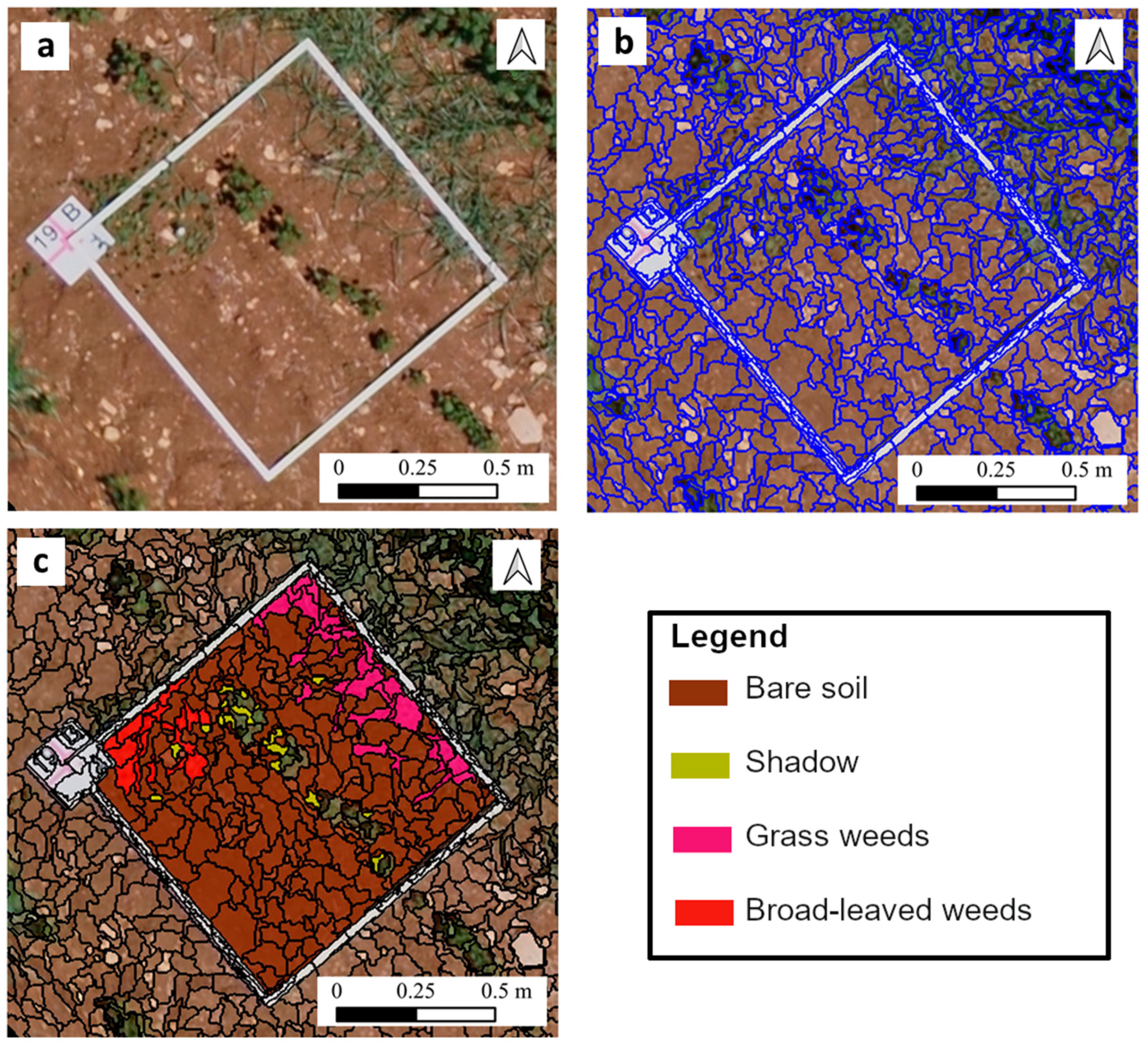
| Crop | Inter-Row Spacing (m) | Location | Area (ha) |
|---|---|---|---|
| Sunflower | 0.75 | Córdoba | 1.14 |
| Cotton | 1.00 | Santaella (Córdoba) | 1.90 |
| Crop | Broad-Leaved Weeds | Grass Weeds |
|---|---|---|
| Sunflower | Amaranthus blitoides | Cyperus rotundus Lolium rigidum |
| Chrozophora tinctoria | ||
| Convolvulus arvensis | ||
| Polygonum aviculare | ||
| Xanthium strumarium | ||
| Cotton | Amaranthus blitoides | Cyperus rotundus Phalaris spp. |
| Amaranthus retroflexus | ||
| Convolvulus arvensis | ||
| Cuscuta spp. | ||
| Datura stramonium | ||
| Ecbalium elaterium | ||
| Mollucella laevis | ||
| Portulaca oleracea | ||
| Xanthium strumarium |
| Crop | Frames | Class | |||
|---|---|---|---|---|---|
| Broad-Leaved Weeds | Grass | Bare Soil | Shadow | ||
| Sunflower | 30 | 635 | 979 | 6665 | 1670 |
| Cotton | 30 | 2023 | 421 | 6437 | 1051 |
| Spectral | RGB values | Red normalized (r): R/(R+G+B) |
| Green normalized (g): G/(R+G+B) | ||
| Blue normalized (b): B/(R+G+B) | ||
| Brightness | ||
| Skewness | ||
| HSI values | Hue | |
| Vegetation indices | R/B [40] | |
| R/G [41] | ||
| Normalized red green difference index NRGDI: (g−r)/(g+r) [42] | ||
| Normalized pigment chlorophyll ratio index NPCI: (r−b)/(r+b) [43] | ||
| Visible atmospherically resistant index VARI: (g−r)/(g+r-b) [44] | ||
| Woebbecke Index WI: (g−b)/(r−g) [45] | ||
| Excess blue ExB: (1.4*b)−g [46] | ||
| Excess green ExG: (2*g)−r−b [45] | ||
| Excess red ExR: (1.4*r)−g [47] | ||
| ExGR: ExG-ExR [48] | ||
| Color index of vegetation CIVE: (0.441*r)−(0.811*g)+(0.385*b)+18.78745 [49] | ||
| Vegetative VEG: g/((r^0.667)*(b^(1−0.667))) [50] | ||
| Combination 1 COMB1: (0.25*ExG)+(0.3*ExGR)+(0.33*CIVE)+(0.12*VEG) [46] | ||
| Combination 2 COMB2: (0.36*ExG)+(0.47*CIVE)+(0.17*VEG) [51] | ||
| Geometric | Shape | Asymmetry |
| Compactness | ||
| Density | ||
| Radius of largest enclosed ellipse | ||
| Radius of smallest enclosing ellipse | ||
| Rectangular fit | ||
| Roundness | ||
| Shape index | ||
| Based on polygons | Compactness (Polygon) | |
| Number of edges (Polygon) | ||
| Perimeter (Polygon) (Pixel) | ||
| Standard deviation of length of edges (Polygon) (Pixel) | ||
| Average area represented by segments (Pixel) | ||
| Number of segments | ||
| Extent | Length/Width | |
| Height derived | CHM: DSM-DTM | |
| Textural | Texture after Haralick | GLCM homogeneity (all directions) |
| GLCM contrast (all directions) | ||
| GLCM dissimilarity (all directions) | ||
| GLCM entropy (all directions) | ||
| GLCM angular second moment (all directions) | ||
| GLCM mean (all directions) | ||
| GLCM standard deviation (all directions) | ||
| GLCM correlation (all directions) | ||
| GLDV angular second moment (all directions) | ||
| GLDV entropy (all directions) | ||
| GLDV mean (all directions) | ||
| GLDV contrast (all directions) |
| Predicted | ||||||
|---|---|---|---|---|---|---|
| Observed | Bare Soil | Broad-Leaved Weeds | Grass Weeds | Shadow | Producer Accuracy (%) | |
| Training | Bare soil | 355 | 6 | 19 | 13 | 90.33 |
| Broad-leaved weeds | 5 | 293 | 71 | 4 | 78.55 | |
| Grass weeds | 23 | 110 | 261 | 4 | 65.58 | |
| Shadow | 13 | 2 | 4 | 365 | 95.05 | |
| User accuracy (%) | 89.65 | 71.29 | 73.52 | 94.56 | OA: 82.30% | |
| Testing | Bare soil | 93 | 1 | 12 | 7 | 82.30 |
| Broad-leaved weeds | 0 | 102 | 35 | 0 | 74.45 | |
| Grass weeds | 10 | 30 | 78 | 5 | 63.41 | |
| Shadow | 5 | 0 | 3 | 126 | 94.03 | |
| User accuracy (%) | 86.11 | 76.69 | 60.94 | 91.30 | OA: 78.70% | |
| Validation | Bare soil | 117 | 2 | 6 | 4 | 90.70 |
| Broad-leaved weeds | 0 | 93 | 32 | 0 | 74.40 | |
| Grass weeds | 5 | 23 | 85 | 1 | 74.56 | |
| Shadow | 3 | 0 | 3 | 109 | 94.78 | |
| User accuracy (%) | 93.60 | 78.81 | 67.46 | 95.61 | OA: 83.64% | |
| Predicted | ||||||
|---|---|---|---|---|---|---|
| Observed | Bare Soil | Broad-Leaved Weeds | Grass Weeds | Shadow | Producer Accuracy (%) | |
| Training | Bare soil | 235 | 5 | 20 | 1 | 90.04 |
| Broad-leaved weeds | 9 | 171 | 70 | 10 | 65.77 | |
| Grass weeds | 32 | 47 | 177 | 10 | 66.54 | |
| Shadow | 3 | 8 | 7 | 231 | 92.77 | |
| User accuracy (%) | 84.23 | 74.03 | 64.60 | 91.67 | OA: 78.57% | |
| Testing | Bare soil | 70 | 1 | 8 | 3 | 85.37 |
| Broad-leaved weeds | 4 | 49 | 21 | 2 | 64.47 | |
| Grass weeds | 13 | 14 | 37 | 4 | 54.41 | |
| Shadow | 1 | 1 | 3 | 69 | 93.24 | |
| User accuracy (%) | 79.55 | 75.38 | 53.62 | 88.46 | OA: 75.00% | |
| Validation | Bare soil | 69 | 1 | 7 | 1 | 88.46 |
| Broad-leaved weeds | 6 | 49 | 23 | 7 | 57.65 | |
| Grass weeds | 9 | 13 | 63 | 2 | 72.41 | |
| Shadow | 1 | 1 | 5 | 91 | 92.86 | |
| User accuracy (%) | 81.18 | 76.56 | 64.29 | 90.10 | OA: 78.16% | |
| Variable | Importance | Normalized Importance (%) |
|---|---|---|
| Brightness | 0.040 | 100.0 |
| COMB1 | 0.040 | 100.0 |
| GLDV Entropy (all dir.) | 0.033 | 82.9 |
| VEG | 0.033 | 81.5 |
| NRGDI | 0.033 | 81.1 |
| Hue | 0.031 | 77.1 |
| r | 0.030 | 75.1 |
| CHM | 0.029 | 73.4 |
| ExGR | 0.029 | 71.7 |
| g | 0.029 | 71.5 |
| Variable | Importance | Normalized Importance (%) |
|---|---|---|
| Brightness | 0.058 | 100.0 |
| GLCM Homogeneity (all dir.) | 0.038 | 65.8 |
| GLCM Ang. 2nd moment (all dir.) | 0.036 | 61.6 |
| ExGR | 0.034 | 58.3 |
| g | 0.033 | 56.9 |
| COMB2 | 0.032 | 55.0 |
| GLCM Entropy (all dir.) | 0.032 | 54.7 |
| GLDV Contrast (all dir.) | 0.030 | 50.9 |
| GLCM SD (all dir.) | 0.030 | 50.6 |
| NRGDI | 0.029 | 49.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Torres-Sánchez, J.; Mesas-Carrascosa, F.J.; Jiménez-Brenes, F.M.; de Castro, A.I.; López-Granados, F. Early Detection of Broad-Leaved and Grass Weeds in Wide Row Crops Using Artificial Neural Networks and UAV Imagery. Agronomy 2021, 11, 749. https://doi.org/10.3390/agronomy11040749
Torres-Sánchez J, Mesas-Carrascosa FJ, Jiménez-Brenes FM, de Castro AI, López-Granados F. Early Detection of Broad-Leaved and Grass Weeds in Wide Row Crops Using Artificial Neural Networks and UAV Imagery. Agronomy. 2021; 11(4):749. https://doi.org/10.3390/agronomy11040749
Chicago/Turabian StyleTorres-Sánchez, Jorge, Francisco Javier Mesas-Carrascosa, Francisco M. Jiménez-Brenes, Ana I. de Castro, and Francisca López-Granados. 2021. "Early Detection of Broad-Leaved and Grass Weeds in Wide Row Crops Using Artificial Neural Networks and UAV Imagery" Agronomy 11, no. 4: 749. https://doi.org/10.3390/agronomy11040749
APA StyleTorres-Sánchez, J., Mesas-Carrascosa, F. J., Jiménez-Brenes, F. M., de Castro, A. I., & López-Granados, F. (2021). Early Detection of Broad-Leaved and Grass Weeds in Wide Row Crops Using Artificial Neural Networks and UAV Imagery. Agronomy, 11(4), 749. https://doi.org/10.3390/agronomy11040749







