Transcriptomic Changes Induced by Drought Stress in Hardneck Garlic during the Bolting/Bulbing Stage
Abstract
1. Introduction
2. Materials and Methods
2.1. Plants and Growth Conditions
2.2. Drought Stress Treatment and Physiological Measurements
2.3. RNA Extraction and Library Construction
2.4. RNA-Seq Data Analysis and Functional Annotation
3. Results
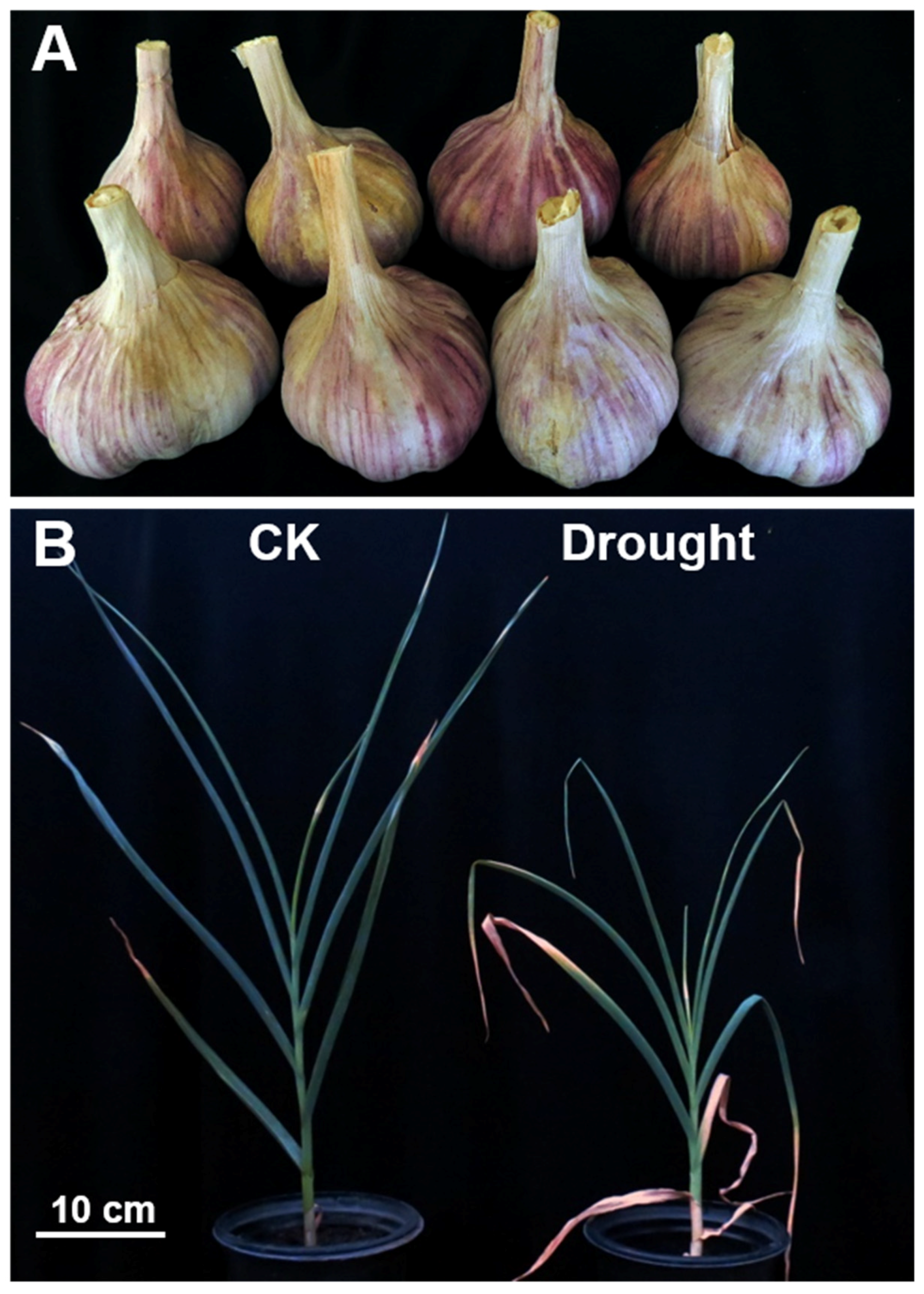
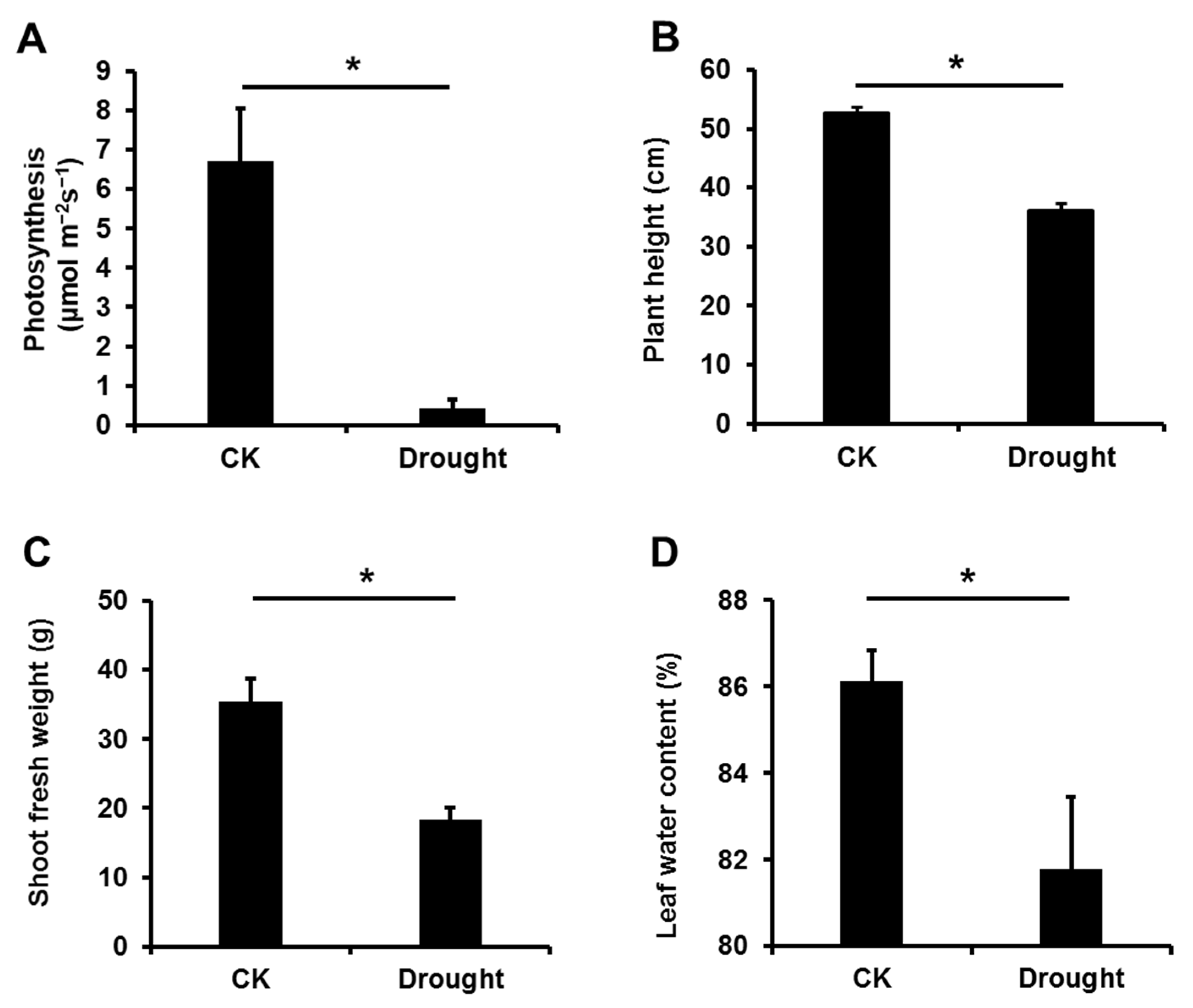
3.1. Drought Stress Significantly Affected the Growth of Garlic Plants
3.2. RNA-Seq Analysis under Normal Conditions and Drought Stress
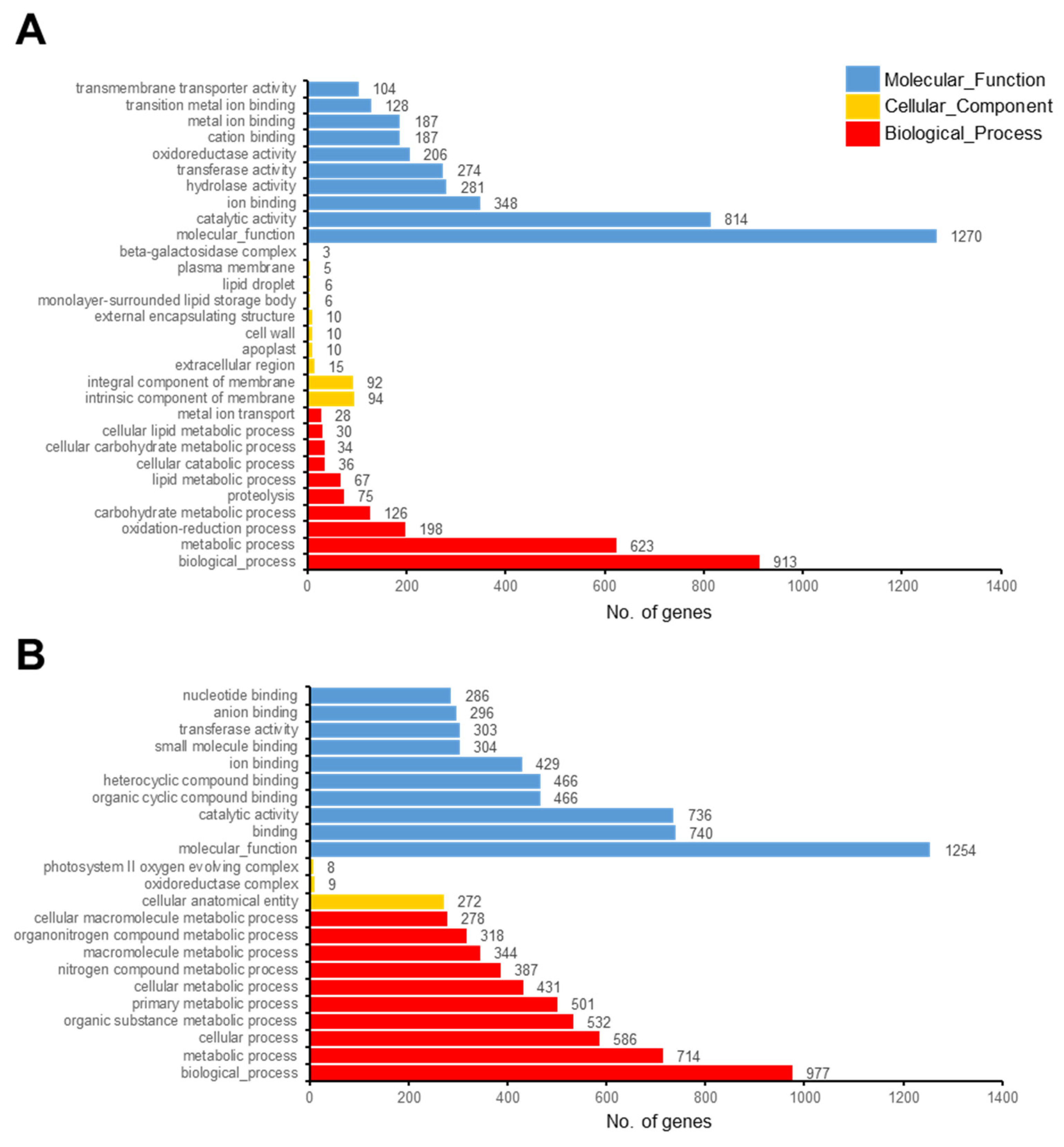
3.3. Functional Categorization of DEGs
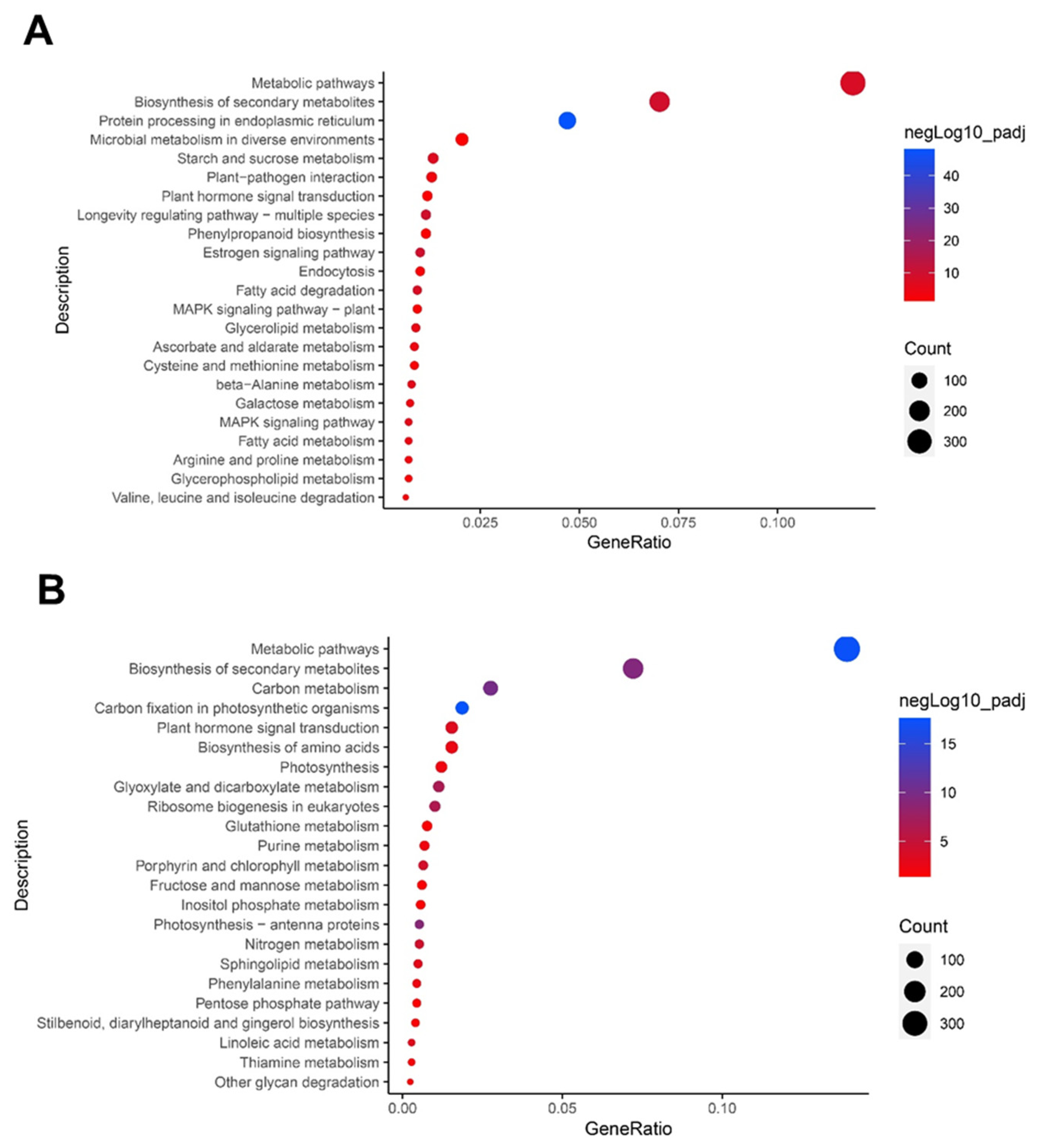
3.4. Altered Pathways in Garlic Leaves by Drought
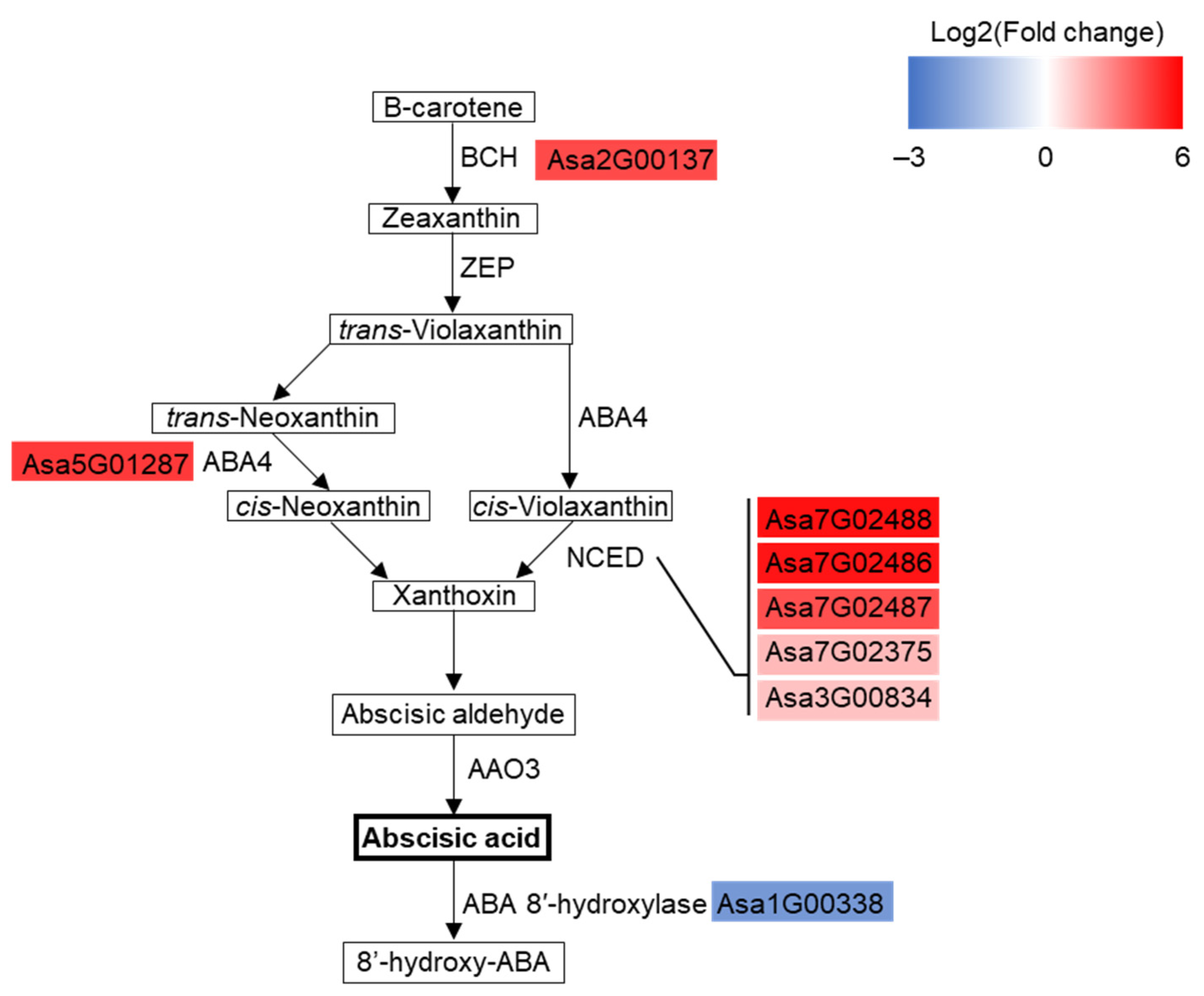
3.5. ABA Biosynthesis Pathway Was Substantially Enhanced in Drought-Treated Garlic Leaves
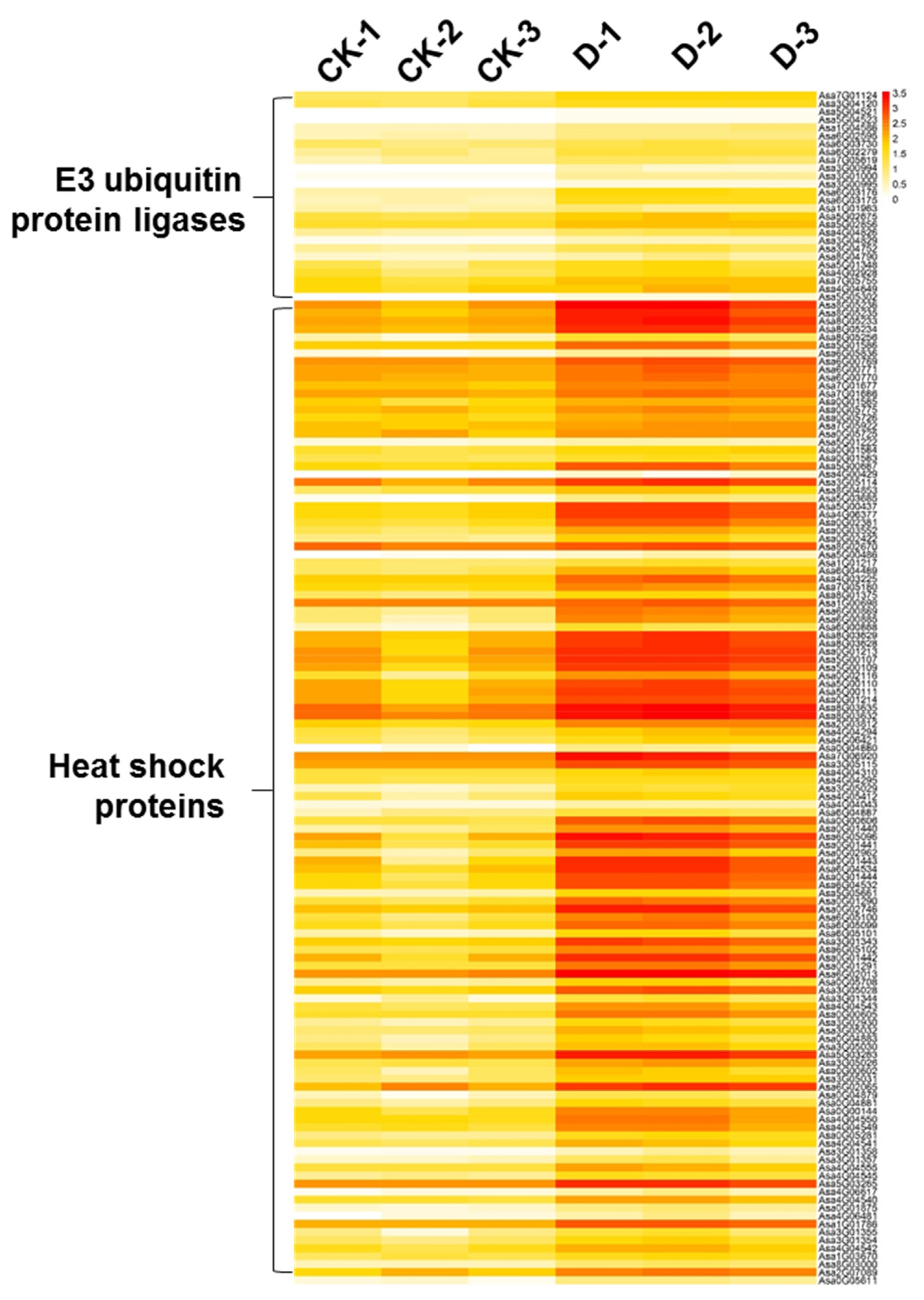
3.6. HSPs and E3 Ubiquitin Ligases Were Significantly Upregulated
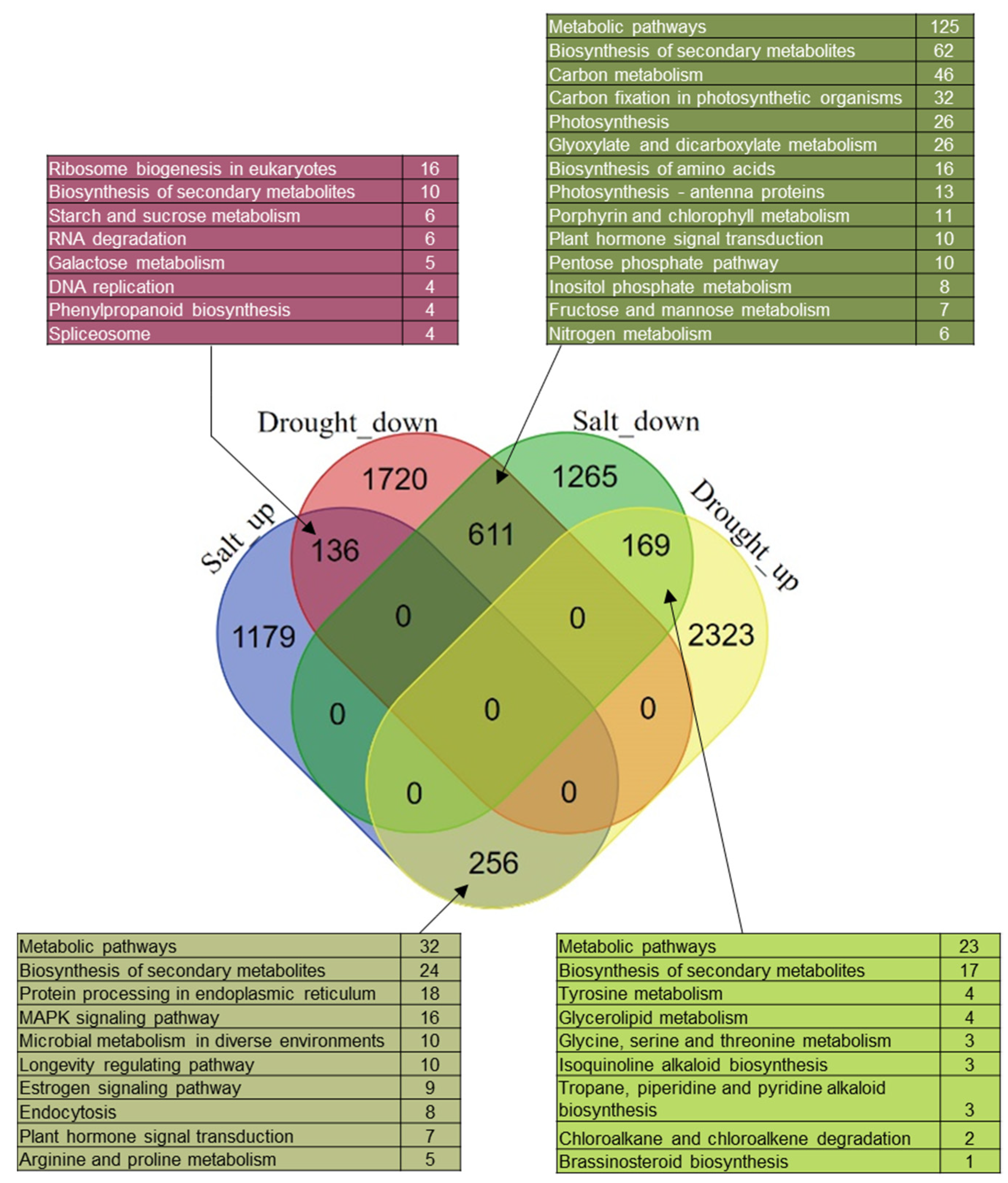
3.7. Commonly and Reversely Regulated-DEGs in Garlic Leaves on Exposure to Drought and Salinity Stresses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cornic, G. Drought stress inhibits photosynthesis by decreasing stomatal aperture-not by affecting ATP synthesis. Trends Plant Sci. 2000, 5, 187–188. [Google Scholar] [CrossRef]
- Parry, A.D.; Horgan, R. Carotenoids and abscisic acid (ABA) biosynthesis in higher plants. Physiol. Plant 1991, 82, 320–326. [Google Scholar] [CrossRef]
- Sah, S.K.; Reddy, K.R.; Li, J. Abscisic acid and abiotic stress tolerance in crop plants. Front. Plant Sci. 2016, 7, 571. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Guo, Y.; Liu, Y.; Zhang, F.; Wang, Z.; Wang, H.; Wang, F.; Li, D.; Mao, D.; Luan, S.; et al. 9-cis-epoxycarotenoid dioxygenase 3 regulates plant growth and enhances multi-abiotic stress tolerance in rice. Front. Plant Sci. 2018, 9, 162. [Google Scholar] [CrossRef]
- Fujita, Y.; Nakashima, K.; Yoshida, T.; Katagiri, T.; Kidokoro, S.; Kanamori, N.; Umezawa, T.; Fujita, M.; Maruyama, K.; Ishiyama, K.; et al. Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol. 2009, 50, 2123–2132. [Google Scholar] [CrossRef]
- Yoshida, T.; Fujita, Y.; Sayama, H.; Kidokoro, S.; Maruyama, K.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 2010, 61, 672–685. [Google Scholar] [CrossRef]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef]
- Dos Reis, S.P.; Lima, A.M.; de Souza, C.R.B. Recent molecular advances on downstream plant responses to abiotic stress. Int. J. Mol. Sci. 2012, 13, 8628–8647. [Google Scholar] [CrossRef]
- Wang, W.; Vinocur, B.; Shoseyov, O.; Altman, A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004, 9, 244–252. [Google Scholar] [CrossRef]
- Singh, R.K.; Jaishankar, J.; Muthamilarasan, M.; Shweta, S.; Dangi, A.; Prasad, M. Genome-wide analysis of heat shock proteins in C4 model, foxtail millet identifies potential candidates for crop improvement under abiotic stress. Sci. Rep. 2016, 6, 32641. [Google Scholar] [CrossRef]
- Qin, F.; Sakuma, Y.; Tran, L.S.; Maruyama, K.; Kidokoro, S.; Fujita, Y.; Fujita, M.; Umezawa, T.; Sawano, Y.; Miyazono, K.; et al. Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell 2008, 20, 1693–1707. [Google Scholar] [CrossRef] [PubMed]
- Guerra, D.; Mastrangelo, A.M.; Lopez-Torrejon, G.; Marzin, S.; Schweizer, P.; Stanca, A.M.; del Pozo, J.C.; Cattivelli, L.; Mazzucotelli, E. Identification of a protein network interacting with TdRF1, a wheat RING ubiquitin ligase with a protective role against cellular dehydration. Plant Physiol. 2012, 158, 777–789. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.K.; Ryu, M.Y.; Song, C.; Kwak, J.M.; Kim, W.T. Arabidopsis PUB22 and PUB23 are homologous U-Box E3 ubiquitin ligases that play combinatory roles in response to drought stress. Plant Cell 2008, 20, 1899–1914. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.K.; Cho, S.K.; Son, O.; Xu, Z.; Hwang, I.; Kim, W.T. Drought stress-induced Rma1H1, a RING membrane-anchor E3 ubiquitin ligase homolog, regulates aquaporin levels via ubiquitination in transgenic Arabidopsis plants. Plant Cell 2009, 21, 622–641. [Google Scholar] [CrossRef] [PubMed]
- Neeraj, S.; Sushila, K.; Neeraj, D.; Milind, P.; Minakashi, P. Garlic: A pungent wonder from nature. Int. Res. J. Pharm. 2014, 5, 523–529. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Langmead, B. Aligning short sequencing reads with bowtie. Curr. Protoc. Bioinform. 2010, 32, 11.17.11–11.17.14. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Sun, X.; Zhu, S.; Li, N.; Cheng, Y.; Zhao, J.; Qiao, X.; Lu, L.; Liu, S.; Wang, Y.; Liu, C.; et al. A chromosome-level genome assembly of garlic (Allium sativum) provides insights into genome evolution and allicin biosynthesis. Mol. Plant 2020, 13, 1328–1339. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.; Binns, D.; Chang, H.Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [PubMed]
- Götz, S.; García-Gómez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talón, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef]
- Wang, X.; Cai, X.; Xu, C.; Wang, Q.; Dai, S. Drought-responsive mechanisms in plant leaves revealed by proteomics. Int. J. Mol. Sci. 2016, 17, 1706. [Google Scholar] [CrossRef]
- Kim, J.; Smith, J.J.; Tian, L.; Dellapenna, D. The evolution and function of carotenoid hydroxylases in Arabidopsis. Plant Cell Physiol. 2009, 50, 463–479. [Google Scholar] [CrossRef]
- North, H.M.; De Almeida, A.; Boutin, J.P.; Frey, A.; To, A.; Botran, L.; Sotta, B.; Marion-Poll, A. The Arabidopsis ABA-deficient mutant aba4 demonstrates that the major route for stress-induced ABA accumulation is via neoxanthin isomers. Plant J. 2007, 50, 810–824. [Google Scholar] [CrossRef]
- Perreau, F.; Frey, A.; Effroy-Cuzzi, D.; Savane, P.; Berger, A.; Gissot, L.; Marion-Poll, A. ABSCISIC ACID-DEFICIENT4 has an essential function in both cis-violaxanthin and cis-neoxanthin synthesis. Plant Physiol. 2020, 184, 1303–1316. [Google Scholar] [CrossRef]
- Qin, X.; Zeevaart, J.A.D. The 9-cis-epoxycarotenoid cleavage reaction is the key regulatory step of abscisic acid biosynthesis in water-stressed bean. Proc. Natl. Acad. Sci. USA 1999, 96, 15354–15361. [Google Scholar] [CrossRef]
- Krochko, J.E.; Abrams, G.D.; Loewen, M.K.; Abrams, S.R.; Cutler, A.J. (+)-Abscisic acid 8′-hydroxylase is a cytochrome P450 monooxygenase. Plant Physiol. 1998, 118, 849–860. [Google Scholar] [CrossRef] [PubMed]
- Park, C.-J.; Seo, Y.-S. Heat shock proteins: A review of the molecular chaperones for plant immunity. Plant Pathol. J. 2015, 31, 323–333. [Google Scholar] [CrossRef] [PubMed]
- Augustine, S.M. Function of heat-shock proteins in drought tolerance regulation of plants. In Drought Stress Tolerance in Plants, Vol 1: Physiol.ogy and Biochemistry; Hossain, M.A., Wani, S.H., Bhattacharjee, S., Burritt, D.J., Tran, L.-S.P., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 163–185. [Google Scholar]
- Chen, L.; Hellmann, H. Plant E3 ligases: Flexible enzymes in a sessile world. Mol. Plant 2013, 6, 1388–1404. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Wang, M.; Cheng, Z.; Meng, H. Response of garlic (Allium sativum L.) bolting and bulbing to temperature and photoperiod treatments. Biol. Open 2016, 5, 507–518. [Google Scholar] [CrossRef]
- Haak, D.C.; Fukao, T.; Grene, R.; Hua, Z.; Ivanov, R.; Perrella, G.; Li, S. Multilevel regulation of abiotic stress responses in plants. Front. Plant Sci. 2017, 8, 1564. [Google Scholar] [CrossRef]
- Zhu, G.; Li, W.; Zhang, F.; Guo, W. RNA-seq analysis reveals alternative splicing under salt stress in cotton, Gossypium davidsonii. BMC Genom. 2018, 19, 73. [Google Scholar] [CrossRef]
- Filichkin, S.A.; Hamilton, M.; Dharmawardhana, P.D.; Singh, S.K.; Sullivan, C.; Ben-Hur, A.; Reddy, A.S.N.; Jaiswal, P. Abiotic stresses modulate landscape of poplar transcriptome via alternative splicing, differential intron retention, and isoform ratio switching. Front. Plant Sci. 2018, 9, 5. [Google Scholar] [CrossRef]
- Dinger, M.E.; Amaral, P.P.; Mercer, T.R.; Mattick, J.S. Pervasive transcription of the eukaryotic genome: Functional indices and conceptual implications. Brief. Funct. Genom. 2009, 8, 407–423. [Google Scholar] [CrossRef]
- Di, C.; Yuan, J.; Wu, Y.; Li, J.; Lin, H.; Hu, L.; Zhang, T.; Qi, Y.; Gerstein, M.B.; Guo, Y.; et al. Characterization of stress-responsive lncRNAs in Arabidopsis thaliana by integrating expression, epigenetic and structural features. Plant J. 2014, 80, 848–861. [Google Scholar] [CrossRef]
- Wang, Y.; Fan, X.; Lin, F.; He, G.; Terzaghi, W.; Zhu, D.; Deng, X.W. Arabidopsis noncoding RNA mediates control of photomorphogenesis by red light. Proc. Natl. Acad. Sci. USA 2014, 111, 10359–10364. [Google Scholar] [CrossRef]
- Wang, D.; Qu, Z.; Yang, L.; Zhang, Q.; Liu, Z.-H.; Do, T.; Adelson, D.L.; Wang, Z.-Y.; Searle, I.; Zhu, J.-K. Transposable elements (TEs) contribute to stress-related long intergenic noncoding RNAs in plants. Plant J. 2017, 90, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Bray, E.A. Drought- and ABA-induced changes in polypeptide and mRNA accumulation in tomato leaves. Plant Physiol. 1988, 88, 1210–1214. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Wang, N.; Cui, F.; Li, X.; Xiao, J.; Xiong, L. Characterization of the beta-carotene hydroxylase gene DSM2 conferring drought and oxidative stress resistance by increasing xanthophylls and abscisic acid synthesis in rice. Plant Physiol. 2010, 154, 1304–1318. [Google Scholar] [CrossRef] [PubMed]
- Iuchi, S.; Kobayashi, M.; Taji, T.; Naramoto, M.; Seki, M.; Kato, T.; Tabata, S.; Kakubari, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 2001, 27, 325–333. [Google Scholar] [CrossRef]
- Iuchi, S.; Kobayashi, M.; Yamaguchi-Shinozaki, K.; Shinozaki, K. A stress-inducible gene for 9-cis-epoxycarotenoid dioxygenase involved in abscisic acid biosynthesis under water stress in drought-tolerant cowpea. Plant Physiol. 2000, 123, 553–562. [Google Scholar] [CrossRef]
- Wan, X.R.; Li, L. Regulation of ABA level and water-stress tolerance of Arabidopsis by ectopic expression of a peanut 9-cis-epoxycarotenoid dioxygenase gene. Biochem. Biophys. Res. Commun. 2006, 347, 1030–1038. [Google Scholar] [CrossRef]
- Tong, S.M.; Xi, H.X.; Ai, K.J.; Hou, H.S. Overexpression of wheat TaNCED gene in Arabidopsis enhances tolerance to drought stress and delays seed germination. Biol. Plant 2017, 61, 64–72. [Google Scholar] [CrossRef]
- Thompson, A.J.; Jackson, A.C.; Parker, R.A.; Morpeth, D.R.; Burbidge, A.; Taylor, I.B. Abscisic acid biosynthesis in tomato: Regulation of zeaxanthin epoxidase and 9-cis-epoxycarotenoid dioxygenase mRNAs by light/dark cycles, water stress and abscisic acid. Plant Mol. Biol. 2000, 42, 833–845. [Google Scholar] [CrossRef]
- Umezawa, T.; Okamoto, M.; Kushiro, T.; Nambara, E.; Oono, Y.; Seki, M.; Kobayashi, M.; Koshiba, T.; Kamiya, Y.; Shinozaki, K. CYP707A3, a major ABA 8′-hydroxylase involved in dehydration and rehydration response in Arabidopsis thaliana. Plant J. 2006, 46, 171–182. [Google Scholar] [CrossRef]
- Iba, K. Acclimative Response to temperature stress in higher plants: Approaches of gene engineering for temperature tolerance. Annu. Rev. Plant Biol. 2002, 53, 225–245. [Google Scholar] [CrossRef]
- Scarpeci, T.E.; Zanor, M.I.; Valle, E.M. Investigating the role of plant heat shock proteins during oxidative stress. Plant Signal. Behav. 2008, 3, 856–857. [Google Scholar] [CrossRef] [PubMed]
- Ye, S.; Yu, S.; Shu, L.; Wu, J.; Wu, A.; Luo, L. Expression profile analysis of 9 heat shock protein genes throughout the life cycle and under abiotic stress in rice. Chin. Sci. Bull. 2012, 57, 336–343. [Google Scholar] [CrossRef]
- Sato, Y.; Yokoya, S. Enhanced tolerance to drought stress in transgenic rice plants overexpressing a small heat-shock protein, sHSP17.7. Plant Cell Rep. 2008, 27, 329–334. [Google Scholar] [CrossRef]
- Xiang, J.; Chen, X.; Hu, W.; Xiang, Y.; Yan, M.; Wang, J. Overexpressing heat-shock protein OsHSP50.2 improves drought tolerance in rice. Plant Cell Rep. 2018, 37, 1585–1595. [Google Scholar] [CrossRef]
- Vierstra, R.D. The ubiquitin–26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 2009, 10, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Stone, S.L. The role of ubiquitin and the 26S proteasome in plant abiotic stress signaling. Front. Plant Sci. 2014, 5, 135. [Google Scholar] [CrossRef]
- Smalle, J.; Vierstra, R.D. The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 2004, 55, 555–590. [Google Scholar] [CrossRef]
- Li, X.-M.; Chao, D.-Y.; Wu, Y.; Huang, X.; Chen, K.; Cui, L.-G.; Su, L.; Ye, W.-W.; Chen, H.; Chen, H.-C.; et al. Natural alleles of a proteasome α2 subunit gene contribute to thermotolerance and adaptation of African rice. Nat. Genet. 2015, 47, 827–833. [Google Scholar] [CrossRef]
- Lyzenga, W.J.; Stone, S.L. Abiotic stress tolerance mediated by protein ubiquitination. J. Exp. Bot. 2012, 63, 599–616. [Google Scholar] [CrossRef]
- Forni, C.; Duca, D.; Glick, B.R. Mechanisms of plant response to salt and drought stress and their alteration by rhizobacteria. Plant Soil 2017, 410, 335–356. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, X.; Condori-Apfata, J.A.; Liu, X.; Condori-Pacsi, S.J.; Valencia, M.V.; Zhang, C. Transcriptomic Changes Induced by Drought Stress in Hardneck Garlic during the Bolting/Bulbing Stage. Agronomy 2021, 11, 246. https://doi.org/10.3390/agronomy11020246
Zhou X, Condori-Apfata JA, Liu X, Condori-Pacsi SJ, Valencia MV, Zhang C. Transcriptomic Changes Induced by Drought Stress in Hardneck Garlic during the Bolting/Bulbing Stage. Agronomy. 2021; 11(2):246. https://doi.org/10.3390/agronomy11020246
Chicago/Turabian StyleZhou, Xiangjun, Jorge Alberto Condori-Apfata, Xiaoqin Liu, Sandro Jhonatan Condori-Pacsi, Maria Valderrama Valencia, and Cankui Zhang. 2021. "Transcriptomic Changes Induced by Drought Stress in Hardneck Garlic during the Bolting/Bulbing Stage" Agronomy 11, no. 2: 246. https://doi.org/10.3390/agronomy11020246
APA StyleZhou, X., Condori-Apfata, J. A., Liu, X., Condori-Pacsi, S. J., Valencia, M. V., & Zhang, C. (2021). Transcriptomic Changes Induced by Drought Stress in Hardneck Garlic during the Bolting/Bulbing Stage. Agronomy, 11(2), 246. https://doi.org/10.3390/agronomy11020246






