Laser Microdissection of Pisum sativum L. Nodules Followed by RNA-Seq Analysis Revealed Crucial Transcriptomic Changes during Infected Cell Differentiation
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material, Inoculation, and Growth Conditions
2.2. Laser Microdissection
2.3. RNA Extraction
2.4. Depletion of Ribosomal RNA and RNA Amplification
2.5. RNA Sequencing and Mapping
2.6. Differential Expression, GO, and KEGG Enrichment Analysis
3. Results
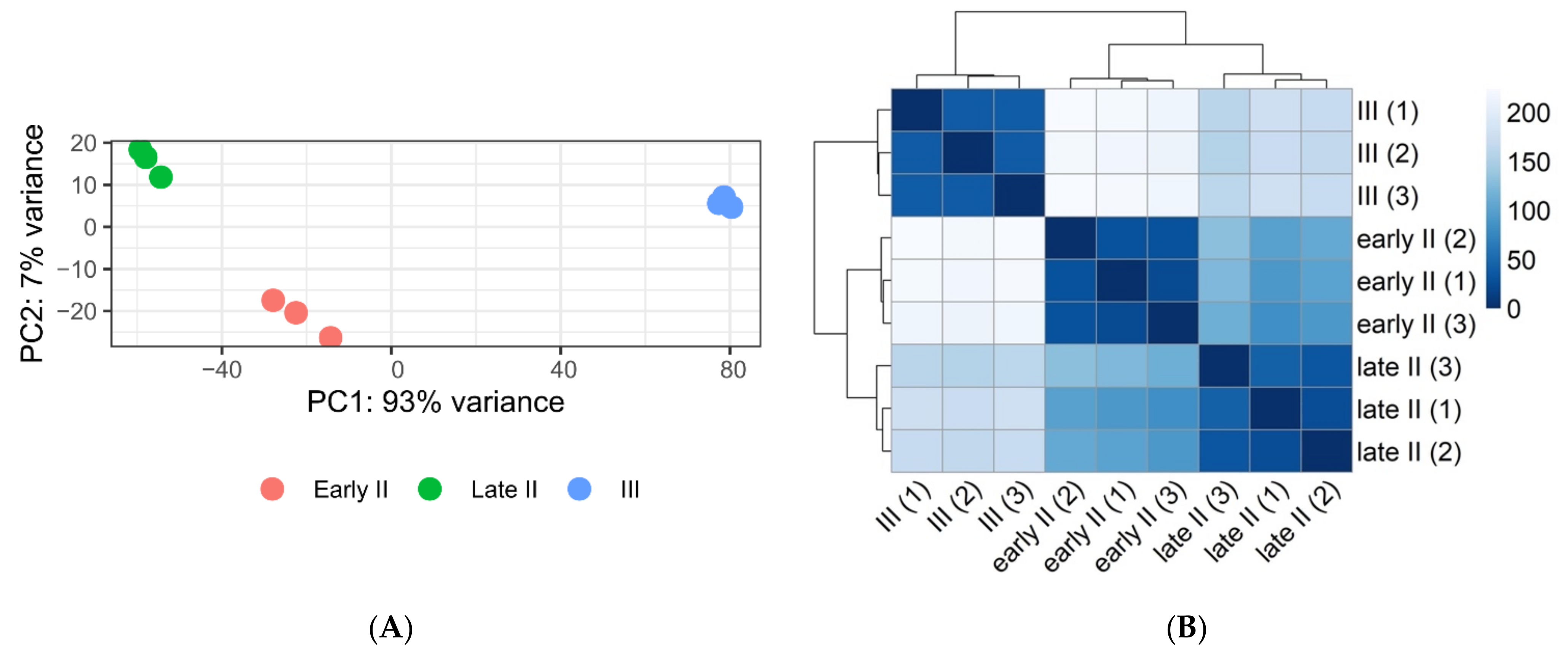
3.1. Laser Microdissection and RNA Sequencing
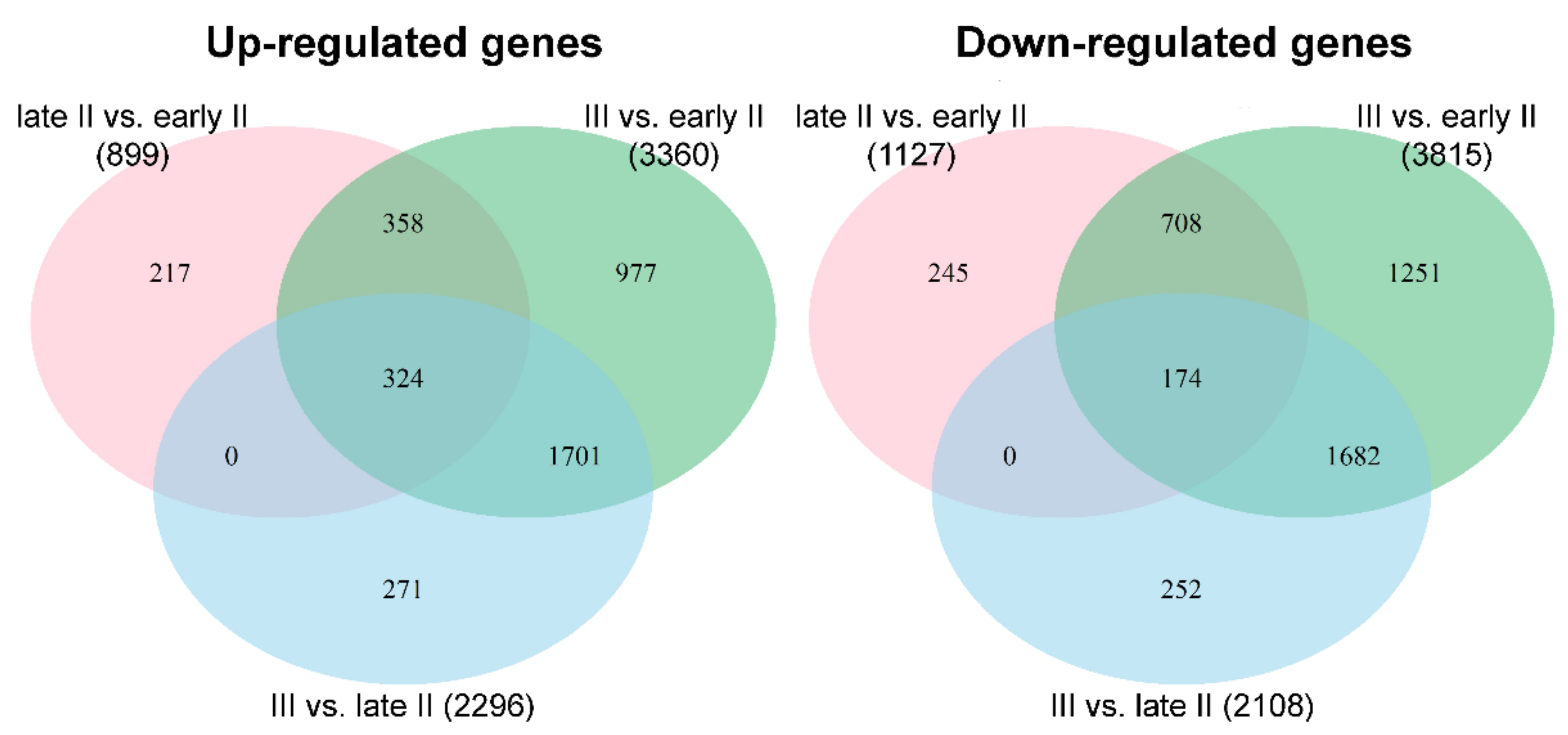
3.2. Analysis of Differentially Expressed Genes
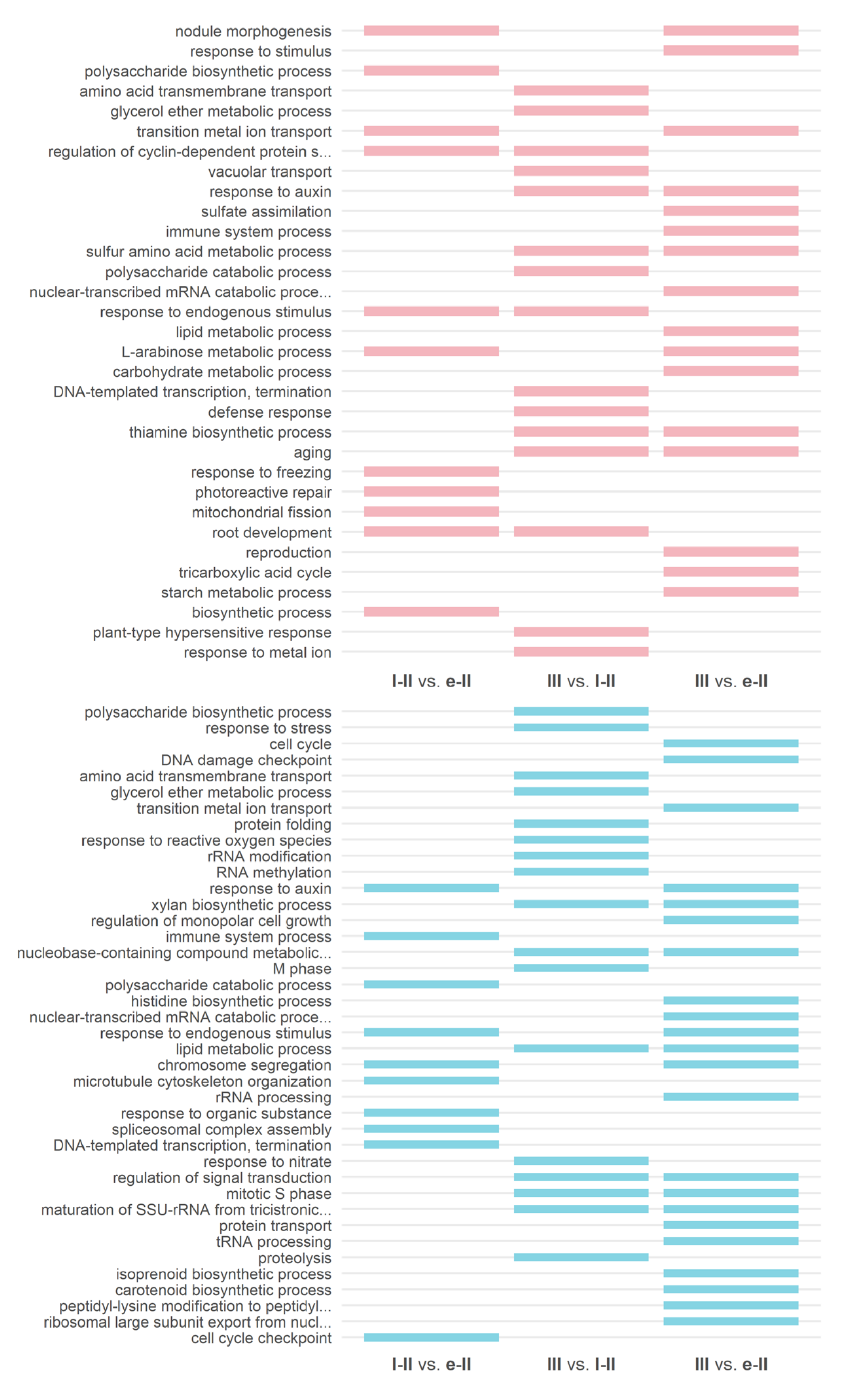
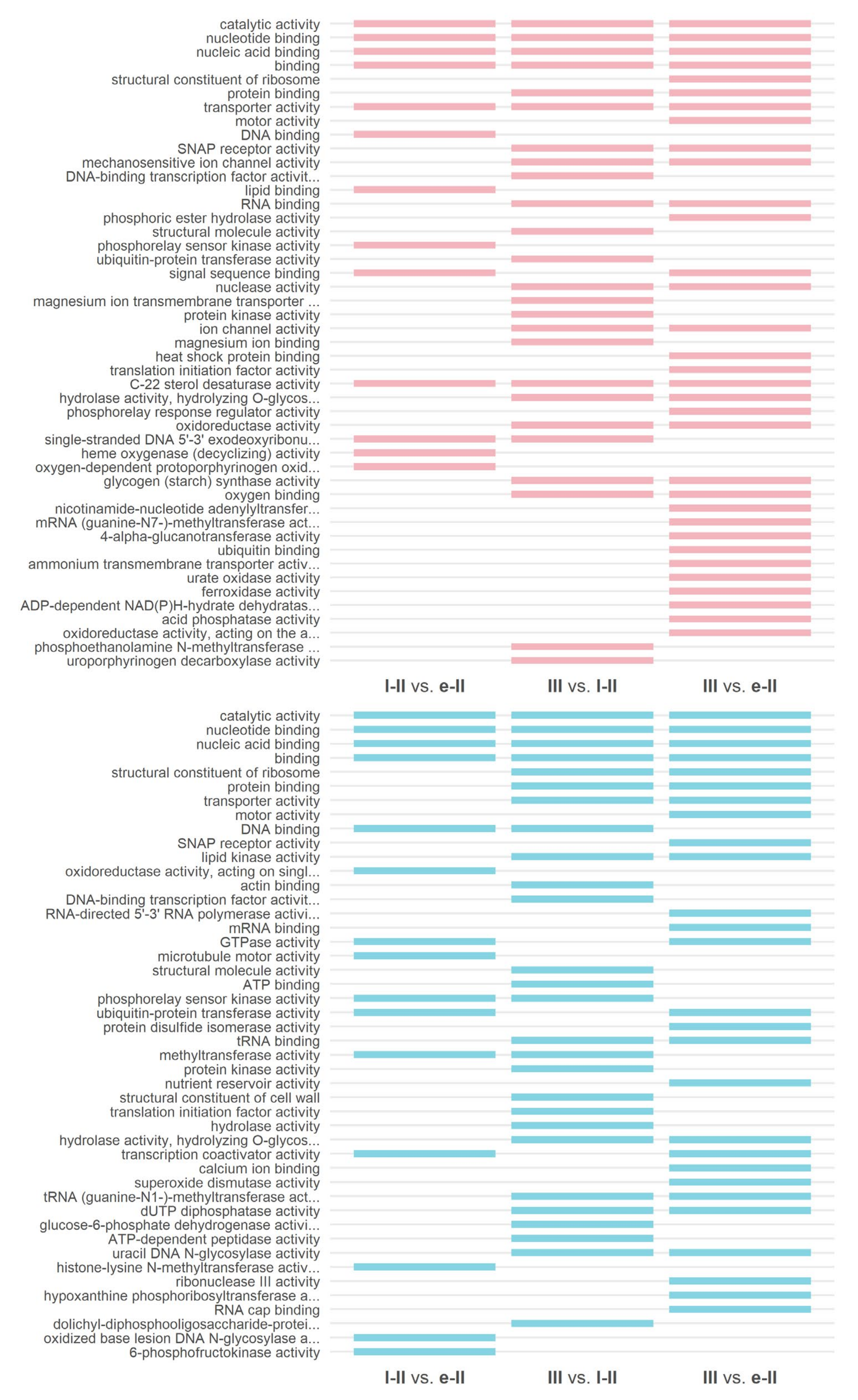
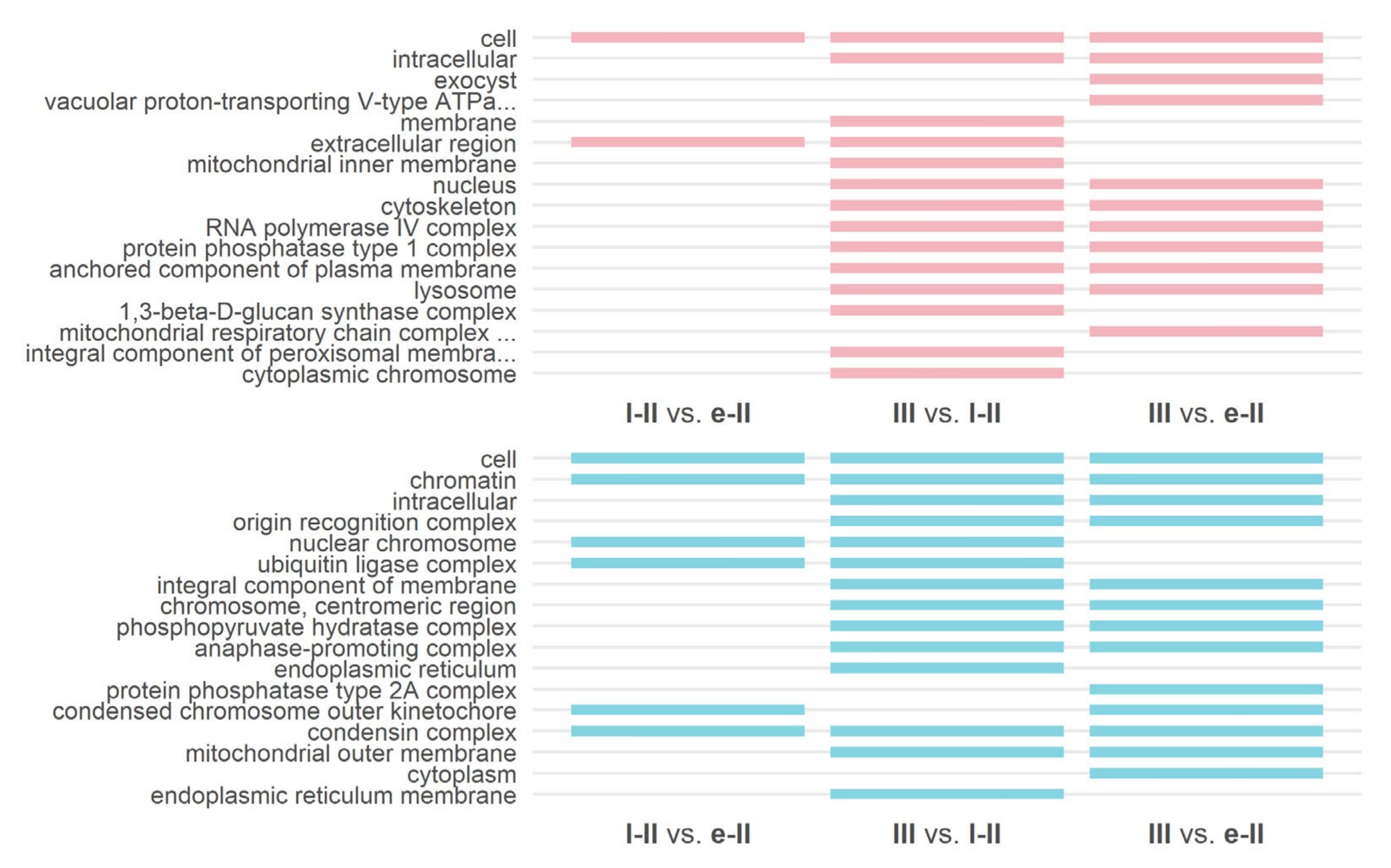
3.3. Gene Ontology and KEGG Enrichment Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Oldroyd, G.E. Speak, friend, and enter: Signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 2013, 11, 252–263. [Google Scholar] [CrossRef] [PubMed]
- Timmers, A.C.J. The role of the plant cytoskeleton in the interaction between legumes and rhizobia. J. Microsc. 2008, 231, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Tsyganova, A.V.; Brewin, N.J.; Tsyganov, V.E. Structure and development of the legume-rhizobial symbiotic interface in infection threads. Cells 2021, 10, 1050. [Google Scholar] [CrossRef] [PubMed]
- Guinel, F.C. Getting around the legume nodule: I. The structure of the peripheral zone in four nodule types. Botany 2009, 87, 1117–1138. [Google Scholar] [CrossRef]
- Brewin, N.J. Plant cell wall remodelling in the Rhizobium–legume symbiosis. Crit. Rev. Plant Sci. 2004, 23, 293–316. [Google Scholar] [CrossRef]
- Tsyganova, A.V.; Kitaeva, A.B.; Tsyganov, V.E. Cell differentiation in nitrogen-fixing nodules hosting symbiosomes. Funct. Plant Biol. 2018, 45, 47–57. [Google Scholar] [CrossRef]
- Van de Velde, W.; Guerra, J.C.P.; Keyser, A.D.; De Rycke, R.; Rombauts, S.; Maunoury, N.; Mergaert, P.; Kondorosi, E.; Holsters, M.; Goormachtig, S. Aging in legume symbiosis. A molecular view on nodule senescence in Medicago truncatula. Plant Physiol. 2006, 141, 711–720. [Google Scholar] [CrossRef]
- Kondorosi, E.; Kondorosi, A. Endoreduplication and activation of the anaphase-promoting complex during symbiotic cell development. FEBS Lett. 2004, 567, 152–157. [Google Scholar] [CrossRef]
- Roy, S.; Liu, W.; Nandety, R.S.; Crook, A.; Mysore, K.S.; Pislariu, C.I.; Frugoli, J.; Dickstein, R.; Udvardi, M.K. Celebrating 20 years of genetic discoveries in legume nodulation and symbiotic nitrogen fixation. Plant Cell 2020, 32, 15–41. [Google Scholar] [CrossRef]
- Tsyganov, V.E.; Tsyganova, A.V. Symbiotic regulatory genes controlling nodule development in Pisum sativum L. Plants 2020, 9, 1741. [Google Scholar] [CrossRef]
- Kitaeva, A.B.; Demchenko, K.N.; Tikhonovich, I.A.; Timmers, A.C.J.; Tsyganov, V.E. Comparative analysis of the tubulin cytoskeleton organization in nodules of Medicago truncatula and Pisum sativum: Bacterial release and bacteroid positioning correlate with characteristic microtubule rearrangements. New Phytol. 2016, 210, 168–183. [Google Scholar] [CrossRef]
- Kitaeva, A.B.; Gorshkov, A.P.; Kirichek, E.A.; Kusakin, P.G.; Tsyganova, A.V.; Tsyganov, V.E. General patterns and species-specific differences in the organization of the tubulin cytoskeleton in indeterminate nodules of three legumes. Cells 2021, 10, 1012. [Google Scholar] [CrossRef]
- Maróti, G.; Kondorosi, É. Nitrogen-fixing Rhizobium-legume symbiosis: Are polyploidy and host peptide-governed symbiont differentiation general principles of endosymbiosis? Front. Microbiol. 2014, 5, 326. [Google Scholar] [CrossRef]
- Stagnari, F.; Maggio, A.; Galieni, A.; Pisante, M. Multiple benefits of legumes for agriculture sustainability: An overview. Chem. Biol. Technol. Agric. 2017, 4, 2. [Google Scholar] [CrossRef]
- Borisov, A.Y.; Danilova, T.N.; Koroleva, T.A.; Kuznetsova, E.V.; Madsen, L.; Mofett, M.; Naumkina, T.S.; Nemankin, T.A.; Ovchinnikova, E.S.; Pavlova, Z.B.; et al. Regulatory genes of garden pea (Pisum sativum L.) controlling the development of nitrogen-fixing nodules and arbuscular mycorrhiza: A review of basic and applied aspects. Appl. Biochem. Microbiol. 2007, 43, 237–243. [Google Scholar] [CrossRef]
- Serova, T.A.; Tsyganova, A.V.; Tikhonovich, I.A.; Tsyganov, V.E. Gibberellins inhibit nodule senescence and stimulate nodule meristem bifurcation in pea (Pisum sativum L.). Front. Plant Sci. 2019, 10, 285. [Google Scholar] [CrossRef]
- Dolgikh, E.A.; Kusakin, P.G.; Kitaeva, A.B.; Tsyganova, A.V.; Kirienko, A.N.; Leppyanen, I.V.; Dolgikh, A.V.; Ilina, E.L.; Demchenko, K.N.; Tikhonovich, I.A.; et al. Mutational analysis indicates that abnormalities in rhizobial infection and subsequent plant cell and bacteroid differentiation in pea (Pisum sativum) nodules coincide with abnormal cytokinin responses and localization. Ann. Bot. 2020, 125, 905–923. [Google Scholar] [CrossRef]
- Sherrier, D.J.; Borisov, A.Y.; Tikhonovich, I.A.; Brewin, N.J. Immunocytological evidence for abnormal symbiosome development in nodules of the pea mutant line Sprint-2Fix− (sym31). Protoplasma 1997, 199, 57–68. [Google Scholar] [CrossRef]
- Dahiya, P.; Sherrier, D.J.; Kardailsky, I.V.; Borisov, A.Y.; Brewin, N.J. Symbiotic gene Sym31 controls the presence of a lectinlike glycoprotein in the symbiosome compartment of nitrogen-fixing pea nodules. Mol. Plant Microbe Interact. 1998, 11, 915–923. [Google Scholar] [CrossRef]
- Ivanova, K.A.; Tsyganova, A.V.; Brewin, N.J.; Tikhonovich, I.A.; Tsyganov, V.E. Induction of host defences by Rhizobium during ineffective nodulation of pea (Pisum sativum L.) carrying symbiotically defective mutations sym40 (PsEFD), sym33 (PsIPD3/PsCYCLOPS) and sym42. Protoplasma 2015, 252, 1505–1517. [Google Scholar] [CrossRef]
- Tsyganova, A.V.; Seliverstova, E.V.; Brewin, N.J.; Tsyganov, V.E. Comparative analysis of remodelling of the plant–microbe interface in Pisum sativum and Medicago truncatula symbiotic nodules. Protoplasma 2019, 256, 983–996. [Google Scholar] [CrossRef]
- Roux, B.; Rodde, N.; Jardinaud, M.-F.; Timmers, T.; Sauviac, L.; Cottret, L.; Carrère, S.; Sallet, E.; Courcelle, E.; Moreau, S.; et al. An integrated analysis of plant and bacterial gene expression in symbiotic root nodules using laser-capture microdissection coupled to RNA sequencing. Plant J. 2014, 77, 817–837. [Google Scholar] [CrossRef]
- Limpens, E.; Moling, S.; Hooiveld, G.; Pereira, P.A.; Bisseling, T.; Becker, J.D.; Küster, H. Cell- and tissue-specific transcriptome analyses of Medicago truncatula root nodules. PLoS ONE 2013, 8, e64377. [Google Scholar] [CrossRef]
- Carrere, S.; Verdier, J.; Gamas, P. MtExpress, a comprehensive and curated rnaseq-based gene expression atlas for the model legume Medicago truncatula. Plant Cell Physiol. 2021, 62, 1494–1500. [Google Scholar] [CrossRef]
- Alves-Carvalho, S.; Aubert, G.; Carrère, S.; Cruaud, C.; Brochot, A.-L.; Jacquin, F.; Klein, A.; Martin, C.; Boucherot, K.; Kreplak, J.; et al. Full-length de novo assembly of RNA-seq data in pea (Pisum sativum L.) provides a gene expression atlas and gives insights into root nodulation in this species. Plant J. 2015, 84, 1–19. [Google Scholar] [CrossRef]
- Kosterin, O.E.; Rozov, S.M. Mapping of the new mutation blb and the problem of integrity of linkage group I. Pisum Genet. 1993, 25, 27–31. [Google Scholar]
- Fåhraeus, G. The infection of clover root hairs by nodule bacteria studied by a simple glass slide technique. J. Gen. Microbiol. 1957, 16, 374–381. [Google Scholar] [CrossRef]
- Glenn, A.R.; Poole, P.S.; Hudman, J.F. Succinate uptake by free-living and bacteroid forms of Rhizobium leguminosarum. Microbiology 1980, 119, 267–271. [Google Scholar] [CrossRef][Green Version]
- Serova, T.A.; Tikhonovich, I.A.; Tsyganov, V.E. Analysis of nodule senescence in pea (Pisum sativum L.) using laser microdissection, real-time PCR, and ACC immunolocalization. J. Plant Physiol. 2017, 212, 29–44. [Google Scholar] [CrossRef]
- Bushnell, B. BBMap: A Fast, Accurate, Splice-Aware Aligner. Available online: https://jgi.doe.gov/data-and-tools/bbtools/ (accessed on 19 February 2014).
- Kreplak, J.; Madoui, M.-A.; Cápal, P.; Novák, P.; Labadie, K.; Aubert, G.; Bayer, P.E.; Gali, K.K.; Syme, R.A.; Main, D.; et al. A reference genome for pea provides insight into legume genome evolution. Nat. Genet. 2019, 51, 1411–1422. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2012, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Alexa, A.; Rahnenfuhrer, J. topGO: Enrichment Analysis for Gene Ontology; R Package Version 2.44.0; R Package: Vienna, Austria, 2021. [Google Scholar]
- Kolde, R. pheatmap: Pretty Heatmaps; R Package Version 1.0.12; R Package: Vienna, Austria, 2019. [Google Scholar]
- Chen, H. VennDiagram: Generate High-Resolution Venn and Euler Plots; R Package Version 1.6.20; R Package: Vienna, Austria, 2018. [Google Scholar]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-mapper v2: Functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 2021. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Mergaert, P.; Kereszt, A.; Kondorosi, E. Gene expression in nitrogen-fixing symbiotic nodule cells in Medicago truncatula and other nodulating plants. Plant Cell 2019, 32, 42–68. [Google Scholar] [CrossRef]
- Zhukov, V.A.; Zhernakov, A.I.; Kulaeva, O.A.; Ershov, N.I.; Borisov, A.Y.; Tikhonovich, I.A. De Novo assembly of the pea (Pisum sativum L.) nodule transcriptome. Int. J. Genomics 2015, 2015, 695947. [Google Scholar] [CrossRef]
- Takeda, N.; Sato, S.; Asamizu, E.; Tabata, S.; Parniske, M. Apoplastic plant subtilases support arbuscular mycorrhiza development in Lotus japonicus. Plant J. 2009, 58, 766–777. [Google Scholar] [CrossRef]
- Laplaze, L.; Ribeiro, A.; Franche, C.; Duhoux, E.; Auguy, F.; Bogusz, D.; Pawlowski, K. Characterization of a Casuarina glauca nodule-specific subtilisin-like protease gene, a homolog of Alnus glutinosa ag12. Mol. Plant Microbe Interact. 2000, 13, 113–117. [Google Scholar] [CrossRef]
- Ribeiro, A.; Akkermans, A.D.; van Kammen, A.; Bisseling, T.; Pawlowski, K. A nodule-specific gene encoding a subtilisin-like protease is expressed in early stages of actinorhizal nodule development. Plant Cell 1995, 7, 785–794. [Google Scholar] [CrossRef][Green Version]
- Taylor, A.; Qiu, Y.-L. Evolutionary history of subtilases in land plants and their involvement in symbiotic interactions. Mol. Plant Microbe Interact. 2017, 30, 489–501. [Google Scholar] [CrossRef]
- Franssen, H.J.; Xiao, T.T.; Kulikova, O.; Wan, X.; Bisseling, T.; Scheres, B.; Heidstra, R. Root developmental programs shape the Medicago truncatula nodule meristem. Development 2015, 142, 2941–2950. [Google Scholar] [CrossRef]
- Schiessl, K.; Lilley, J.L.S.; Lee, T.; Tamvakis, I.; Kohlen, W.; Bailey, P.C.; Thomas, A.; Luptak, J.; Ramakrishnan, K.; Carpenter, M.D.; et al. NODULE INCEPTION recruits the lateral root developmental program for symbiotic nodule organogenesis in Medicago truncatula. Curr. Biol. 2019, 29, 3657–3668. [Google Scholar] [CrossRef]
- Lee, H.W.; Cho, C.; Kim, J. Lateral organ boundaries domain16 and 18 act downstream of the AUXIN1 and LIKE-AUXIN3 auxin influx carriers to control lateral root development in Arabidopsis. Plant Physiol. 2015, 168, 1792–1806. [Google Scholar] [CrossRef]
- Ballas, N.; Wong, L.-M.; Theologis, A. Identification of the auxin-responsive element, AuxRE, in the primary indoleacetic acid-inducible gene, PS-IAA4/5, of pea (Pisum sativum). J. Mol. Biol. 1993, 233, 580–596. [Google Scholar] [CrossRef]
- Parry, G.; Estelle, M. Auxin receptors: A new role for F-box proteins. Curr. Opin. Cell Biol. 2006, 18, 152–156. [Google Scholar] [CrossRef]
- Azarakhsh, M.; Kirienko, A.N.; Zhukov, V.A.; Lebedeva, M.A.; Dolgikh, E.A.; Lutova, L.A. KNOTTED1-LIKE HOMEOBOX 3: A new regulator of symbiotic nodule development. J. Exp. Bot. 2015, 66, 7181–7195. [Google Scholar] [CrossRef]
- Sánchez-Rodríguez, C.; Estévez, J.M.; Llorente, F.; Hernández-Blanco, C.; Jordá, L.; Pagán, I.; Berrocal, M.; Marco, Y.; Somerville, S.; Molina, A. The ERECTA receptor-like kinase regulates cell wall–mediated resistance to pathogens in Arabidopsis thaliana. Mol. Plant Microbe Interact. 2009, 22, 953–963. [Google Scholar] [CrossRef]
- Mithöfer, A. Suppression of plant defence in rhizobia–legume symbiosis. Trends Plant Sci. 2002, 7, 440–444. [Google Scholar] [CrossRef]
- Mitra, R.M.; Long, S.R. Plant and bacterial symbiotic mutants define three transcriptionally distinct stages in the development of the Medicago truncatula/Sinorhizobium meliloti symbiosis. Plant Physiol. 2004, 134, 595–604. [Google Scholar] [CrossRef]
- Tsyganova, A.V.; Seliverstova, E.V.; Brewin, N.J.; Tsyganov, V.E. Bacterial release is accompanied by ectopic accumulation of cell wall material around the vacuole in nodules of Pisum sativum sym33-3 allele encoding transcription factor PsCYCLOPS/PsIPD3. Protoplasma 2019, 256, 1449–1453. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 2005, 6, 850–861. [Google Scholar] [CrossRef]
- Giordano, W.; Hirsch, A.M. The expression of MaEXP1, a Melilotus alba expansin gene, is upregulated during the sweetclover-Sinorhizobium meliloti interaction. Mol. Plant Microbe Interact. 2004, 17, 613–622. [Google Scholar] [CrossRef]
- Sujkowska, M.; Borucki, W.; Golinowski, W. Localization of expansin-like protein in apoplast of pea (Pisum sativum L.) root nodules during interaction with Rhizobium leguminosarum bv.viciae 248. Acta Soc. Bot. Pol. 2007, 76, 17–26. [Google Scholar] [CrossRef][Green Version]
- Li, X.; Zhao, J.; Tan, Z.; Zeng, R.; Liao, H. GmEXPB2, a cell wall β-expansin, affects soybean nodulation through modifying root architecture and promoting nodule formation and development. Plant Physiol. 2015, 169, 2640–2653. [Google Scholar] [CrossRef]
- Smith, P.M.C.; Atkins, C.A. Purine biosynthesis. Big in cell division, even bigger in nitrogen assimilation. Plant Physiol. 2002, 128, 793–802. [Google Scholar] [CrossRef]
- Garneau, M.G.; Tan, Q.; Tegeder, M. Function of pea amino acid permease AAP6 in nodule nitrogen metabolism and export, and plant nutrition. J. Exp. Bot. 2018, 69, 5205–5219. [Google Scholar] [CrossRef]
- Udvardi, M.; Poole, P.S. Transport and metabolism in legume-rhizobia symbioses. Annu. Rev. Plant Biol. 2013, 64, 781–805. [Google Scholar] [CrossRef]
- Valkov, V.T.; Sol, S.; Rogato, A.; Chiurazzi, M. The functional characterization of LjNRT2.4 indicates a novel, positive role of nitrate for an efficient nodule N2-fixation activity. New Phytol. 2020, 228, 682–696. [Google Scholar] [CrossRef]
- Pratelli, R.; Voll, L.M.; Horst, R.J.; Frommer, W.B.; Pilot, G. Stimulation of nonselective amino acid export by glutamine dumper proteins. Plant Physiol. 2009, 152, 762–773. [Google Scholar] [CrossRef] [PubMed]
- Peiter, E.; Yan, F.; Schubert, S. Amino acid export from infected cells of Vicia faba root nodules: Evidence for an apoplastic step in the infected zone. Physiol. Plant. 2004, 122, 107–114. [Google Scholar] [CrossRef]
- White, J.; Prell, J.; James, E.K.; Poole, P. Nutrient sharing between symbionts. Plant Physiol. 2007, 144, 604–614. [Google Scholar] [CrossRef] [PubMed]
- Borisov, A.Y.; Madsen, L.H.; Tsyganov, V.E.; Umehara, Y.; Voroshilova, V.A.; Batagov, A.O.; Sandal, N.; Mortensen, A.; Schauser, L.; Ellis, N.; et al. The Sym35 gene required for root nodule development in pea is an ortholog of Nin from Lotus japonicus. Plant Physiol. 2003, 131, 1009–1017. [Google Scholar] [CrossRef]
- Liu, J.; Bisseling, T. Evolution of NIN and NIN-like genes in relation to nodule symbiosis. Genes 2020, 11, 777. [Google Scholar] [CrossRef]
- De Zélicourt, A.; Diet, A.; Marion, J.; Laffont, C.; Ariel, F.; Moison, M.; Zahaf, O.; Crespi, M.; Gruber, V.; Frugier, F. Dual involvement of a Medicago truncatula NAC transcription factor in root abiotic stress response and symbiotic nodule senescence. Plant J. 2012, 70, 220–230. [Google Scholar] [CrossRef]
- Burgeff, C.; Liljegren, S.J.; Tapia-López, R.; Yanofsky, M.F.; Alvarez-Buylla, E.R. MADS-box gene expression in lateral primordia, meristems and differentiated tissues of Arabidopsis thaliana roots. Planta 2002, 214, 365–372. [Google Scholar] [CrossRef]
- Vinardell, J.M.; Fedorova, E.; Cebolla, A.; Kevei, Z.; Horvath, G.; Kelemen, Z.; Tarayre, S.; Roudier, F.; Mergaert, P.; Kondorosi, A.; et al. Endoreduplication mediated by the anaphase-promoting complex activator CCS52A is required for symbiotic cell differentiation in Medicago truncatula nodules. Plant Cell 2003, 15, 2093–2105. [Google Scholar] [CrossRef]
- Barkan, A.; Small, I. Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 2014, 65, 415–442. [Google Scholar] [CrossRef]
- Ohnishi, T.; Szatmari, A.-M.; Watanabe, B.; Fujita, S.; Bancos, S.; Koncz, C.; Lafos, M.; Shibata, K.; Yokota, T.; Sakata, K.; et al. C-23 hydroxylation by Arabidopsis CYP90C1 and CYP90D1 reveals a novel shortcut in brassinosteroid biosynthesis. Plant Cell 2006, 18, 3275–3288. [Google Scholar] [CrossRef]
- McGuiness, P.N.; Reid, J.B.; Foo, E. Brassinosteroids play multiple roles in nodulation of pea via interactions with ethylene and auxin. Planta 2020, 252, 70. [Google Scholar] [CrossRef]
- Khatoon, A.; Rehman, S.; Salavati, A.; Komatsu, S. A comparative proteomics analysis in roots of soybean to compatible symbiotic bacteria under flooding stress. Amino Acids 2012, 43, 2513–2525. [Google Scholar] [CrossRef]
- Doidy, J.; Vidal, U.; Lemoine, R. Sugar transporters in Fabaceae, featuring SUT MST and SWEET families of the model plant Medicago truncatula and the agricultural crop Pisum sativum. PLoS ONE 2019, 14, e0223173. [Google Scholar] [CrossRef]
- Kryvoruchko, I.S.; Sinharoy, S.; Torres-Jerez, I.; Sosso, D.; Pislariu, C.I.; Guan, D.; Murray, J.; Benedito, V.A.; Frommer, W.B.; Udvardi, M.K. MtSWEET11, a nodule-specific sucrose transporter of Medicago truncatula. Plant Physiol. 2016, 171, 554–565. [Google Scholar] [CrossRef]
- Flemetakis, E.; Dimou, M.; Cotzur, D.; Efrose, R.C.; Aivalakis, G.; Colebatch, G.; Udvardi, M.; Katinakis, P. A sucrose transporter, LjSUT4, is up-regulated during Lotus japonicus nodule development. J. Exp. Bot. 2003, 54, 1789–1791. [Google Scholar] [CrossRef]
- Osipova, M.A.; Mortier, V.; Demchenko, K.N.; Tsyganov, V.E.; Tikhonovich, I.A.; Lutova, L.A.; Dolgikh, E.A.; Goormachtig, S. WUSCHEL-RELATED HOMEOBOX5 gene expression and interaction of CLE peptides with components of the systemic control add two pieces to the puzzle of autoregulation of nodulation. Plant Physiol. 2012, 158, 1329–1341. [Google Scholar] [CrossRef]
- Alloing, G.; Mandon, K.; Boncompagni, E.; Montrichard, F.; Frendo, P. Involvement of glutaredoxin and thioredoxin systems in the nitrogen-fixing symbiosis between legumes and rhizobia. Antioxidants 2018, 7, 182. [Google Scholar] [CrossRef]
- Ivanova, K.A.; Chernova, E.N.; Kulaeva, O.A.; Tsyganova, A.V.; Kusakin, P.G.; Russkikh, I.V.; Tikhonovich, I.A.; Tsyganov, V.E. The regulation of pea (Pisum sativum L.) symbiotic nodule infection and defense responses by glutathione, homoglutathione, and their ratio. Front. Plant Sci. 2021. submitted. [Google Scholar]
- Kijne, J.W. The fine structure of pea root nodules. 2. Senescence and disintegration of the bacteroid tissue. Physiological Plant Pathology 1975, 7, 17–21. [Google Scholar] [CrossRef]
- Kneen, B.E.; LaRue, T.A.; Hirsch, A.M.; Smith, C.A.; Weeden, N.F. sym 13—A gene conditioning ineffective nodulation in Pisum sativum. Plant Physiol. 1990, 94, 899–905. [Google Scholar] [CrossRef]
- Serova, T.A.; Tsyganova, A.V.; Tsyganov, V.E. Early nodule senescence is activated in symbiotic mutants of pea (Pisum sativum L.) forming ineffective nodules blocked at different nodule developmental stages. Protoplasma 2018, 255, 1443–1459. [Google Scholar] [CrossRef]
- Fedorova, E.E.; Coba de la Peña, T.; Lara-Dampier, V.; Trifonova, N.A.; Kulikova, O.; Pueyo, J.J.; Lucas, M.M. Potassium content diminishes in infected cells of Medicago truncatula nodules due to the mislocation of channels MtAKT1 and MtSKOR/GORK. J. Exp. Bot. 2021, 72, 1336–1348. [Google Scholar] [CrossRef]
- Fedorova, E.E.; de Felipe, M.R.; Pueyo, J.J.; Lucas, M.M. Conformation of cytoskeletal elements during the division of infected Lupinus albus L. nodule cells. J. Exp. Bot. 2007, 58, 2225–2236. [Google Scholar] [CrossRef]
- Gavrin, A.; Jansen, V.; Ivanov, S.; Bisseling, T.; Fedorova, E. ARP2/3-mediated actin nucleation associated with symbiosome membrane is essential for the development of symbiosomes in infected cells of Medicago truncatula root nodules. Mol. Plant Microbe Interact. 2015, 28, 605–614. [Google Scholar] [CrossRef]
- Zhang, X.; Han, L.; Wang, Q.; Zhang, C.; Yu, Y.; Tian, J.; Kong, Z. The host actin cytoskeleton channels rhizobia release and facilitates symbiosome accommodation during nodulation in Medicago truncatula. New Phytol. 2019, 221, 1049–1059. [Google Scholar] [CrossRef]
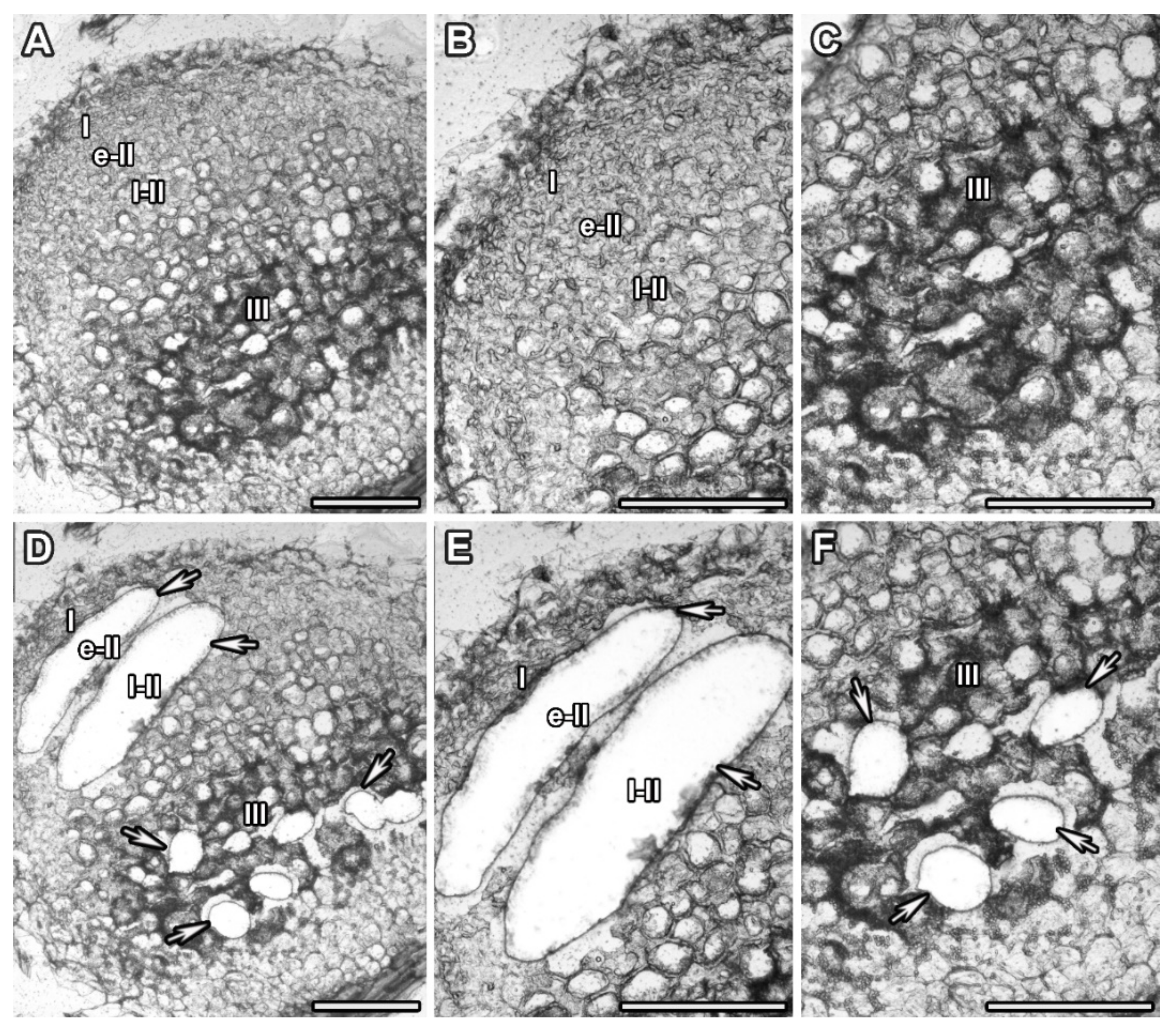
| Replicate | Nodules | Nodule Zone | Number of Sections | Number of Areas Cut 1 |
|---|---|---|---|---|
| 1 | 40 | Early II | 594 | 594 |
| Late II | 594 | |||
| III | 6799 | |||
| 2 | 38 | Early II | 662 | 662 |
| Late II | 662 | |||
| III | 6315 | |||
| 3 | 46 | Early II | 616 | 616 |
| Late II | 616 | |||
| III | 5984 |
| Tissue Sample | Biological Replicate | Raw Reads | Filtered Reads | % of Mapped Reads |
|---|---|---|---|---|
| Early II | 1 | 22, 003, 587 | 21, 953, 422 | 90.27 |
| 2 | 30, 443, 307 | 30, 376, 438 | 86.93 | |
| 3 | 34, 177, 880 | 34, 120, 254 | 88.73 | |
| Late II | 1 | 26, 368, 470 | 26, 309, 716 | 90.07 |
| 2 | 20, 855, 263 | 20, 826, 237 | 87.41 | |
| 3 | 23, 261, 468 | 23, 225, 423 | 89.36 | |
| III | 1 | 23, 401, 941 | 23, 345, 915 | 83.55 |
| 2 | 25, 659, 354 | 25, 618, 921 | 81.65 | |
| 3 | 29, 106, 470 | 29, 062, 966 | 82.84 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kusakin, P.G.; Serova, T.A.; Gogoleva, N.E.; Gogolev, Y.V.; Tsyganov, V.E. Laser Microdissection of Pisum sativum L. Nodules Followed by RNA-Seq Analysis Revealed Crucial Transcriptomic Changes during Infected Cell Differentiation. Agronomy 2021, 11, 2504. https://doi.org/10.3390/agronomy11122504
Kusakin PG, Serova TA, Gogoleva NE, Gogolev YV, Tsyganov VE. Laser Microdissection of Pisum sativum L. Nodules Followed by RNA-Seq Analysis Revealed Crucial Transcriptomic Changes during Infected Cell Differentiation. Agronomy. 2021; 11(12):2504. https://doi.org/10.3390/agronomy11122504
Chicago/Turabian StyleKusakin, Pyotr G., Tatiana A. Serova, Natalia E. Gogoleva, Yuri V. Gogolev, and Viktor E. Tsyganov. 2021. "Laser Microdissection of Pisum sativum L. Nodules Followed by RNA-Seq Analysis Revealed Crucial Transcriptomic Changes during Infected Cell Differentiation" Agronomy 11, no. 12: 2504. https://doi.org/10.3390/agronomy11122504
APA StyleKusakin, P. G., Serova, T. A., Gogoleva, N. E., Gogolev, Y. V., & Tsyganov, V. E. (2021). Laser Microdissection of Pisum sativum L. Nodules Followed by RNA-Seq Analysis Revealed Crucial Transcriptomic Changes during Infected Cell Differentiation. Agronomy, 11(12), 2504. https://doi.org/10.3390/agronomy11122504







