Rendering Multivariate Statistical Models for Genetic Diversity Assessment in A-Genome Diploid Wheat Population
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Experimental Design
2.2. Data Collection
2.3. Statistical Analysis
3. Results
3.1. Pearson Correlation Coefficients
3.2. Principal Component Analysis (PCA)
3.3. Cluster Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Girma, E. Genetic erosion of wheat (Triticum spp.): Concept, research results and challenges. J. Nat. Sci. Res. 2017, 7, 72–81. [Google Scholar]
- Cardinale, B.J.; Duffy, J.E.; Gonzalez, A.; Hooper, D.U.; Perrings, C.; Venail, P.; Narwani, A.; Mace, G.M.; Tilman, D.; Wardle, D.A. Biodiversity loss and its impact on humanity. Nature 2012, 486, 59–67. [Google Scholar] [CrossRef]
- Zavaleta, E.S.; Pasari, J.R.; Hulvey, K.B.; Tilman, G.D. Sustaining multiple ecosystem functions in grassland communities requires higher biodiversity. Proc. Natl. Acad. Sci. USA 2010, 107, 1443–1446. [Google Scholar] [CrossRef] [Green Version]
- Choudhary, S.K.; Kumar, V.; Singhal, R.K.; Bose, B.; Chauhan, J.; Alamri, S.; Siddiqui, M.H.; Javed, T.; Shabbir, R.; Rajendran, K.; et al. Seed Priming with Mg(NO3)2 and ZnSO4 Salts Triggers the Germination and Growth Attributes Synergistically in Wheat Varieties. Agronomy 2021, 11, 2110. [Google Scholar] [CrossRef]
- Abhinandan, K.; Skori, L.; Stanic, M.; Hickerson, N.M.N.; Jamshed, M.; Samuel, M.A. Abiotic Stress Signaling in Wheat An Inclusive Overview of Hormonal Interactions During Abiotic Stress Responses in Wheat. Front. Plant Sci. 2018, 9, 734. [Google Scholar] [CrossRef]
- Zaheer, M.S.; Ali, H.H.; Soufan, W.; Iqbal, R.; Habib-Ur-Rahman, M.; Iqbal, J.; Israr, M.; El Sabagh, A. Potential Effects of Biochar Application for Improving Wheat (Triticum aestivum L.) Growth and Soil Biochemical Properties under Drought Stress Conditions. Land 2021, 10, 1125. [Google Scholar] [CrossRef]
- Chen, X. Pathogens which threaten food security: Puccinia striiformis, the wheat stripe rust pathogen. Food Secur. 2020, 12, 239–251. [Google Scholar] [CrossRef]
- Brook, B.W.; Sodhi, N.S.; Bradshaw, C.J.A. Synergies among extinction drivers under global change. Trends Ecol. Evol. 2008, 23, 453–460. [Google Scholar] [CrossRef]
- Dunn, R.R.; Harris, N.C.; Colwell, R.K.; Koh, L.P.; Sodhi, N.S. The sixth mass coextinction: Are most endangered species parasites and mutualists? Proc. R. Soc. B Biol. Sci. 2009, 276, 3037–3045. [Google Scholar] [CrossRef] [Green Version]
- Shmelev, S.E.; Salnikov, V.; Turulina, G.; Polyakova, S.; Tazhibayeva, T.; Schnitzler, T.; Shmeleva, I. Climate Change and Food Security: The Impact of Some Key Variables on Wheat Yield in Kazakhstan. Sustainability 2021, 13, 8583. [Google Scholar] [CrossRef]
- Gupta, P.K.; Mir, R.R.; Mohan, A.; Kumar, J. Wheat Genomics: Present Status and Future Prospects. Int. J. Plant Genom. 2008, 2008, 1–36. [Google Scholar] [CrossRef]
- Sarfraz, Z.; Shah, M.M.; Iqbal, M.S.; Nazir, M.F.; Fatima, S.A. Identification of Valuable Traits Through Molecular and Morphological Markers in Diploid Wheat. Pak. J. Biotechnol. 2020, 17. [Google Scholar] [CrossRef]
- Endo, T.R.; Gill, B.S. The Deletion Stocks of Common Wheat. J. Hered. 1996, 87, 295–307. [Google Scholar] [CrossRef]
- Sears, E.R. Nullisomic-tetrasomic combinations in hexaploid wheat. In Chromosome Manipulations and Plant Genetics; Springer: Berlin/Heidelberg, Germany, 1966; pp. 29–45. [Google Scholar]
- Dhaliwal, H.S.; Singh, H.; Gill, K.S.; Randhawa, H.S. Evaluation and cataloguing of wheat germplasm for disease resistance and quality. In Biodiversity and Wheat Improvement; Damania, A.B., Ed.; John Wiley and Sons: Chichester, UK, 1993; pp. 123–140. [Google Scholar]
- Feldman, M.; Sears, E.R. The Wild Gene Resources of Wheat. Sci. Am. 1981, 244, 102–112. [Google Scholar] [CrossRef]
- Hussien, T.; Bowden, R.L.; Gill, B.S.; Cox, T.S.; Marshall, D.S. Performance of Four New Leaf Rust Resistance Genes Transferred to Common Wheat from Aegilops tauschii and Triticum monococcum. Plant Dis. 1997, 81, 582–586. [Google Scholar] [CrossRef] [Green Version]
- Qiu, Y.C.; Zhou, R.H.; Kong, X.Y.; Zhang, S.S.; Jia, J.Z. Microsatellite mapping of a Triticum urartu Tum. derived powdery mildew resistance gene transferred to common wheat (Triticum aestivum L.). Theor. Appl. Genet. 2005, 111, 1524–1531. [Google Scholar] [CrossRef]
- Dvořák, J.; Di Terlizzi, P.; Zhang, H.-B.; Resta, P. The evolution of polyploid wheats: Identification of the A genome donor species. Genome 1993, 36, 21–31. [Google Scholar] [CrossRef]
- Singh, K.; Ghai, M.; Garg, M.; Chhuneja, P.; Kaur, P.; Schnurbusch, T.; Keller, B.; Dhaliwal, H.S. An integrated molecular linkage map of diploid wheat based on a Triticum boeoticum × T. monococcum RIL population. Theor. Appl. Genet. 2007, 115, 301–312. [Google Scholar] [CrossRef]
- Cox, T.S.; Harrell, L.G.; Chen, P.; Gill, B.S. Reproductive Behavior of Hexaploid/Diploid Wheat Hybrids 1. Plant Breed. 1991, 107, 105–118. [Google Scholar] [CrossRef]
- Plamenov, D.; Belchev, I.; Kiryakova, V.; Spetsov, P. Fungal resistance of Triticum durum—T. monococcum ssp. aegilopoides amphiploid. J. Plant Dis. Prot. 2009, 116, 60–62. [Google Scholar] [CrossRef]
- The, T.; Baker, E. Basic Studies relating to the Transference of Genetic Characters From Triticum Monococcum L. to Hexaploid Wheat. Aust. J. Biol. Sci. 1975, 28, 189. [Google Scholar] [CrossRef] [Green Version]
- Bhagyalakshmi, K.; Vinod, K.K.; Kumar, M.; Arumugachamy, S.; Prabhakaran, A.J.; Raveendran, T.S. Interspecific Hybrids from Wild × Cultivated Triticum Crosses—A Study on the Cytological Behaviour and Molecular Relations. J. Crop. Sci. Biotechnol. 2008, 11, 257–262. [Google Scholar]
- Nazir, M.; Sarfraz, Z.; Mangi, N.; Shah, M.N.; Mahmood, T.; Mahmood, T.; Iqbal, M.; Rehmani, M.I.A.; El-Sharnouby, M.; Shabaan, M.; et al. Post-Anthesis Mobilization of Stem Assimilates in Wheat under Induced Stress. Sustainability 2021, 13, 5940. [Google Scholar] [CrossRef]
- Dorofeev, V.F.; Udachin, R.A.; Semenova, L.V.; Novikova, M.V.; Grazhdaninova, O.D.; Shitova, I.P.; Merezhko, A.F.; Filatenko, A.A. World Wheat; Agropromizdat: Saint Petersburg, Russia, 1987; p. 560. (In Russian) [Google Scholar]
- Hao, C.; Wang, L.; Ge, H.; Dong, Y.; Zhang, X. Genetic Diversity and Linkage Disequilibrium in Chinese Bread Wheat (Triticum aestivum L.) Revealed by SSR Markers. PLoS ONE 2011, 6, e17279. [Google Scholar] [CrossRef] [Green Version]
- Ladizinsky, G. Founder effect in crop-plant evolution. Econ. Bot. 1985, 39, 191–199. [Google Scholar] [CrossRef]
- Liu, K.; Xu, H.; Liu, G.; Guan, P.; Zhou, X.; Peng, H.; Yao, Y.; Ni, Z.; Sun, Q.; Du, J. QTL mapping of flag leaf-related traits in wheat (Triticum aestivum L.). Theor. Appl. Genet. 2018, 131, 839–849. [Google Scholar] [CrossRef] [Green Version]
- Petronaitis, T.; Simpfendorfer, S.; Hüberli, D. Importance of Fusarium spp. in Wheat to Food Security: A Global Perspective. In Plant Disease Food Security 21st Century; Springer: Berlin/Heidelberg, Germany, 2021; pp. 127–159. [Google Scholar]
- Mengistu, D.D.; Degaga, D.T.; Tsehay, A.S. Analyzing the contribution of crop diversification in improving household food security among wheat dominated rural households in Sinana District, Bale Zone, Ethiopia. Agric. Food Secur. 2021, 10, 1–15. [Google Scholar] [CrossRef]
- Mirzaghaderi, G.; Mason, A.S. Broadening the bread wheat D genome. Theor. Appl. Genet. 2019, 132, 1295–1307. [Google Scholar] [CrossRef]
- Singh, S.; Vikram, P.; Sehgal, D.; Burgueño, J.; Sharma, A.; Singh, S.K.; Sansaloni, C.P.; Joynson, R.; Brabbs, T.; Ortiz, C.; et al. Harnessing genetic potential of wheat germplasm banks through impact-oriented-prebreeding for future food and nutritional security. Sci. Rep. 2018, 8, 12527. [Google Scholar] [CrossRef] [Green Version]
- Pour-Aboughadareh, A.; Ahmadi, J.; Mehrabi, A.A.; Etminan, A.; Moghaddam, M. Insight into the genetic variability analysis and relationships among some Aegilops and Triticum species, as genome progenitors of bread wheat, using SCoT markers. Plant Biosyst. Int. J. Deal. Asp. Plant Biol. 2017, 152, 694–703. [Google Scholar] [CrossRef]
- Badr, S.S.M. Evaluation of some Egyptian cotton varieties by the yield and seven methods of earliness of crop maturity measurements. Egypt. J. Agric. Res. 2003, 81, 671–688. [Google Scholar]
- Batool, S.; Khan, N.U.; Makhdoom, K.; Bibi, Z.; Hassan, G.; Marwat, K.B.; Farhatullah, F.; Mohammad, R.; Khan, I.A. Heritability and genetic potential of upland cotton genotypes for morpho-yield traits. Pak. J. Bot. 2010, 42, 1057–1064. [Google Scholar]
- Soomro, Z.A.; Larik, A.S.; Kumbhar, M.B.; Khan, N.U.; Panhwar, N.A. Correlation and path analysis in hybrid cotton. Sabrao J. Breed. Genet. 2008, 40, 49–56. [Google Scholar]
- Roostaei, M.; Kamali, M.R.J.; Roohi, E.; Mohammadi, R. Evaluation of Sardari bread wheat ecotypes under the rainfed cold conditions of Iran. J. Agric. Sci. 2018, 156, 504–514. [Google Scholar] [CrossRef]
- Abbasov, M.; Akparov, Z.; Gross, T.; Babayeva, S.; Izzatullayeva, V.; Hajiyev, E.; Rustamov, K.; Gross, P.; Tekin, M.; Akar, T.; et al. Genetic relationship of diploid wheat (Triticum spp.) species assessed by SSR markers. Genet. Resour. Crop. Evol. 2018, 65, 1441–1453. [Google Scholar] [CrossRef]
- Singh, P.; Narayanam, S.S. Biometical Techniques in Plant Breeding; Kalyani Publishers: New Delhi, India, 2007. [Google Scholar]
- Ihsan, M.; Nazir, N.; Ghafoor, A.; Khalil, A.A.K.; Zahoor, M.; Nisar, M.; Khames, A.; Ullah, R.; Shah, A.B. Genetic Diversity in Local and Exotic Avena sativa L. (Oat) Germplasm Using Multivariate Analysis. Agronomy 2021, 11, 1713. [Google Scholar] [CrossRef]
- Murube, E.; Beleggia, R.; Pacetti, D.; Nartea, A.; Frascarelli, G.; Lanzavecchia, G.; Bellucci, E.; Nanni, L.; Gioia, T.; Marciello, U.; et al. Characterization of Nutritional Quality Traits of a Common Bean Germplasm Collection. Foods 2021, 10, 1572. [Google Scholar] [CrossRef]
- Johnson, R.A.; Wichern, D.W. Applied Multivariate Statistical Analysis; Prentice Hall: Upper Saddle River, NJ, USA, 2002; Volume 5, pp. 426–475. [Google Scholar]
- International Board for Plant Genetic Resources (IBPGR). Descriptors for Wheat (Revised); International Board for Plant Genetic Resources Secretariat: Rome, Italy, 1985; p. 12. [Google Scholar]
- Steel, R. Analysis of variance II: Multiway classifications. In Principles and Procedures of Statistics: A Biometrical Approach; McGraw-Hill Co. Inc.: New York, NY, USA, 1997; pp. 204–252. [Google Scholar]
- Mujeeb-Kazi, A.; Kimber, G. The Production, Cytology and Practically of Wide Hybrids in the Triticeae. Cereal Res. Commun. 1985, 13, 111–124. [Google Scholar]
- Bernhardt, N.; Brassac, J.; Dong, X.; Willing, E.M.; Poskar, C.H.; Kilian, B.; Blattner, F.R. Genome-wide sequence information reveals recurrent hybridization among diploid wheat wild relatives. Plant J. 2019, 102, 493–506. [Google Scholar] [CrossRef]
- Gill, B.S.; Browder, L.E.; Hatchett, J.H.; Harvey, T.L.; Martin, T.J.; Raupp, W.J.; Sharma, H.C.; Waines, J.G. Disease and Insect Resistance in Wild Wheats. In Proceedings of the Sixth International Wheat Genetics Symposium, Kyoto, Japan, 28 November–3 December 1983. [Google Scholar]
- Marshall, D.R.; Brown, A.H.D. Optimum sampling strategies in genetic conservation. In Crop Genetic Resources for Today and Tomorrow; Frankel, O.H., Hawkes, J.G., Eds.; Cambridge University Press: Cambridge, UK, 1975; pp. 53–80. [Google Scholar]
- Sharma, H.C.; Waines, J.G.; Foster, K.W. Variability in Primitive and Wild Wheats for Useful Genetic Characters 1. Crop. Sci. 1981, 21, 555–559. [Google Scholar] [CrossRef]
- Waines, J.G. Genetic Resources in Diploid Wheats: The Case for Diploid Commercial Wheats. In Proceedings of the Sixth International Wheat Genetics Symposium, Kyoto, Japan, 28 November–3 December 1983. [Google Scholar]
- Heun, M.; Schäfer-Pregl, R.; Klawan, D.; Castagna, R.; Accerbi, M.; Borghi, B.; Salamini, F. Site of Einkorn Wheat Domestication Identified by DNA Fingerprinting. Science 1997, 278, 1312–1314. [Google Scholar] [CrossRef] [Green Version]
- Dvorak, J.; McGuire, P.E.; Cassidy, B. Apparent sources of the A genomes of wheats inferred from polymorphism in abundance and restriction fragment length of repeated nucleotide sequences. Genome 1988, 30, 680–689. [Google Scholar] [CrossRef]
- Zhao, F.; Li, Y.; Yang, B.; Yuan, H.; Jin, C.; Zhou, L.; Pei, H.; Zhao, L.; Li, Y.; Zhou, Y.; et al. Powdery mildew disease resistance and marker-assisted screening at the Pm60 locus in wild diploid wheat Triticum urartu. Crop. J. 2019, 8, 252–259. [Google Scholar] [CrossRef]
- Chhuneja, P.; Kaur, S.; Garg, T.; Ghai, M.; Kaur, S.; Prashar, M.; Bains, N.S.; Goel, R.K.; Keller, B.; Dhaliwal, H.S.; et al. Mapping of adult plant stripe rust resistance genes in diploid A genome wheat species and their transfer to bread wheat. Theor. Appl. Genet. 2007, 116, 313–324. [Google Scholar] [CrossRef]
- Aharizad, S.; Sabzi, M.; Mohammadi, S.A.; Khodadadi, E. Multivariate analysis of genetic diversity in wheat (Triticum aestivum L.) recombinant inbred lines using agronomic traits. Ann. Biol. Res. 2012, 3, 2118–2126. [Google Scholar]
- Qaseem, M.F.; Qureshi, R.; Illyas, N.; Jalal-Ud-Din, S.G. Multivariate statistical analysis for yield and yield components in bread wheat planted under rainfed conditions. Pak. J. Bot. 2017, 49, 2445–2450. [Google Scholar]
- Blum, A.; Shpiler, L.; Golan, G.; Mayer, J. Yield stability and canopy temperature of wheat genotypes under drought-stress. Field Crop. Res. 1989, 22, 289–296. [Google Scholar] [CrossRef]
- Jaradat, A.A. Phenotypic divergence for morphological and yield-related traits among landrace genotypes of durum wheat from Jordan. Euphytica 1991, 52, 155–164. [Google Scholar] [CrossRef]
- Takumi, S.; Mitta, S.; Komura, S.; Ikeda, T.M.; Matsunaka, H.; Sato, K.; Yoshida, K.; Murai, K. Introgression of chromosomal segments conferring early heading date from wheat diploid progenitor, Aegilops tauschii Coss., into Japanese elite wheat cultivars. PLoS ONE 2020, 15, e0228397. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Lin, Y.; Gao, S.; Li, Z.; Ma, J.; Deng, M.; Chen, G.; Wei, Y.; Zheng, Y. A genome-wide association study of 23 agronomic traits in Chinese wheat landraces. Plant J. 2017, 91, 861–873. [Google Scholar] [CrossRef] [Green Version]
- Picascia, S.; Camarca, A.; Malamisura, M.; Mandile, R.; Galatola, M.; Cielo, D.; Gazza, L.; Mamone, G.; Auricchio, S.; Troncone, R.; et al. In Celiac Disease Patients the In Vivo Challenge with the Diploid Triticum monococcum Elicits a Reduced Immune Response Compared to Hexaploid Wheat. Mol. Nutr. Food Res. 2020, 64, e1901032. [Google Scholar] [CrossRef]
- Prasad, P.; Bhardwaj, S.C.; Gangwar, O.P.; Kumar, S.; Khan, H.; Kumar, S.; Rawal, H.C.; Sharma, T.R. Population Differentiation of Wheat Leaf Rust Fungus Puccinia triticina in South Asia. Curr. Sci. 2017, 112. [Google Scholar] [CrossRef]
- Sakhare, S.B.; Ghawat, N.P. Correlation and path analysis in durum wheat. PKV. Res. J. 2011, 35, 23–25. [Google Scholar]
- Sarfraz, Z.; Shah, M.M.; Iqbal, M.S.; Nazir, M.F.; Fatima, S.A. Cause and effect relationship of morphological attributes with yield in a-genome Wheat. J. Appl. Res. Plant Sci. 2020, 1, 13–19. [Google Scholar]
- Wang, X.; Luo, G.; Yang, W.; Li, Y.; Sun, J.; Zhan, K.; Liu, D.; Zhang, A. Genetic diversity, population structure and marker-trait associations for agronomic and grain traits in wild diploid wheat Triticum urartu. BMC Plant Biol. 2017, 17, 112. [Google Scholar] [CrossRef]
- Marza, F.; Bai, G.-H.; Carver, B.F.; Zhou, W.-C. Quantitative trait loci for yield and related traits in the wheat population Ning7840 × Clark. Theor. Appl. Genet. 2005, 112, 688–698. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.H.; Dar, A.N. Correlation and path coefficient analysis of some quantitative traits in wheat. Afr. Crop. Sci. J. 2010, 18. [Google Scholar] [CrossRef] [Green Version]
- Kumar, R.; Gaurav, S.S.; Bhushan, B.; Pal, R. Study of genetic parameters and genetic divergence for yield and yield components of bread wheat (Triticum aestivum L.). J. Wheat Res. 2013, 5, 39–42. [Google Scholar]
- Muhammad, K.; Khalil, I.H.; Abdul, B.; Mohammad, T.; Shahid, A.; Sajjad, A.; Amjad, A.; Muhammad, I. Assessment of heritability estimates for some yield traits in winter wheat (Triticum aestivum L.). Pak. J. Bot. 2011, 43, 2733–2736. [Google Scholar]
- Sarfraz, Z.; Shah, M.M.; Iqbal, M.S. Estimation of components of variation of morphological traits and yield in A-genome wheat populations. SABRAO J. Breed. Genet. 2016, 48, 105–109. [Google Scholar]
- Sheikh, S.; Singh, I. Studies on path co-efficient analysis of harvest index and its related traits in wheat. Indian J. Agric. Res. 2001, 35, 127–129. [Google Scholar]
- Sarfraz, Z.; Shah, M.M.; Iqbal, M.S. Genetic variability, heritability and genetic advance for agronomic traits among A-genome donor wheat genotypes. J. Agric. Res. 2016, 54, 15–20. [Google Scholar]
- Lee, I.; Yang, J. Common Clustering Algorithms. In Comprehensive Chemometrics; Brown, S.D., Tauler, R., Walczak, B., Eds.; Chemical and Biochemical Data Analysis; Elsevier: Amsterdam, The Netherlands, 2009; Volume 2, pp. 577–618. [Google Scholar]
- Massart, D.L.; Vandeginste, B.G.; Buydens, L.M.; Lewi, P.J.; Smeyers-Verbeke, J.; Jong, S.D. Handbook of Chemometrics and Qualimetrics; Elsevier Science Inc.: Amsterdam, The Netherlands, 1998. [Google Scholar]
- Kaiser, H.F. The Application of Electronic Computers to Factor Analysis. Educ. Psychol. Meas. 1960, 20, 141–151. [Google Scholar] [CrossRef]
- Abdi, H.; Williams, L.J. Principal component analysis. Wiley Interdiscip. Rev. Comput. Stat. 2010, 2, 433–459. [Google Scholar] [CrossRef]
- Chairi, F.; Sanchez-Bragado, R.; Serret, M.D.; Aparicio, N.; Nieto-Taladriz, M.T.; Araus, J.L. Agronomic and physiological traits related to the genetic advance of semi-dwarf durum wheat: The case of Spain. Plant Sci. 2020, 295, 110210. [Google Scholar] [CrossRef]
- Dutamo, D.; Alamerew, S.; Eticha, F.; Fikre, G. Genetic Variability in Bread Wheat (Triticum aestivum L.) Germplasm for Yield and Yield Component Traits. J. Biol. Agric. Healthc. 2015, 5, 39–46. [Google Scholar]
- Getachew, A.; Worede, F.; Alamerew, S. Multivariate analysis of phenotypic diversity of bread wheat (Triticum aestivum L.) in the highlands of northern Ethiopia. Adv. Crop. Sci. Technol. 2017, 5, 1–7. [Google Scholar]
- Wolde, T.; Eticha, F.; Alamerew, S.; Assefa, E.; Dutamo, D. Multivariate Analysis of Some Metric Traits in Durum Wheat (Triticum durum L.) Accessions. Food Sci. Qual. Manag. 2016, 74, 26–31. [Google Scholar]
- Hailu, F.; Merker, A.; Singh, H.; Belay, G.; Johansson, E. Multivariate Analysis of Diversity of Tetraploid Wheat Germplasm from Ethiopia. Genet. Resour. Crop. Evol. 2006, 53, 1089–1098. [Google Scholar] [CrossRef]
- Vaishnav, G.; Shukla, R.; Pandey, S. Assessment of principal component analysis for yield and its attributing traits in bread wheat (Triticum astivum L.) for normal and late sown conditions. J. Pharmacogn. Phytochem. 2020, 9, 1706–1709. [Google Scholar]
- Faris, J.D. Wheat Domestication: Key to Agricultural Revolutions Past and Future. In Genomics of Plant Genetic Resources; Springer: Berlin/Heidelberg, Germany, 2014; pp. 3, 439–464. [Google Scholar] [CrossRef]
- Anker, C.C.; Buntjer, J.B.; Niks, R.E. Morphological and molecular characterisation confirm that Triticum monococcum s.s. is resistant to wheat leaf rust. Theor. Appl. Genet. 2001, 103, 1093–1098. [Google Scholar] [CrossRef]
- Malaki, M.; Naghavi, M.R.; Alizadeh, H.; Potki, P.; Kazemi, M.; Pirseyedi, S.M.; Mardi, M.; Fakhre-Tabatabaei, S. Study of Genetic Variation in Wild Diploid Wheat (Triticum boeoticum) from Iran Using AFLP Markers. Iran. J. Biotechnol. 2006, 4, 269–274. [Google Scholar]
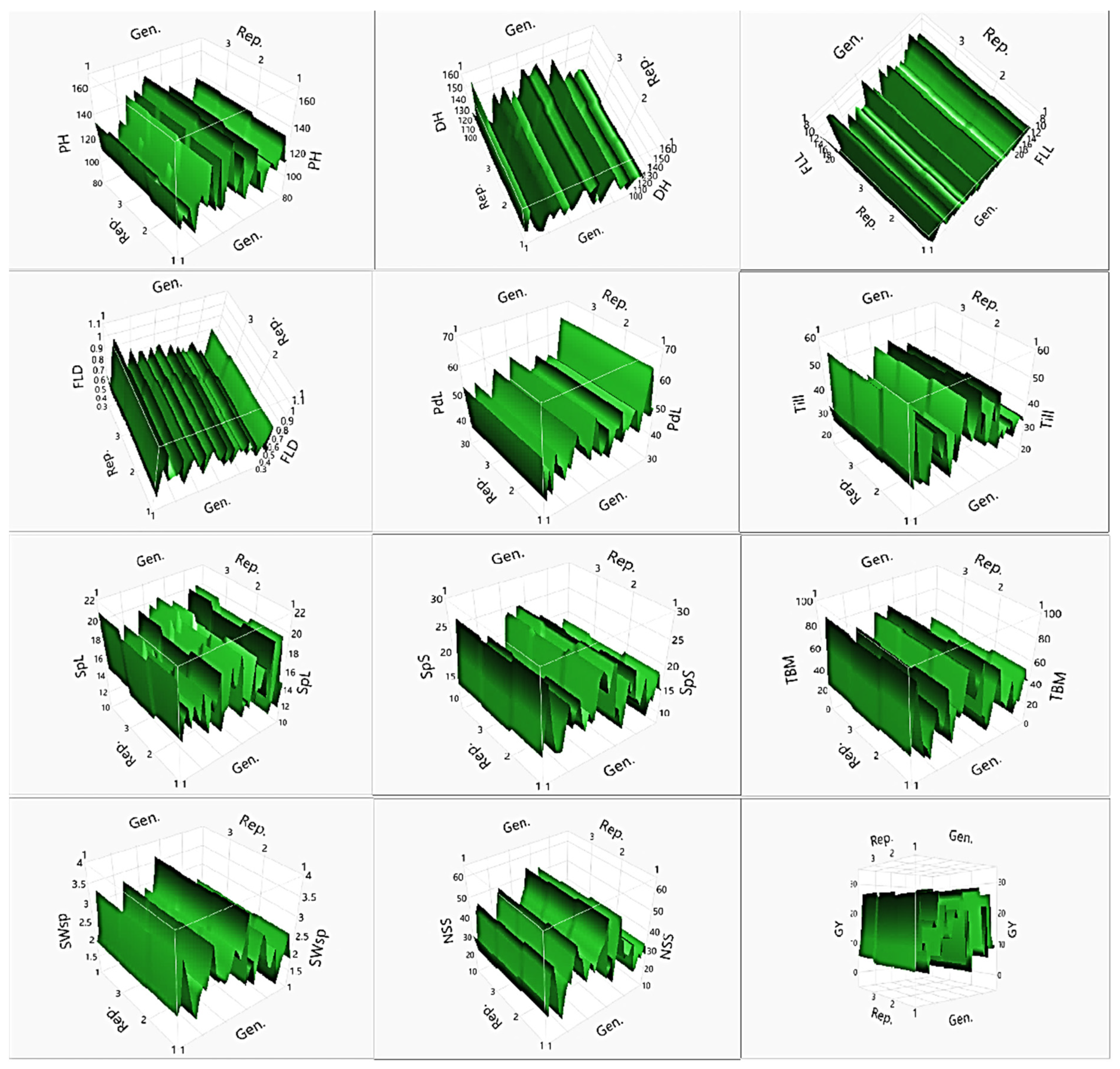
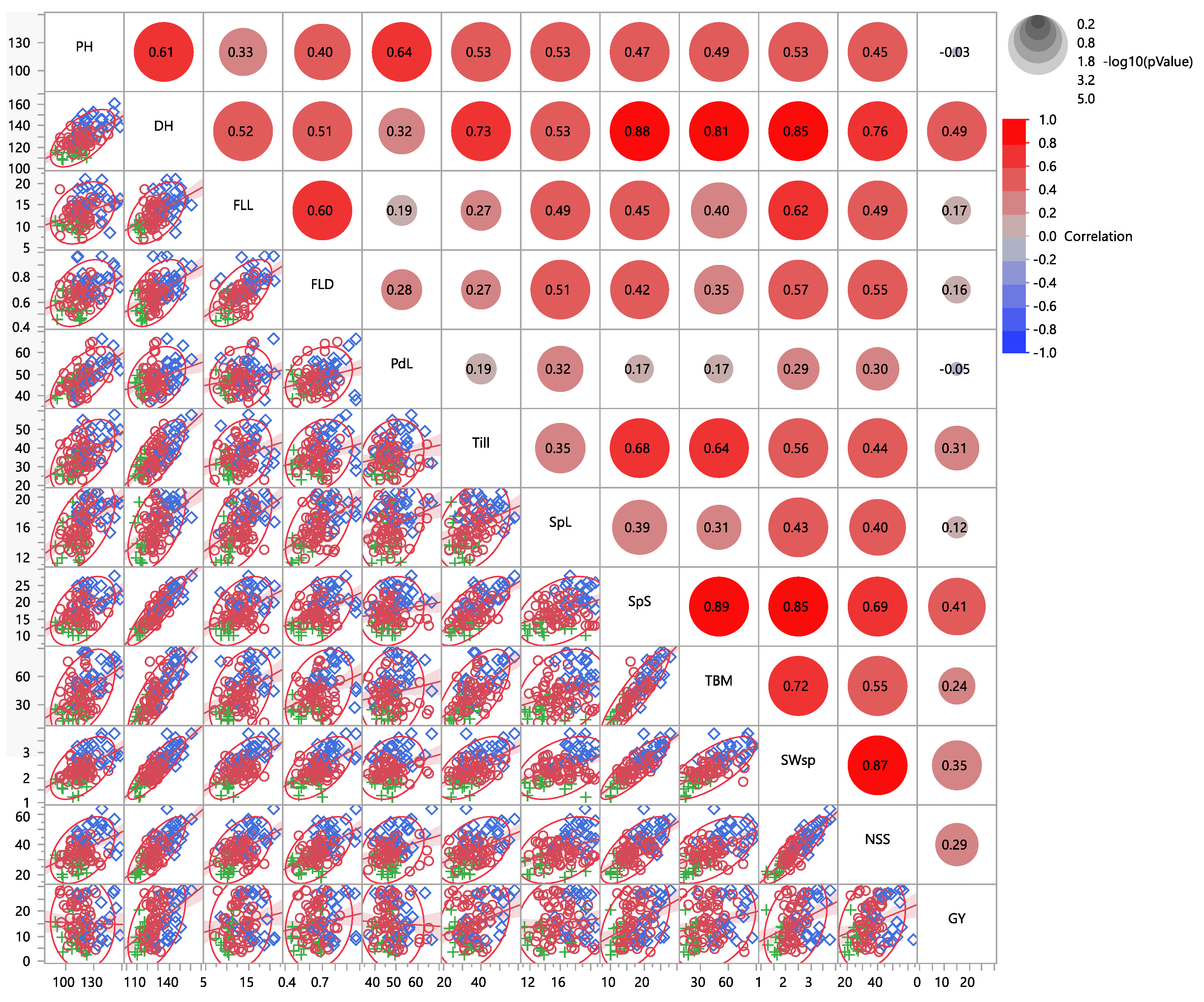
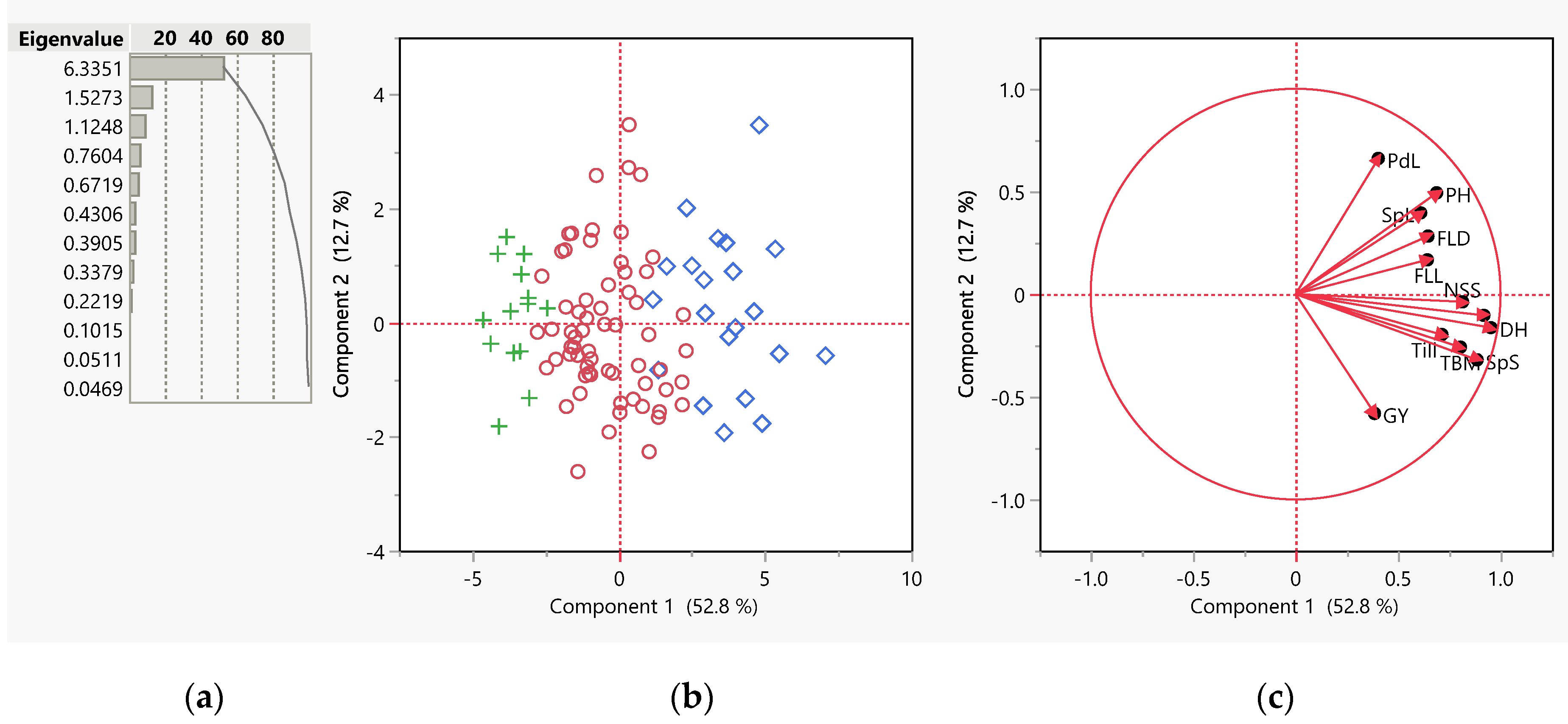

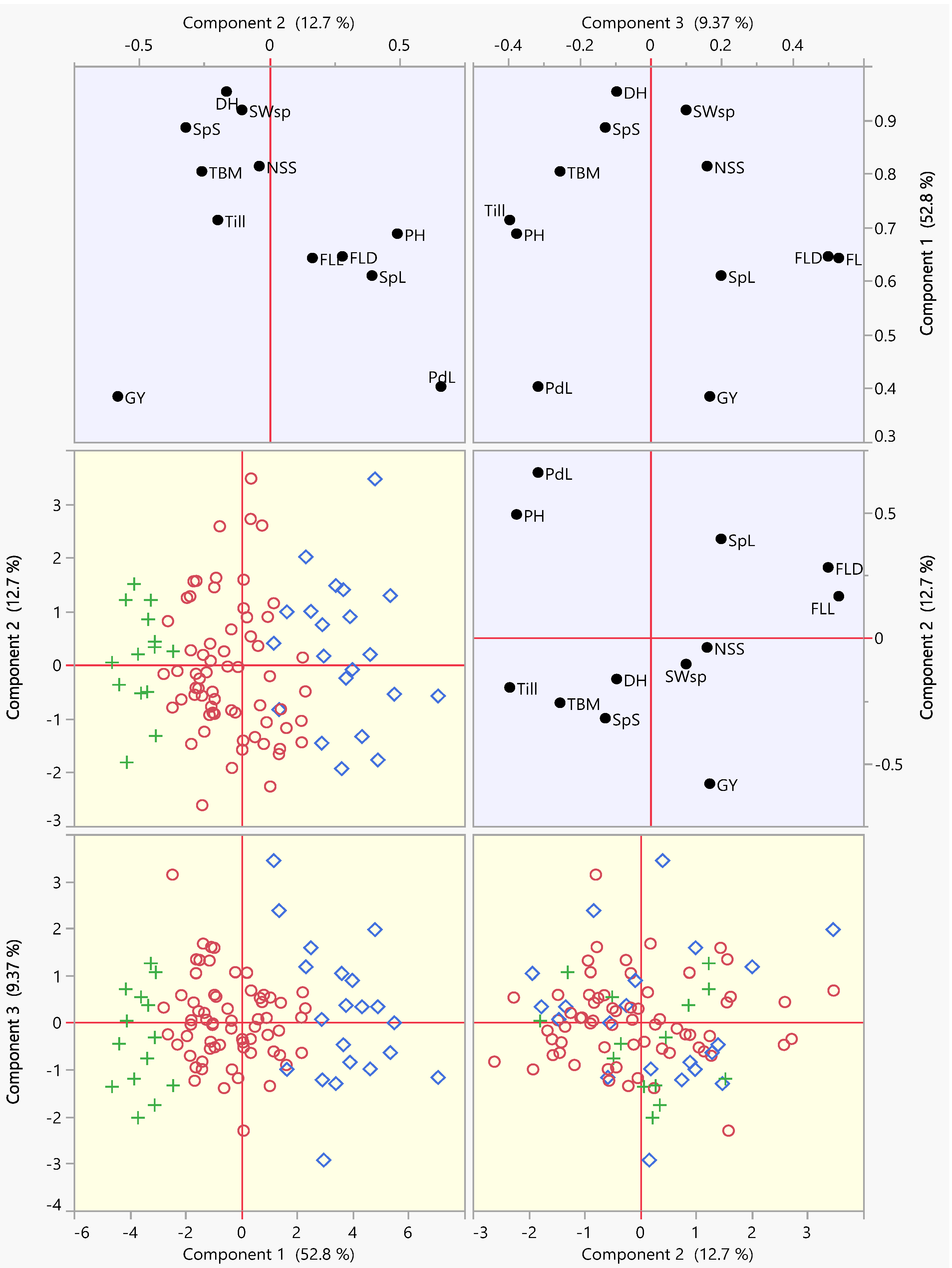
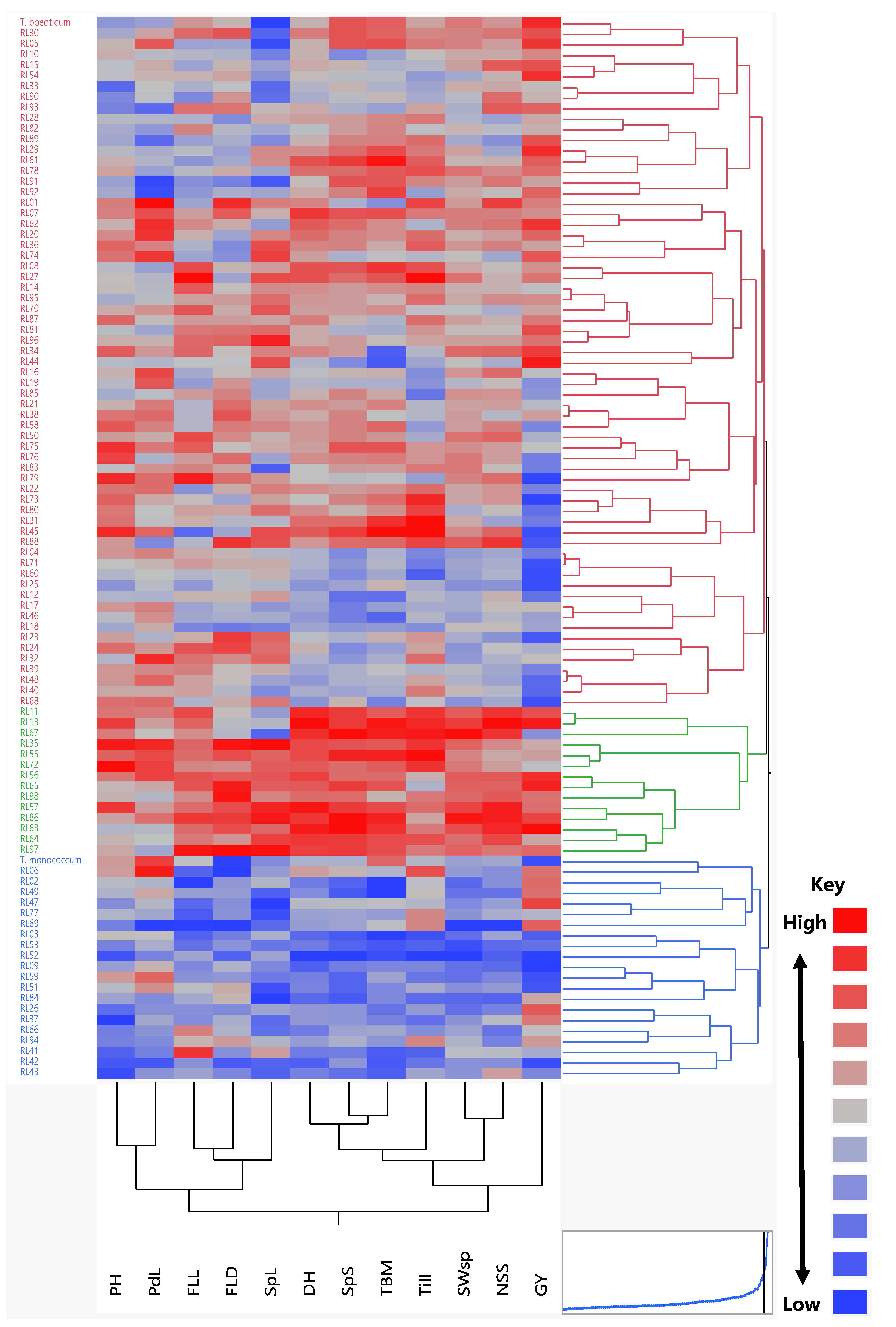
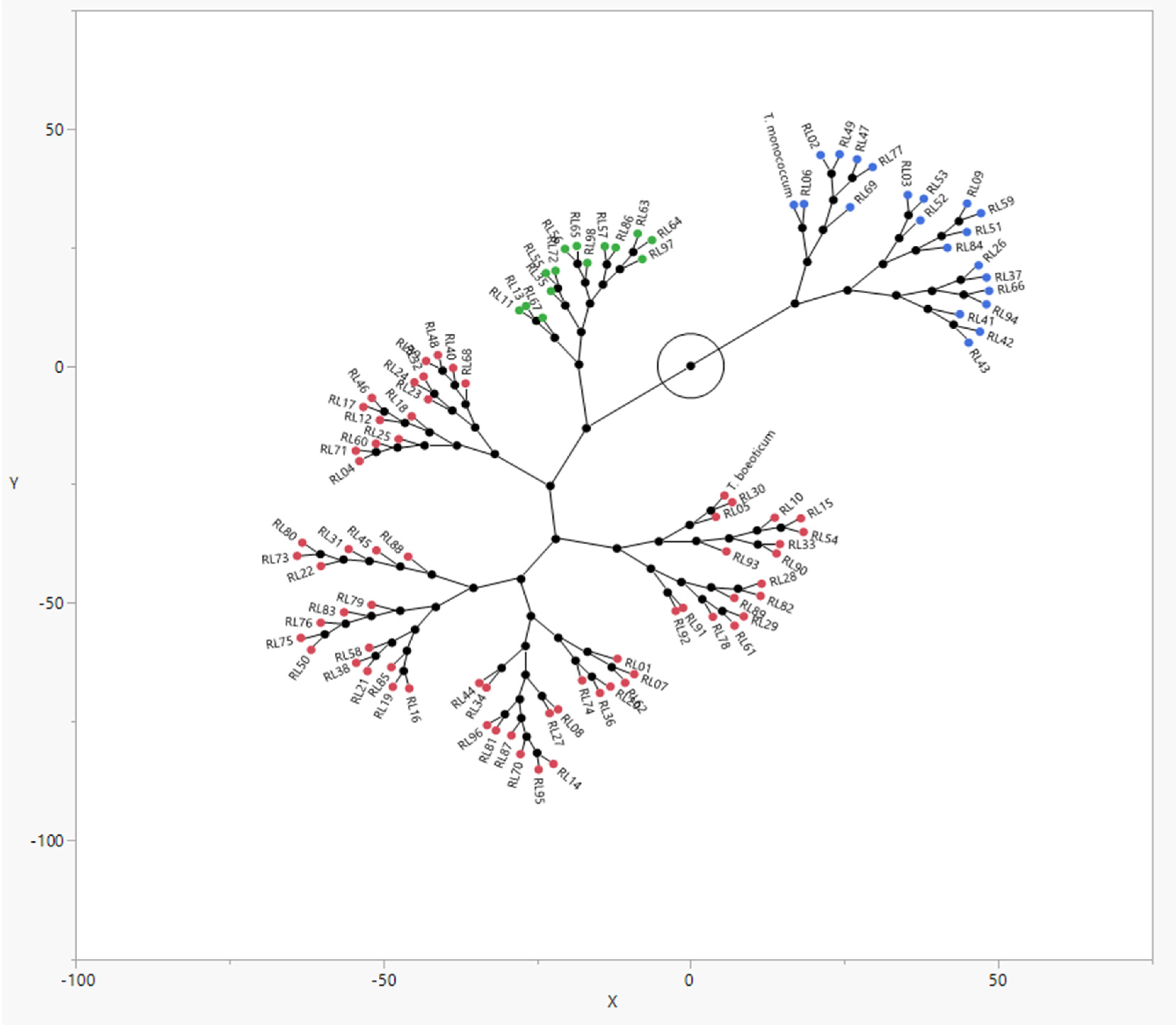
| Pop | PH | DH | FLL | FLD | PdL | Till | SpL | SpS | TBM | SWsp | NSS | GY | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | Parents | 122.2 | 130 | 12.6 | 0.81 | 46.6 | 36 | 19.4 | 16 | 26.25 | 2.34 | 37.00 | 15.90 |
| RILS | 118.0 | 128 | 13.2 | 0.67 | 48.0 | 36 | 16.1 | 17 | 42.78 | 2.39 | 36.24 | 15.28 | |
| Max | Parents | 132.0 | 133 | 13.4 | 0.96 | 53.0 | 38 | 20.6 | 19 | 27.00 | 2.75 | 42.00 | 27.20 |
| RILS | 158.2 | 161 | 21.1 | 0.97 | 66.6 | 58 | 20.7 | 28 | 86.00 | 3.78 | 64.00 | 28.90 | |
| Min | Parents | 112.4 | 127 | 11.8 | 0.66 | 40.1 | 34 | 18.1 | 13 | 25.50 | 1.93 | 32.00 | 4.60 |
| RILS | 88.9 | 108 | 7.1 | 0.45 | 36.8 | 23 | 11.2 | 10 | 10.83 | 1.22 | 18.00 | 2.50 | |
| SD | Parents | 9.80 | 3 | 0.8 | 0.15 | 6.5 | 2 | 1.3 | 3 | 0.75 | 0.41 | 5.00 | 11.30 |
| RILS | 14.0 | 11.0 | 3.0 | 0.11 | 6.1 | 8 | 2.4 | 4 | 19.10 | 0.52 | 9.18 | 7.47 |
| Trait | Source | DF | Sum of Square | F-Ratio | Log Worth | Summary Graph | p Value |
|---|---|---|---|---|---|---|---|
| PH | Gen. | 99 | 59,007.095 | 3.3718 | 12.730 |  | <0.0001 * |
| Rep. | 2 | 868.505 | 2.4566 | 1.054 | 0.0883 | ||
| DH | Gen. | 99 | 36,247.237 | 7.0711 | 30.775 |  | <0.0001 * |
| Rep. | 2 | 281.84700 | 2.7217 | 1.166 | 0.0682 | ||
| FLL | Gen. | 99 | 2658.4515 | 3.9113 | 15.757 |  | <0.0001 * |
| Rep. | 2 | 4.6242000 | 0.3368 | 0.146 | 0.7145 | ||
| FLD | Gen. | 99 | 3.6148147 | 3.1578 | 11.495 |  | <0.0001 * |
| Rep. | 2 | 0.0209040 | 0.9039 | 0.391 | 0.4066 | ||
| PdL | Gen. | 99 | 11,141.533 | 3.4734 | 13.310 |  | <0.0001 * |
| Rep. | 2 | 90.825000 | 1.4016 | 0.604 | 0.2486 | ||
| Till | Gen. | 99 | 18,548.319 | 3.7377 | 14.798 |  | <0.0001 * |
| Rep. | 2 | 217.09700 | 2.1655 | 0.930 | 0.1174 | ||
| SpL | Gen. | 99 | 1761.9510 | 3.2953 | 12.291 |  | <0.0001 * |
| Rep. | 2 | 32.275700 | 2.9880 | 1.278 | 0.0527 | ||
| SpS | Gen. | 99 | 5231.9644 | 5.8634 | 25.553 |  | <0.0001 * |
| Rep. | 2 | 43.038300 | 2.3875 | 1.025 | 0.0945 | ||
| TBM | Gen. | 99 | 108,816.51 | 6.5285 | 28.500 |  | <0.0001 * |
| Rep. | 2 | 727.57000 | 2.1607 | 0.928 | 0.1180 | ||
| SWsp | Gen. | 99 | 79.446467 | 4.0812 | 16.682 |  | <0.0001 * |
| Rep. | 2 | 0.0630190 | 0.1602 | 0.070 | 0.8520 | ||
| NSS | Gen. | 99 | 25,167.903 | 3.1422 | 11.404 |  | <0.0001 * |
| Rep. | 2 | 263.84700 | 1.6306 | 0.702 | 0.1984 | ||
| GY | Gen. | 99 | 17,190.157 | 7.7570 | 33.500 |  | <0.0001 * |
| Rep. | 2 | 111.46100 | 2.4897 | 1.068 | 0.0855 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sarfraz, Z.; Shah, M.M.; Iqbal, M.S.; Nazir, M.F.; Al-Ashkar, I.; Rehmani, M.I.A.; Shahid Iqbal, M.; Ullah, N.; El Sabagh, A. Rendering Multivariate Statistical Models for Genetic Diversity Assessment in A-Genome Diploid Wheat Population. Agronomy 2021, 11, 2339. https://doi.org/10.3390/agronomy11112339
Sarfraz Z, Shah MM, Iqbal MS, Nazir MF, Al-Ashkar I, Rehmani MIA, Shahid Iqbal M, Ullah N, El Sabagh A. Rendering Multivariate Statistical Models for Genetic Diversity Assessment in A-Genome Diploid Wheat Population. Agronomy. 2021; 11(11):2339. https://doi.org/10.3390/agronomy11112339
Chicago/Turabian StyleSarfraz, Zareen, Mohammad Maroof Shah, Muhammad Sajid Iqbal, Mian Faisal Nazir, Ibrahim Al-Ashkar, Muhammad Ishaq Asif Rehmani, Muhammad Shahid Iqbal, Najeeb Ullah, and Ayman El Sabagh. 2021. "Rendering Multivariate Statistical Models for Genetic Diversity Assessment in A-Genome Diploid Wheat Population" Agronomy 11, no. 11: 2339. https://doi.org/10.3390/agronomy11112339
APA StyleSarfraz, Z., Shah, M. M., Iqbal, M. S., Nazir, M. F., Al-Ashkar, I., Rehmani, M. I. A., Shahid Iqbal, M., Ullah, N., & El Sabagh, A. (2021). Rendering Multivariate Statistical Models for Genetic Diversity Assessment in A-Genome Diploid Wheat Population. Agronomy, 11(11), 2339. https://doi.org/10.3390/agronomy11112339









