Phenylalanine Ammonia-Lyase (PAL) Genes Family in Wheat (Triticum aestivum L.): Genome-Wide Characterization and Expression Profiling
Abstract
:1. Introduction
2. Materials and Methods
2.1. Retrieval of Protein Sequences Containing the PAL Gene Family in Triticum aestivum
2.2. Gene Structure and Conserved Domain Analysis of TaPAL Genes
2.3. Phylogenetic Identification
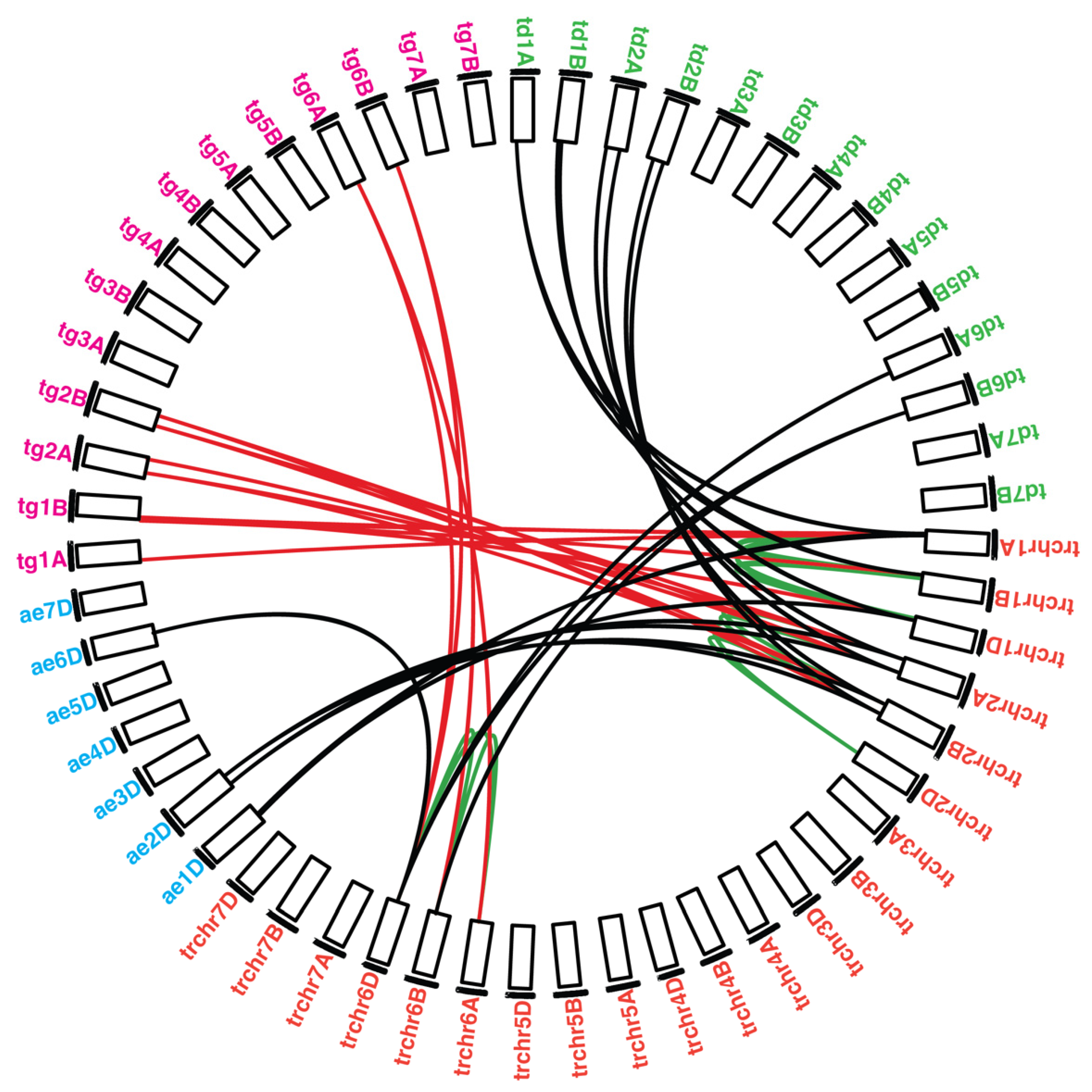
2.4. Synteny Analysis
2.5. miRNA Prediction in Wheat PAL Family Genes
2.6. Promoter and Gene Ontology (GO) Enrichment Analysis
2.7. Protein–Protein Interaction
2.8. Analysis of RNA-Seq Base expression profiling
3. Results
3.1. Common Wheat PAL Gene Characterization and Identification
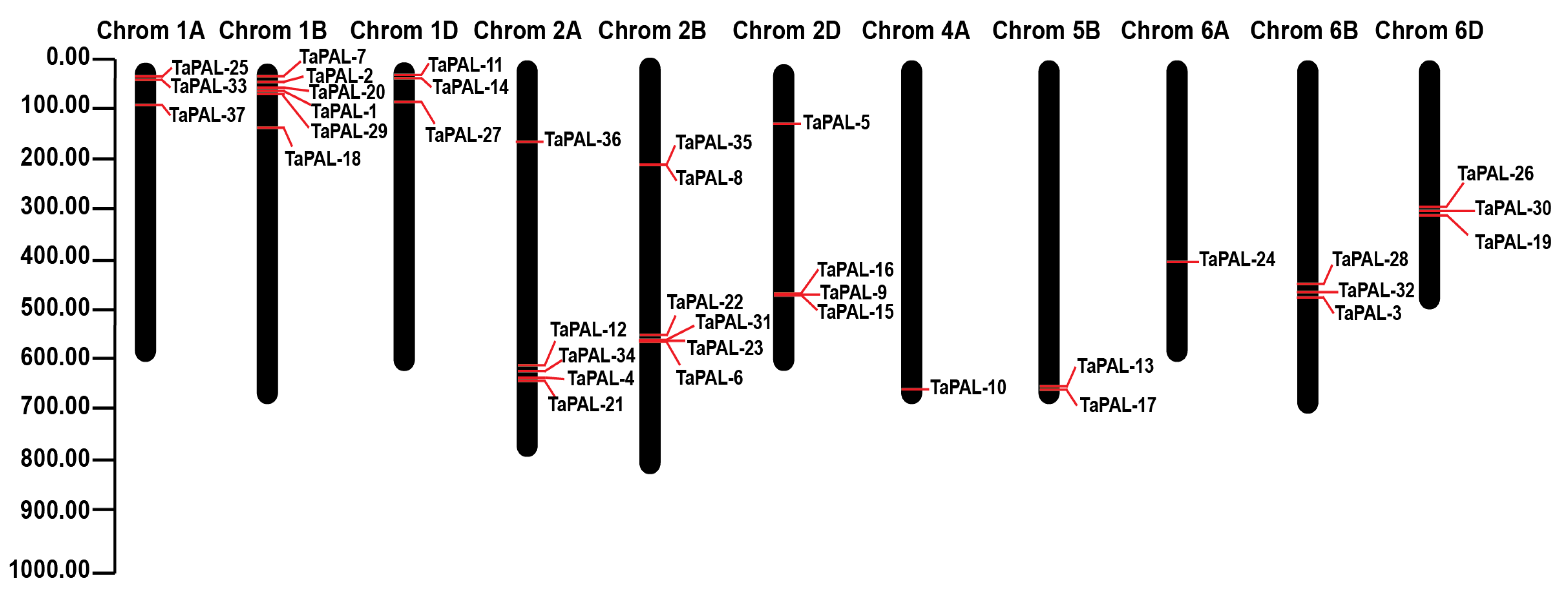
3.2. Localization of TaPAL Genes on the Chromosomes
3.3. Identification of Conserved Protein Domains and Motifs in TaPAL Proteins
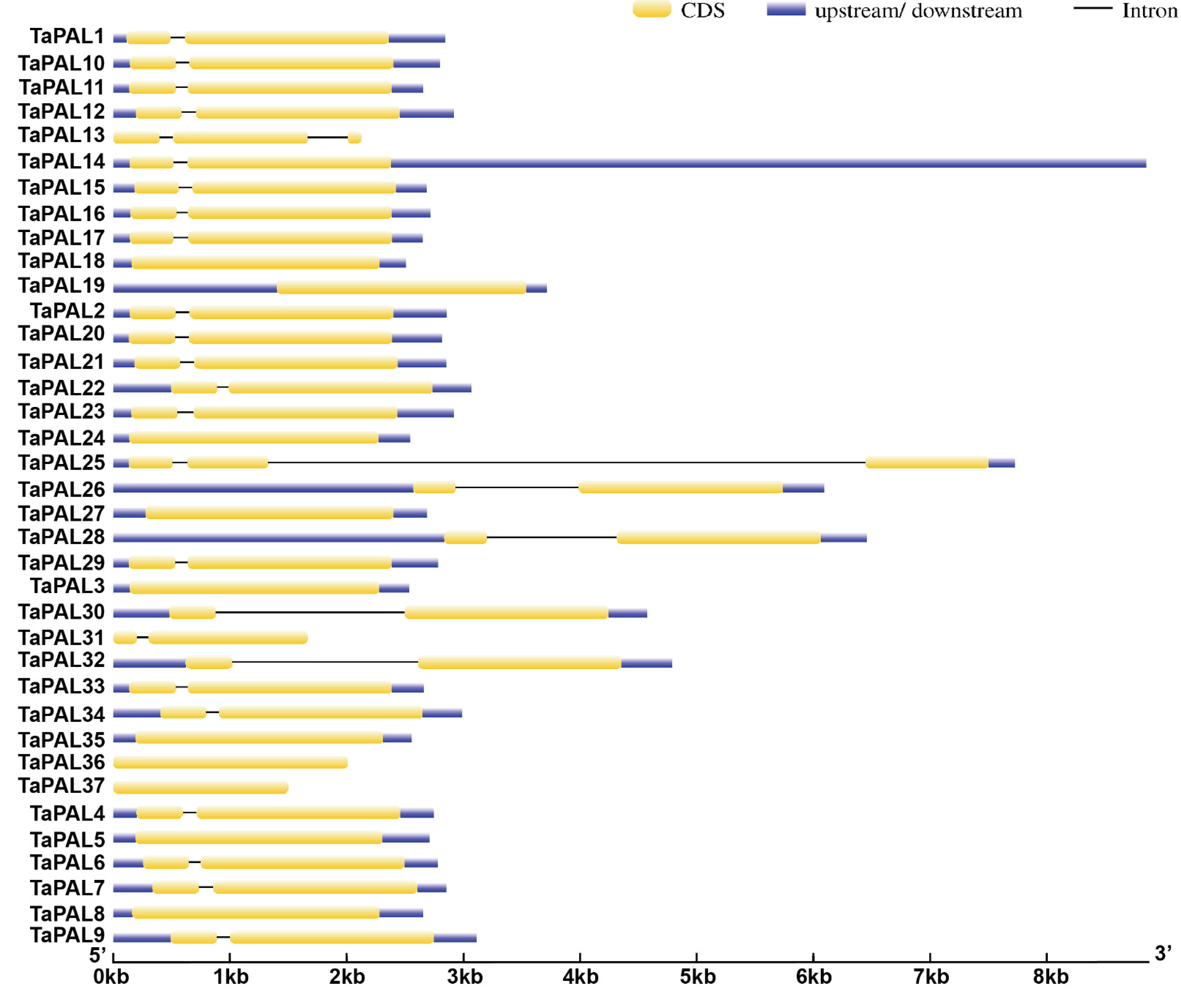
3.4. Gene Structure Analysis
3.5. Gene Ontology of PAL Genes
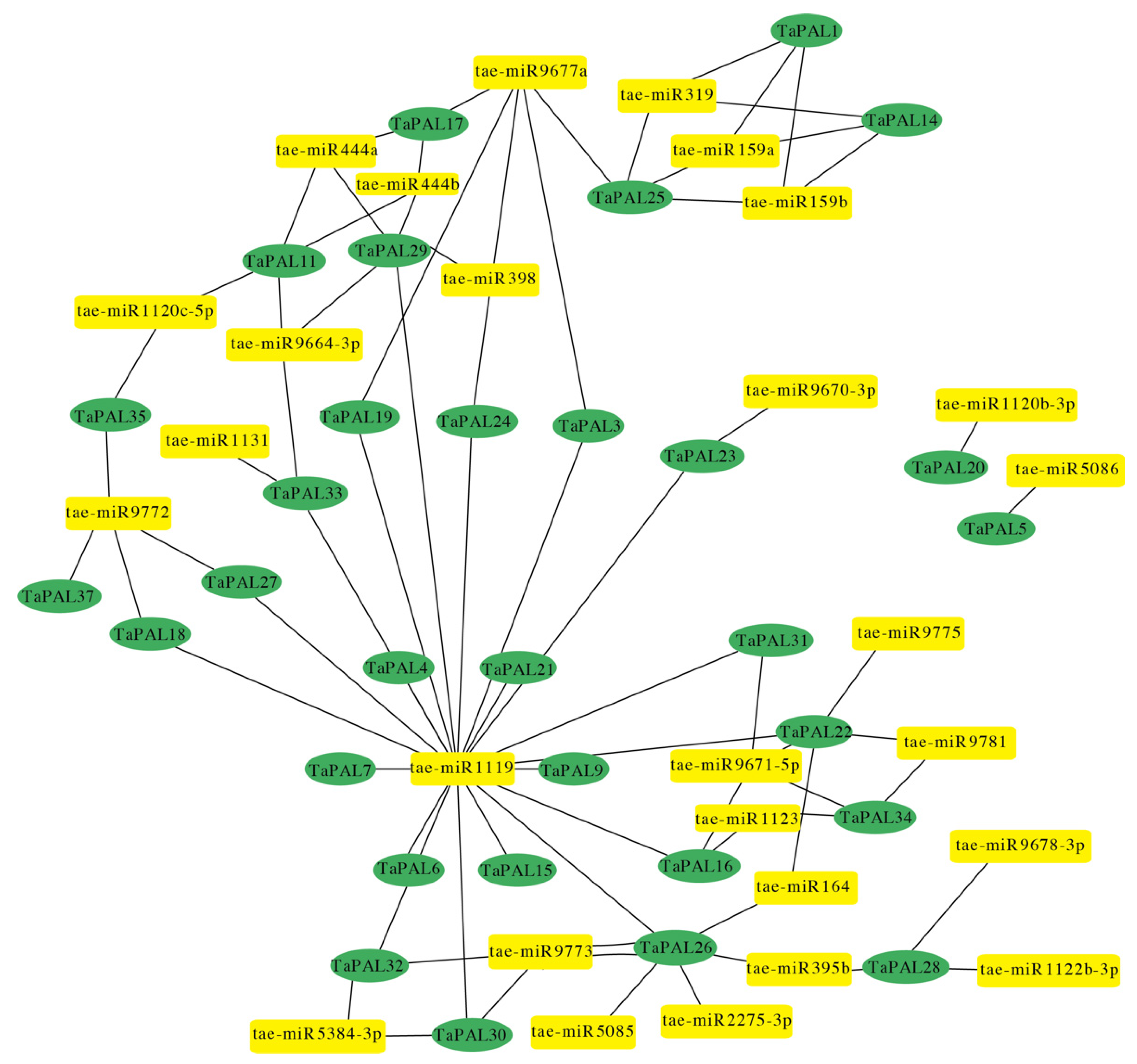
3.6. MicroRNA-Targeting TaPAL Genes
3.7. Promoter Analysis
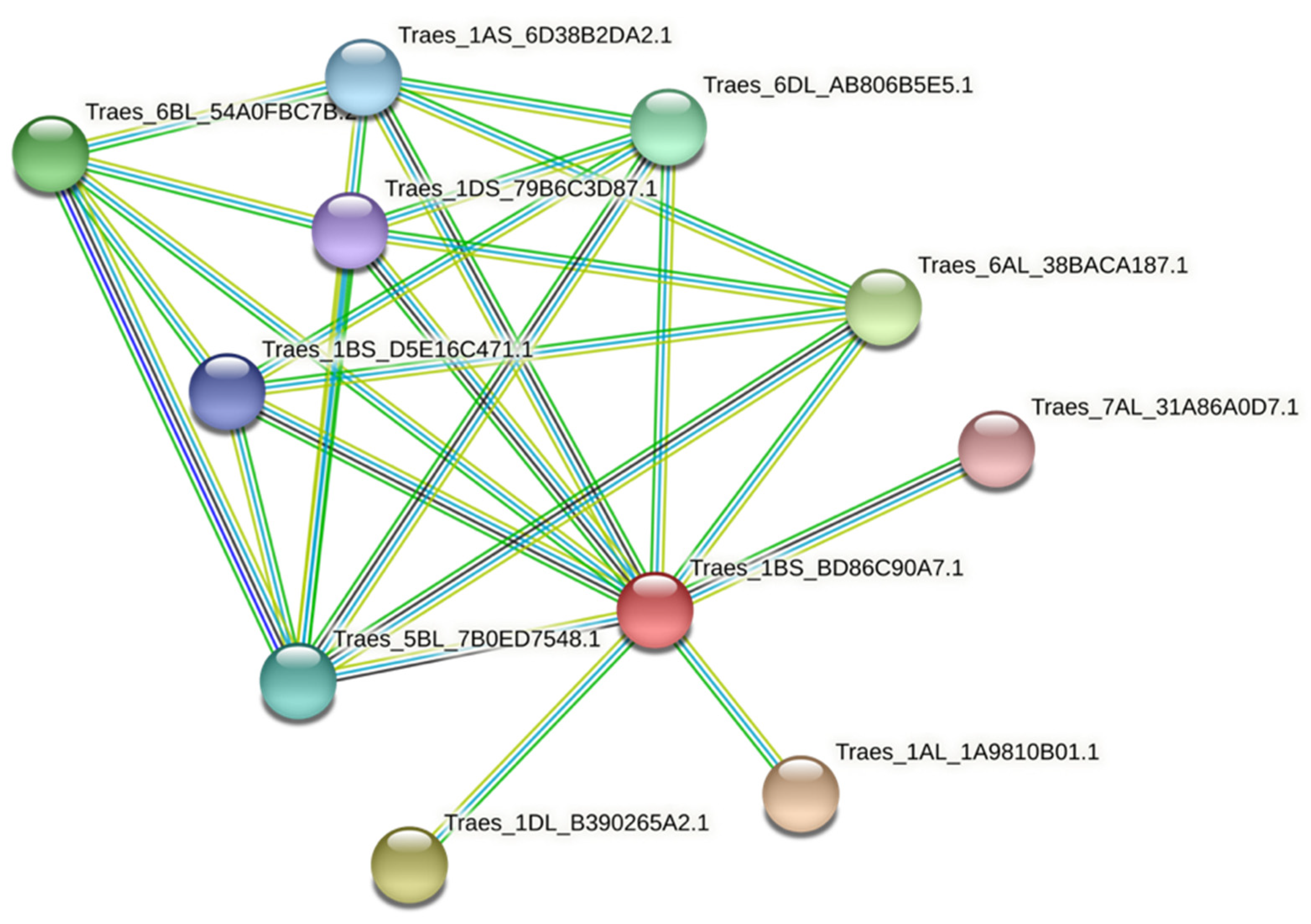
3.8. Protein–Protein Interaction of TaPAL
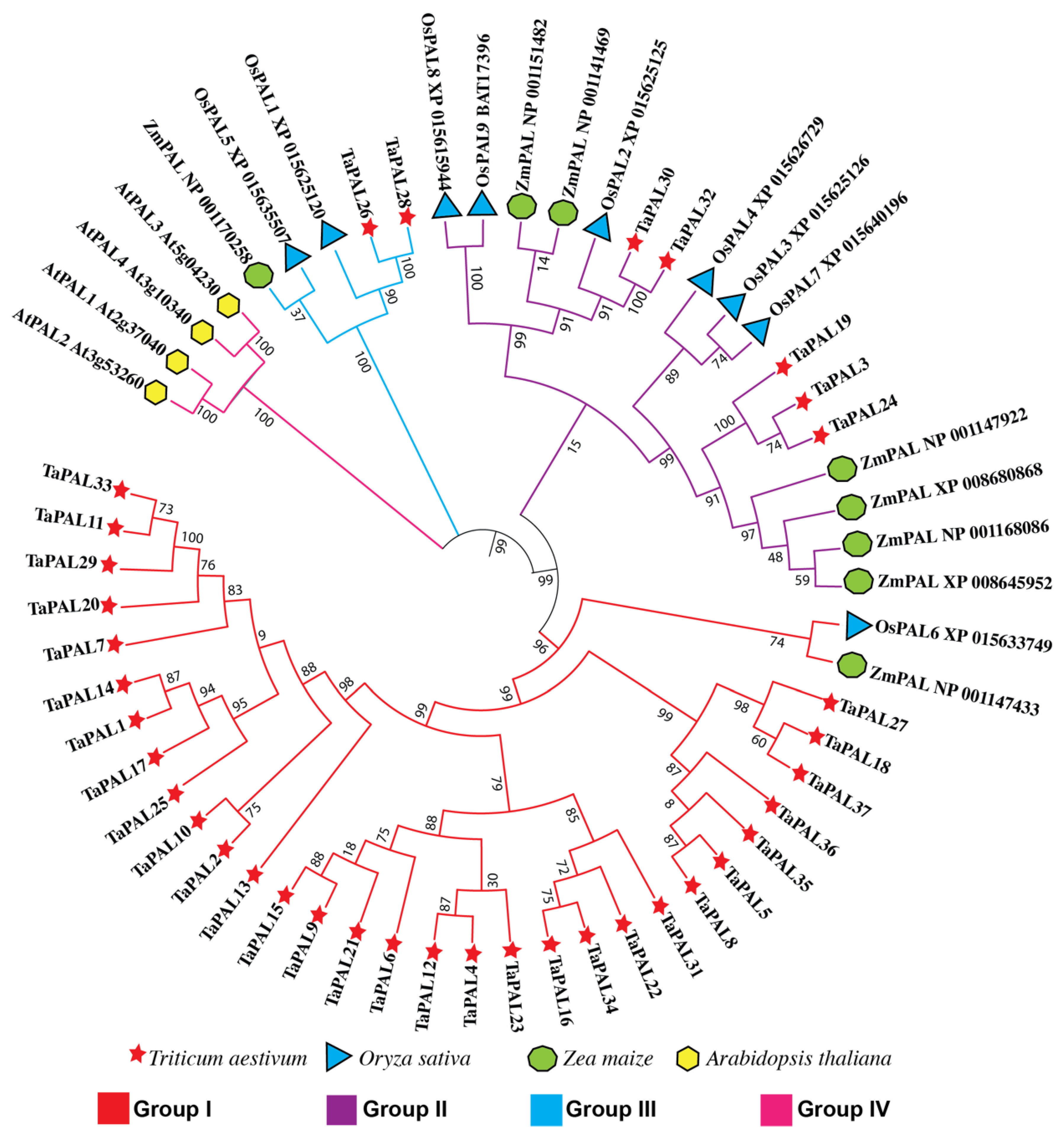
3.9. Phylogenetic Analysis of the PAL Gene Family
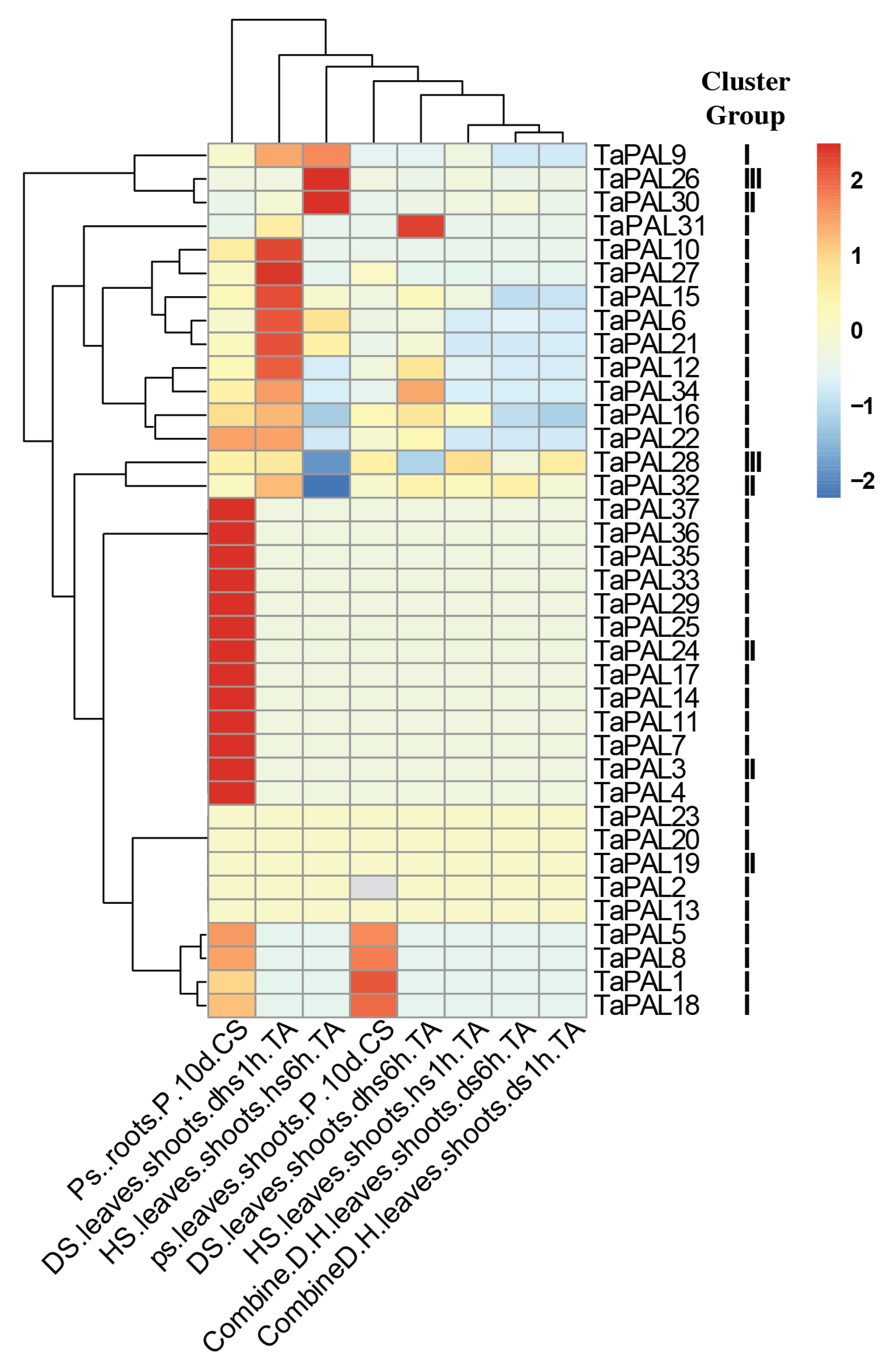
3.10. In Silico Expression Profile Analysis of PAL Gene Family in Six Genotypes of Wheat
3.11. Expression of PAL Gene Family under Abiotic Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huang, J.; Gu, M.; Lai, Z.; Fan, B.; Shi, K.; Zhou, Y.-H.; Yu, J.-Q.; Chen, Z. Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol. 2010, 153, 1526–1538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, P.; Guo, Q.-q.; Qin, Z.-w. The fungicide propamocarb increases lignin by activating the phenylpropanoid pathway in Cucumis sativus L. Hortic. Environ. Biotechnol. 2016, 57, 511–518. [Google Scholar] [CrossRef]
- Raza, A.; Su, W.; Hussain, M.A.; Mehmood, S.S.; Zhang, X.; Cheng, Y.; Zou, X.; Lv, Y. Integrated Analysis of Metabolome and Transcriptome Reveals Insights for Cold Tolerance in Rapeseed (Brassica napus L.). Front. Plant Sci. 2021, 12, 1796. [Google Scholar] [CrossRef]
- Schwede, T.F.; Rétey, J.; Schulz, G.E. Crystal structure of histidine ammonia-lyase revealing a novel polypeptide modification as the catalytic electrophile. Biochemistry 1999, 38, 5355–5361. [Google Scholar] [CrossRef]
- Vogt, T. Phenylpropanoid biosynthesis. Mol. Plant 2010, 3, 2–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cochrane, F.C.; Davin, L.B.; Lewis, N.G. The Arabidopsis phenylalanine ammonia lyase gene family: Kinetic characterization of the four PAL isoforms. Phytochemistry 2004, 65, 1557–1564. [Google Scholar] [CrossRef]
- Pascual, M.B.; El-Azaz, J.; de la Torre, F.N.; Cañas, R.A.; Avila, C.; Cánovas, F.M. Biosynthesis and metabolic fate of phenylalanine in conifers. Front. Plant Sci. 2016, 7, 1030. [Google Scholar] [CrossRef] [Green Version]
- Lei, L.; Zhou, S.-L.; Ma, H.; Zhang, L.-S. Expansion and diversification of the SET domain gene family following whole-genome duplications in Populus trichocarpa. BMC Evol. Biol. 2012, 12, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Dixon, R.A.; Paiva, N.L. Stress-induced phenylpropanoid metabolism. Plant Cell 1995, 7, 1085. [Google Scholar] [CrossRef]
- La Camera, S.; Gouzerh, G.; Dhondt, S.; Hoffmann, L.; Fritig, B.; Legrand, M.; Heitz, T. Metabolic reprogramming in plant innate immunity: The contributions of phenylpropanoid and oxylipin pathways. Immunol. Rev. 2004, 198, 267–284. [Google Scholar] [CrossRef]
- Kim, D.S.; Hwang, B.K. An important role of the pepper phenylalanine ammonia-lyase gene (PAL1) in salicylic acid-dependent signalling of the defence response to microbial pathogens. J. Exp. Bot. 2014, 65, 2295–2306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ritter, H.; Schulz, G.E. Structural basis for the entrance into the phenylpropanoid metabolism catalyzed by phenylalanine ammonia-lyase. Plant Cell 2004, 16, 3426–3436. [Google Scholar] [CrossRef]
- Purwar, S.; Sundaram, S.; Sinha, S.; Gupta, A.; Dobriyall, N.; Kumar, A. Expression and in silico characterization of Phenylalanine ammonium lyase against karnal bunt (Tilletia indica) in wheat (Triticum aestivum). Bioinformation 2013, 9, 1013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rawal, H.; Singh, N.; Sharma, T. Conservation, divergence, and genome-wide distribution of PAL and POX A gene families in plants. Int. J. Genom. 2013, 2013, 678969. [Google Scholar]
- MacDonald, M.J.; D’Cunha, G.B. A modern view of phenylalanine ammonia lyase. Biochem. Cell Biol. 2007, 85, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Gui, S.; Wang, S.; Ding, Y. Molecular evolution and functional characterisation of an ancient phenylalanine ammonia-lyase gene (NnPAL1) from Nelumbo nucifera: Novel insight into the evolution of the PAL family in angiosperms. BMC Evol. Biol. 2014, 14, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Chaw, S.-M.; Zharkikh, A.; Sung, H.-M.; Lau, T.-C.; Li, W.-H. Molecular phylogeny of extant gymnosperms and seed plant evolution: Analysis of nuclear 18S rRNA sequences. Mol. Biol. Evol. 1997, 14, 56–68. [Google Scholar] [CrossRef]
- de Jong, F.; Hanley, S.J.; Beale, M.H.; Karp, A. Characterisation of the willow phenylalanine ammonia-lyase (PAL) gene family reveals expression differences compared with poplar. Phytochemistry 2015, 117, 90–97. [Google Scholar] [CrossRef] [Green Version]
- Wanner, L.A.; Li, G.; Ware, D.; Somssich, I.E.; Davis, K.R. The phenylalanine ammonia-lyase gene family in Arabidopsis thaliana. Plant Mol. Biol. 1995, 27, 327–338. [Google Scholar] [CrossRef]
- Raes, J.; Rohde, A.; Christensen, J.H.; Van de Peer, Y.; Boerjan, W. Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol. 2003, 133, 1051–1071. [Google Scholar] [CrossRef] [Green Version]
- Shi, R.; Sun, Y.-H.; Li, Q.; Heber, S.; Sederoff, R.; Chiang, V.L. Towards a systems approach for lignin biosynthesis in Populus trichocarpa: Transcript abundance and specificity of the monolignol biosynthetic genes. Plant Cell Physiol. 2010, 51, 144–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, H.; Park, N.I.; Li, X.; Kim, Y.K.; Lee, S.Y.; Park, S.U. Molecular cloning and characterization of phenylalanine ammonia-lyase, cinnamate 4-hydroxylase and genes involved in flavone biosynthesis in Scutellaria baicalensis. Bioresour. Technol. 2010, 101, 9715–9722. [Google Scholar] [CrossRef] [PubMed]
- Lepelley, M.; Mahesh, V.; McCarthy, J.; Rigoreau, M.; Crouzillat, D.; Chabrillange, N.; de Kochko, A.; Campa, C. Characterization, high-resolution mapping and differential expression of three homologous PAL genes in Coffea canephora Pierre (Rubiaceae). Planta 2012, 236, 313–326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bagal, U.R.; Leebens-Mack, J.H.; Lorenz, W.W.; Dean, J.F. The phenylalanine ammonia lyase (PAL) gene family shows a gymnosperm-specific lineage. In Proceedings of the BMC Genomics; Springer: Berlin/Heidelberg, Germany, 2012; pp. 1–9. [Google Scholar]
- Dong, C.-j.; Ning, C.; ZHANG, Z.-g.; SHANG, Q.-m. Phenylalanine ammonia-lyase gene families in cucurbit species: Structure, evolution, and expression. J. Integr. Agric. 2016, 15, 1239–1255. [Google Scholar] [CrossRef] [Green Version]
- Shang, Q.-M.; Li, L.; Dong, C.-J. Multiple tandem duplication of the phenylalanine ammonia-lyase genes in Cucumis sativus L. Planta 2012, 236, 1093–1105. [Google Scholar] [CrossRef]
- Souri, Z.; Karimi, N.; Sandalio, L.M. Arsenic hyperaccumulation strategies: An overview. Front. Cell Dev. Biol. 2017, 5, 67. [Google Scholar] [CrossRef] [Green Version]
- Bolser, D.; Staines, D.M.; Pritchard, E.; Kersey, P. Ensembl plants: Integrating tools for visualizing, mining, and analyzing plant genomics data. In Plant Bioinformatics; Springer: Berlin/Heidelberg, Germany, 2016; pp. 115–140. [Google Scholar]
- Madden, T. The BLAST sequence analysis tool. In NCBI Handbook, 2nd ed.; National Center for Biotechnology Information (US): Bethesda, MD, USA, 2013; Volume 2, pp. 425–436. [Google Scholar]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [Green Version]
- Bateman, A.; Coin, L.; Durbin, R.; Finn, R.D.; Hollich, V.; Griffiths-Jones, S.; Khanna, A.; Marshall, M.; Moxon, S.; Sonnhammer, E.L. The Pfam protein families database. Nucleic Acids Res. 2004, 32, D138–D141. [Google Scholar] [CrossRef]
- Schultz, J.; Copley, R.R.; Doerks, T.; Ponting, C.P.; Bork, P. SMART: A web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 2000, 28, 231–234. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [Green Version]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Consortium, U. UniProt: A hub for protein information. Nucleic Acids Res. 2015, 43, D204–D212. [Google Scholar] [CrossRef] [PubMed]
- Okonechnikov, K.; Golosova, O.; Fursov, M.; Team, U. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blum, M.; Chang, H.-Y.; Chuguransky, S.; Grego, T.; Kandasaamy, S.; Mitchell, A.; Nuka, G.; Paysan-Lafosse, T.; Qureshi, M.; Raj, S. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 2021, 49, D344–D354. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Tamura, K.; Nei, M. MEGA: Molecular evolutionary genetics analysis software for microcomputers. Bioinformatics 1994, 10, 189–191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-h.; Jin, H.; Marler, B.; Guo, H. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.; Chen, H.; He, Y.; Xia, R. TBtools, a toolkit for biologists integrating various biological data handling tools with a user-friendly interface. BioRxiv 2018, 289660. [Google Scholar] [CrossRef]
- Bektas, Y.; Eulgem, T. Synthetic plant defense elicitors. Front. Plant Sci. 2015, 5, 804. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Li, H.; Zhao, P. Genome-Wide Identification and transcriptional expression of the PAL Gene family in common Walnut (Juglans regia L.). Genes 2019, 10, 46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, X.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server. Nucleic Acids Res. 2011, 39, W155–W159. [Google Scholar] [CrossRef] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Higo, K.; Ugawa, Y.; Iwamoto, M.; Korenaga, T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999, 27, 297–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g: Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mering, C.v.; Huynen, M.; Jaeggi, D.; Schmidt, S.; Bork, P.; Snel, B. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 2003, 31, 258–261. [Google Scholar] [CrossRef]
- Iquebal, M.A.; Sharma, P.; Jasrotia, R.S.; Jaiswal, S.; Kaur, A.; Saroha, M.; Angadi, U.; Sheoran, S.; Singh, R.; Singh, G. RNAseq analysis reveals drought-responsive molecular pathways with candidate genes and putative molecular markers in root tissue of wheat. Sci. Rep. 2019, 9, 1–18. [Google Scholar]
- Zou, C.; Wang, P.; Xu, Y. Bulked sample analysis in genetics, genomics and crop improvement. Plant Biotechnol. J. 2016, 14, 1941–1955. [Google Scholar] [CrossRef] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [Green Version]
- Tarazona, S.; Furió-Tarı, P.; Ferrer, A.; Conesa, A. NOISeq: Differential Expression in RNA-Seq—Bioconductor; Department of Statistics: TU Dortmund, Germany, 2013. [Google Scholar]
- Borrill, P.; Ramirez-Gonzalez, R.; Uauy, C. expVIP: A customizable RNA-seq data analysis and visualization platform. Plant Physiol. 2016, 170, 2172–2186. [Google Scholar] [CrossRef] [Green Version]
- Kolde, R.; Kolde, M.R. Package ‘pheatmap’. R Package 2015, 1, 790. [Google Scholar]
- Hossain, M.S.; Rasel Ahmed, M.; Ullah, W.; Honi, U.; Tareq, M.Z.; Sarker, M.S.A.; Ahmed, B.; Islam, M.S. Phenylalanine ammonia-lyase gene family (PAL): Genome wide characterization and transcriptional expression in jute (Corchorus olitorius). J. Biosci. Agric. Res. 2020, 26, 2185–2191. [Google Scholar] [CrossRef]
- Darzentas, N. Circoletto: Visualizing sequence similarity with Circos. Bioinformatics 2010, 26, 2620–2621. [Google Scholar] [CrossRef] [PubMed]
- Jin, Q.; Yao, Y.; Cai, Y.; Lin, Y. Molecular cloning and sequence analysis of a phenylalanine ammonia-lyase gene from Dendrobium. PLoS ONE 2013, 8, e62352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, G.; Wang, H.; Cheng, X.; Su, X.; Zhao, Y.; Jiang, T.; Jin, Q.; Lin, Y.; Cai, Y. Comparative genomic analysis of the PAL genes in five Rosaceae species and functional identification of Chinese white pear. PeerJ 2019, 7, e8064. [Google Scholar] [CrossRef] [Green Version]
- Dong, C.-J.; Shang, Q.-M. Genome-wide characterization of phenylalanine ammonia-lyase gene family in watermelon (Citrullus lanatus). Planta 2013, 238, 35–49. [Google Scholar] [CrossRef]
- Rushton, P.J.; Reinstadler, A.; Lipka, V.; Lippok, B.; Somssich, I.E. Synthetic plant promoters containing defined regulatory elements provide novel insights into pathogen-and wound-induced signaling. Plant Cell 2002, 14, 749–762. [Google Scholar] [CrossRef] [Green Version]
- Kaur, A.; Pati, P.K.; Pati, A.M.; Nagpal, A.K. In-silico analysis of cis-acting regulatory elements of pathogenesis-related proteins of Arabidopsis thaliana and Oryza sativa. PLoS ONE 2017, 12, e0184523. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Dong, T.; Li, J.; Wang, M. Molecular cloning, expression, and subcellular localization of a PAL gene from Citrus reticulata under iron deficiency. Biol. Plant. 2016, 60, 482–488. [Google Scholar] [CrossRef] [Green Version]
- Kong, W.; Ding, L.; Cheng, J.; Wang, B. Identification and expression analysis of genes with pathogen-inducible cis-regulatory elements in the promoter regions in Oryza sativa. Rice 2018, 11, 1–12. [Google Scholar] [CrossRef]
- Ren, W.; Wang, Y.; Xu, A.; Zhao, Y. Genome-wide identification and characterization of the Phenylalanine Ammonia-lyase (PAL) gene family in Medicago truncatula. Legume Res. Int. J. 2019, 42, 461–466. [Google Scholar] [CrossRef]
- Yuan, W.; Jiang, T.; Du, K.; Chen, H.; Cao, Y.; Xie, J.; Li, M.; Carr, J.P.; Wu, B.; Fan, Z. Maize phenylalanine ammonia-lyases contribute to resistance to Sugarcane mosaic virus infection, most likely through positive regulation of salicylic acid accumulation. Mol. Plant Pathol. 2019, 20, 1365–1378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukasawa-Akada, T.; Kung, S.-d.; Watson, J.C. Phenylalanine ammonia-lyase gene structure, expression, and evolution in Nicotiana. Plant Mol. Biol. 1996, 30, 711–722. [Google Scholar] [CrossRef]
- Han, H.; Woeste, K.E.; Hu, Y.; Dang, M.; Zhang, T.; Gao, X.-X.; Zhou, H.; Feng, X.; Zhao, G.; Zhao, P. Genetic diversity and population structure of common walnut (Juglans regia) in China based on EST-SSRs and the nuclear gene phenylalanine ammonia-lyase (PAL). Tree Genet. Genomes 2016, 12, 1–12. [Google Scholar] [CrossRef]
- Cheng, X.; Wang, S.; Xu, D.; Liu, X.; Li, X.; Xiao, W.; Cao, J.; Jiang, H.; Min, X.; Wang, J. Identification and analysis of the GASR gene family in common wheat (Triticum aestivum L.) and characterization of TaGASR34, a gene associated with seed dormancy and germination. Front. Genet. 2019, 10, 980. [Google Scholar] [CrossRef] [PubMed]
- Reichert, A.I.; He, X.-Z.; Dixon, R.A. Phenylalanine ammonia-lyase (PAL) from tobacco (Nicotiana tabacum): Characterization of the four tobacco PAL genes and active heterotetrameric enzymes. Biochem. J. 2009, 424, 233–242. [Google Scholar] [CrossRef] [Green Version]
- Hamberger, B.; Ellis, M.; Friedmann, M.; de Azevedo Souza, C.; Barbazuk, B.; Douglas, C.J. Genome-wide analyses of phenylpropanoid-related genes in Populus trichocarpa, Arabidopsis thaliana, and Oryza sativa: The Populus lignin toolbox and conservation and diversification of angiosperm gene families. Botany 2007, 85, 1182–1201. [Google Scholar]
- Kaur, H.; Salh, P.; Singh, B. Role of defense enzymes and phenolics in resistance of wheat crop (Triticum aestivum L.) towards aphid complex. J. Plant Interact. 2017, 12, 304–311. [Google Scholar] [CrossRef] [Green Version]
- Kumar, A.; Batra, R.; Gahlaut, V.; Gautam, T.; Kumar, S.; Sharma, M.; Tyagi, S.; Singh, K.P.; Balyan, H.S.; Pandey, R. Genome-wide identification and characterization of gene family for RWP-RK transcription factors in wheat (Triticum aestivum L.). PLoS ONE 2018, 13, e0208409. [Google Scholar] [CrossRef] [Green Version]
- Chang, A.; Lim, M.-H.; Lee, S.-W.; Robb, E.J.; Nazar, R.N. Tomato phenylalanine ammonia-lyase gene family, highly redundant but strongly underutilized. J. Biol. Chem. 2008, 283, 33591–33601. [Google Scholar] [CrossRef] [Green Version]
- Akdogan, G.; Tufekci, E.D.; Uranbey, S.; Unver, T. miRNA-based drought regulation in wheat. Funct. Integr. Genom. 2016, 16, 221–233. [Google Scholar] [CrossRef]
- Feng, H.; Xu, M.; Gao, Y.; Liang, J.; Guo, F.; Guo, Y.; Huang, L. Vm-milR37 contributes to pathogenicity by regulating glutathione peroxidase gene VmGP in Valsa mali. Mol. Plant Pathol. 2021, 22, 243–254. [Google Scholar] [CrossRef]
- Shan, T.; Fu, R.; Xie, Y.; Chen, Q.; Wang, Y.; Li, Z.; Song, X.; Li, P.; Wang, B. Regulatory mechanism of maize (Zea mays L.) miR164 in salt stress response. Russ. J. Genet. 2020, 56, 835–842. [Google Scholar] [CrossRef]
- Yamada, T.; Matsuda, F.; Kasai, K.; Fukuoka, S.; Kitamura, K.; Tozawa, Y.; Miyagawa, H.; Wakasa, K. Mutation of a rice gene encoding a phenylalanine biosynthetic enzyme results in accumulation of phenylalanine and tryptophan. Plant Cell 2008, 20, 1316–1329. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Wang, W.; Li, Y.; Dai, X.; Ma, G.; Xing, D.; Zhu, M.; Gao, L.; Xia, T. Six phenylalanine ammonia-lyases from Camellia sinensis: Evolution, expression, and kinetics. Plant Physiol. Biochem. 2017, 118, 413–421. [Google Scholar] [CrossRef] [PubMed]
- Wada, K.C.; Mizuuchi, K.; Koshio, A.; Kaneko, K.; Mitsui, T.; Takeno, K. Stress enhances the gene expression and enzyme activity of phenylalanine ammonia-lyase and the endogenous content of salicylic acid to induce flowering in pharbitis. J. Plant Physiol. 2014, 171, 895–902. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-B.; Chen, X.; Wang, W.; Cheng, K.-D.; Kong, J.-Q. Transcriptome-wide identification and characterization of Ornithogalum saundersiae phenylalanine ammonia lyase gene family. RSC Adv. 2014, 4, 27159–27175. [Google Scholar] [CrossRef]
- Cao, Y.; Meng, D.; Abdullah, M.; Jin, Q.; Lin, Y.; Cai, Y. Genome wide identification, evolutionary, and expression analysis of VQ genes from two Pyrus species. Genes 2018, 9, 224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tufail, M.A.; Touceda-González, M.; Pertot, I.; Ehlers, R.-U. Gluconacetobacter diazotrophicus PAL5 Enhances Plant Robustness Status under the Combination of Moderate Drought and Low Nitrogen Stress in Zea mays L. Microorganisms 2021, 9, 870. [Google Scholar] [CrossRef]
- Christopoulos, M.V.; Tsantili, E. Participation of phenylalanine ammonia-lyase (PAL) in increased phenolic compounds in fresh cold stressed walnut (Juglans regia L.) kernels. Postharvest Biol. Technol. 2015, 104, 17–25. [Google Scholar] [CrossRef]
- Kervinen, T.; Peltonen, S.; Utriainen, M.; Kangasjärvi, J.; Teeri, T.H.; Karjalainen, R. Cloning and characterization of cDNA clones encoding phenylalanine ammonia-lyase in barley. Plant Sci. 1997, 123, 143–150. [Google Scholar] [CrossRef]
- Joos, H.J.; Hahlbrock, K. Phenylalanine ammonia-lyase in potato (Solanum tuberosum L.) Genomic complexity, structural comparison of two selected genes and modes of expression. Eur. J. Biochem. 1992, 204, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Deng, G.; Cheng, S.; Zhang, W.; Huang, X.; Li, L.; Cheng, H.; Rong, X.; Li, J. Molecular cloning, characterization and expression of the phenylalanine ammonia-lyase gene from Juglans regia. Molecules 2012, 17, 7810–7823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dixon, R.A.; Maxwell, C.A.; Ni, W.; Oommen, A.; Paiva, N.L. Genetic manipulation of lignin and phenylpropanoid compounds involved in interactions with microorganisms. In Genetic Engineering of Plant Secondary Metabolism; Springer: Berlin/Heidelberg, Germany, 1994; pp. 153–178. [Google Scholar]
- Pearce, S.; Vazquez-Gross, H.; Herin, S.Y.; Hane, D.; Wang, Y.; Gu, Y.Q.; Dubcovsky, J. WheatExp: An RNA-seq expression database for polyploid wheat. BMC Plant Biol. 2015, 15, 299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, J.; Ding, P.; Qin, P.; Liu, Y.-X.; Xie, Q.; Chen, G.; Li, W.; Jiang, Q.; Chen, G.; Lan, X.-J. Structure and expression of the TaGW7 in bread wheat (Triticum aestivum L.). Plant Growth Regul. 2017, 82, 281. [Google Scholar] [CrossRef]



| Genotypes | Characteristics |
|---|---|
| Batis | Winter wheat (drought-susceptible) from Germany (INRES) |
| Blue Silver | Spring wheat (drought-susceptible) was released in Pakistan in 1971 by the NARC |
| Chakwal-50 | Released in 2008 as drought-tolerant, high-yielding, and disease-resistant wheat grown in rain-fed areas of Pakistan by BARI |
| Local White | Drought-tolerant land race, grown in dryer areas of Pakistan (NARC) |
| Syn-22 | Synthetic wheat (drought-tolerant) from The Netherlands (INRES) |
| UZ-11-CWA-8 | Drought-tolerant line collected from Uzbekistan (INRES) |
| Sr.# | Gene ID | Name | Subcellular Location | AA | MW (g/mol) | IP | Charge | Chrom | Gene Position Start–End (bp) | St |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | TraesCS1B02G048400 | TaPAL1 | Cytoplasm | 707 | 76,625.34 | 6.2111 | −3.5 | 1B | 28,442,914–28,445,758 | R |
| 2 | TraesCS1B02G048300 | TaPAL2 | Cytoplasm | 713 | 77,090.98 | 6.0532 | −5.0 | 1B | 28,373,087–28,375,944 | R |
| 3 | TraesCS6B02G258600 | TaPAL3 | Cytoplasm | 712 | 76,561.87 | 6.2119 | −3.0 | 6B | 465,682,950–465,685,480 | R |
| 4 | TraesCS2A02G381000 | TaPAL4 | Cytoplasm | 713 | 77,163.92 | 5.9516 | −6.0 | 2A | 624,446,955–624,449,699 | F |
| 5 | TraesCS2D02G204700 | TaPAL5 | Cytoplasm | 707 | 76,132.96 | 6.2107 | −3.0 | 2D | 156,707,284–156,709,989 | F |
| 6 | TraesCS2B02G398400 | TaPAL6 | Cytoplasm | 713 | 77,233.31 | 6.5058 | 0 | 2B | 565,209,774–565,212,553 | F |
| 7 | TraesCS1B02G048100 | TaPAL7 | Cytoplasm | 714 | 77,341.05 | 5.9303 | −6.5 | 1B | 28,305,997–28,308,847 | R |
| 8 | TraesCS2B02G224300 | TaPAL8 | Cytoplasm | 706 | 76,220.04 | 6.3114 | −2.0 | 2B | 214,312,779–214,315,434 | F |
| 9 | TraesCS2D02G377500 | TaPAL9 | Cytoplasm | 713 | 77,188.05 | 5.9739 | −5.5 | 2D | 481,960,998–481,964,109 | F |
| 10 | TraesCS4A02G401300 | TaPAL10 | Cytoplasm | 713 | 77,087.87 | 6.1487 | −4.0 | 4A | 675,283,030–675,285,827 | R |
| 11 | TraesCS1D02G039500 | TaPAL11 | Cytoplasm | 714 | 77,180.97 | 6.3467 | −2.0 | 1D | 19,022,361–19,025,014 | F |
| 12 | TraesCS2A02G380900 | TaPAL12 | Cytoplasm | 713 | 77,261.17 | 6.1497 | −4.0 | 2A | 624,394,141–624,397,057 | F |
| 13 | TraesCS5B02G468400 | TaPAL13 | Cytoplasm | 552 | 58,944.16 | 6.5575 | 0.5 | 5B | 641,833,347–641,835,469 | F |
| 14 | TraesCS1D02G039400 | TaPAL14 | Cytoplasm | 707 | 76,681.33 | 5.9509 | −6.0 | 1D | 19,012,091–19,020,939 | R |
| 15 | TraesCS2D02G377600 | TaPAL15 | Cytoplasm | 709 | 76,866.84 | 6.0796 | −4.5 | 2D | 481,988,232–481,990,914 | F |
| 16 | TraesCS2D02G377200 | TaPAL16 | Cytoplasm | 713 | 77,097.96 | 6.2412 | −3.0 | 2D | 481,599,475–481,602,192 | F |
| 17 | TraesCS5B02G468300 | TaPAL17 | Cytoplasm | 707 | 76,488.16 | 6.4644 | −0.5 | 5B | 641,822,078–641,824,730 | F |
| 18 | TraesCS1B02G122800 | TaPAL18 | Cytoplasm | 708 | 76,047.86 | 6.4086 | −1.0 | 1B | 148,413,936–148,416,442 | R |
| 19 | TraesCS6D02G212500 | TaPAL19 | Cytoplasm | 712 | 76,757.91 | 5.9498 | −6.0 | 6D | 300,896,671–300,900,381 | R |
| 20 | TraesCS1B02G048200 | TaPAL20 | Cytoplasm | 714 | 77,250.05 | 6.2977 | −2.5 | 1B | 28,322,038–28,324,849 | R |
| 21 | TraesCS2A02G381100 | TaPAL21 | Cytoplasm | 713 | 77,133.01 | 6.0528 | −5.0 | 2A | 624,458,106–624,460,956 | F |
| 22 | TraesCS2B02G398000 | TaPAL22 | Cytoplasm | 713 | 77,170.99 | 6.1485 | −4.0 | 2B | 564,920,332–564,923,399 | F |
| 23 | TraesCS2B02G398200 | TaPAL23 | Cytoplasm | 713 | 77,216.16 | 6.0265 | −5.5 | 2B | 565,076,277–565,079,194 | F |
| 24 | TraesCS6A02G222700 | TaPAL24 | Cytoplasm | 712 | 76,580.83 | 6.3123 | −2.0 | 6A | 416,648,337–416,650,876 | F |
| 25 | TraesCS1A02G037700 | TaPAL25 | Cytoplasm | 707 | 76,566.32 | 6.0782 | −4.5 | 1A | 20,924,887–20,932,614 | R |
| 26 | TraesCS6D02G212200 | TaPAL26 | Cytoplasm | 704 | 75,619.36 | 6.211 | −3.0 | 6D | 300,326,934–300,333,023 | R |
| 27 | TraesCS1D02G103500 | TaPAL27 | Cytoplasm | 708 | 76,233.04 | 6.211 | −3.0 | 1D | 93,102,756–93,105,441 | R |
| 28 | TraesCS6B02G258400 | TaPAL28 | Cytoplasm | 704 | 75,605.29 | 6.1061 | −4.0 | 6B | 465,148,031–465,154,487 | R |
| 29 | TraesCS1B02G048500 | TaPAL29 | Cytoplasm | 714 | 77,314.11 | 6.096 | −5.0 | 1B | 28,483,566–28,486,349 | F |
| 30 | TraesCS6D02G212400 | TaPAL30 | Cytoplasm | 714 | 77,167.39 | 6.1081 | −4.0 | 6D | 300,888,767–300,893,342 | R |
| 31 | TraesCS2B02G398100 | TaPAL31 | Cytoplasm | 520 | 56,049.19 | 7.5761 | 8.5 | 2B | 565,039,719–565,041,383 | F |
| 32 | TraesCS6B02G258500 | TaPAL32 | Cytoplasm | 714 | 77,108.33 | 6.2111 | −3.0 | 6B | 465,675,009–465,679,798 | R |
| 33 | TraesCS1A02G037800 | TaPAL33 | Cytoplasm | 714 | 77,228.05 | 6.3463 | −2.0 | 1A | 20,935,423–20,938,081 | R |
| 34 | TraesCS2A02G380800 | TaPAL34 | Cytoplasm | 713 | 77,015.89 | 6.2406 | −3.0 | 2A | 624,359,166–624,362,152 | F |
| 35 | TraesCS2B02G224000 | TaPAL35 | Cytoplasm | 707 | 76,207.83 | 5.7689 | −7.0 | 2B | 214,280,956–214,283,508 | F |
| 36 | TraesCS2A02G196400 | TaPAL36 | Cytoplasm | 668 | 71,935.46 | 7.4015 | 7.0 | 2A | 166,254,570–166,256,576 | F |
| 37 | TraesCS1A02G094900 | TaPAL37 | Cytoplasm | 498 | 52,784.16 | 6.7764 | 2.0 | 1A | 90,162,001–90,163,497 | R |
| Site Name | Functions | |
|---|---|---|
| Hormone | ABRE | cis-acting element involved in abscisic acid responsiveness |
| ACE | cis-acting element involved in light responsiveness | |
| CCAAT-box | MYBHv1 binding site | |
| CGTCA-motif | cis-acting regulatory element involved in MeJA-responsiveness | |
| GARE-motif | Gibberellin-responsive element | |
| GC-motif | Enhancer-like element involved in anoxic-specific inducibility | |
| P-box | Gibberellin-responsive element and part of a light-responsive element | |
| TCA-element | cis-acting element involved in salicylic acid responsiveness | |
| TGA-element | Auxin-responsive element | |
| TGACG-motif | cis-acting regulatory element involved in MeJA-responsiveness | |
| Stress and Growth | A-box | cis-acting regulatory element |
| AE-box | Part of a module for light response | |
| Box 4 | Part of a conserved DNA module involved in light responsiveness | |
| ARE | cis-acting regulatory element essential for anaerobic induction | |
| ATC-motif | Part of a conserved DNA module involved in light responsiveness | |
| ATCT-motif | Part of a conserved DNA module involved in light responsiveness | |
| C-box | cis-acting regulatory element involved in light responsiveness | |
| CAAT-box | Common cis-acting element in promoter and enhancer regions | |
| CAG-motif | Part of a light response element | |
| CAT-box | cis-acting regulatory element related to meristem expression | |
| chs-CMA2a | Part of a light responsive element | |
| chs-Unit 1 ml | Part of a light responsive element | |
| Circadian | cis-acting regulatory element involved in circadian control | |
| G-box | cis-acting regulatory element involved in light responsiveness | |
| GATA-motif | Part of a light-responsive element | |
| GCN4-motif | cis-regulatory element involved in endosperm expression | |
| GT1-motif | Light-responsive element | |
| GTGGC-motif | Part of a light-responsive element | |
| I-box | Part of a light-responsive element | |
| LTR | cis-acting element involved in low-temperature responsiveness | |
| MBS | MYB binding site involved in drought-inducibility | |
| O2-site | cis-acting regulatory element involved in zein metabolism regulation | |
| Sp1 | Light-responsive element | |
| TATA-box | Core promoter element around -30 of transcription start | |
| TC-rich repeats | cis-acting element involved in defense and stress responsiveness | |
| TCT-motif | Part of a light responsive element |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rasool, F.; Uzair, M.; Naeem, M.K.; Rehman, N.; Afroz, A.; Shah, H.; Khan, M.R. Phenylalanine Ammonia-Lyase (PAL) Genes Family in Wheat (Triticum aestivum L.): Genome-Wide Characterization and Expression Profiling. Agronomy 2021, 11, 2511. https://doi.org/10.3390/agronomy11122511
Rasool F, Uzair M, Naeem MK, Rehman N, Afroz A, Shah H, Khan MR. Phenylalanine Ammonia-Lyase (PAL) Genes Family in Wheat (Triticum aestivum L.): Genome-Wide Characterization and Expression Profiling. Agronomy. 2021; 11(12):2511. https://doi.org/10.3390/agronomy11122511
Chicago/Turabian StyleRasool, Fatima, Muhammad Uzair, Muhammad Kashif Naeem, Nazia Rehman, Amber Afroz, Hussain Shah, and Muhammad Ramzan Khan. 2021. "Phenylalanine Ammonia-Lyase (PAL) Genes Family in Wheat (Triticum aestivum L.): Genome-Wide Characterization and Expression Profiling" Agronomy 11, no. 12: 2511. https://doi.org/10.3390/agronomy11122511
APA StyleRasool, F., Uzair, M., Naeem, M. K., Rehman, N., Afroz, A., Shah, H., & Khan, M. R. (2021). Phenylalanine Ammonia-Lyase (PAL) Genes Family in Wheat (Triticum aestivum L.): Genome-Wide Characterization and Expression Profiling. Agronomy, 11(12), 2511. https://doi.org/10.3390/agronomy11122511







