Single-Species Artificial Grasslands Decrease Soil Multifunctionality in a Temperate Steppe on the Qinghai–Tibet Plateau
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Experiment Design
2.3. Soil Sampling and Analysis
2.4. Microbial DNA Extraction, Sequencing, and Data Processing
2.5. Assessing Soil Multifunctionality
2.6. Data Analysis
3. Results
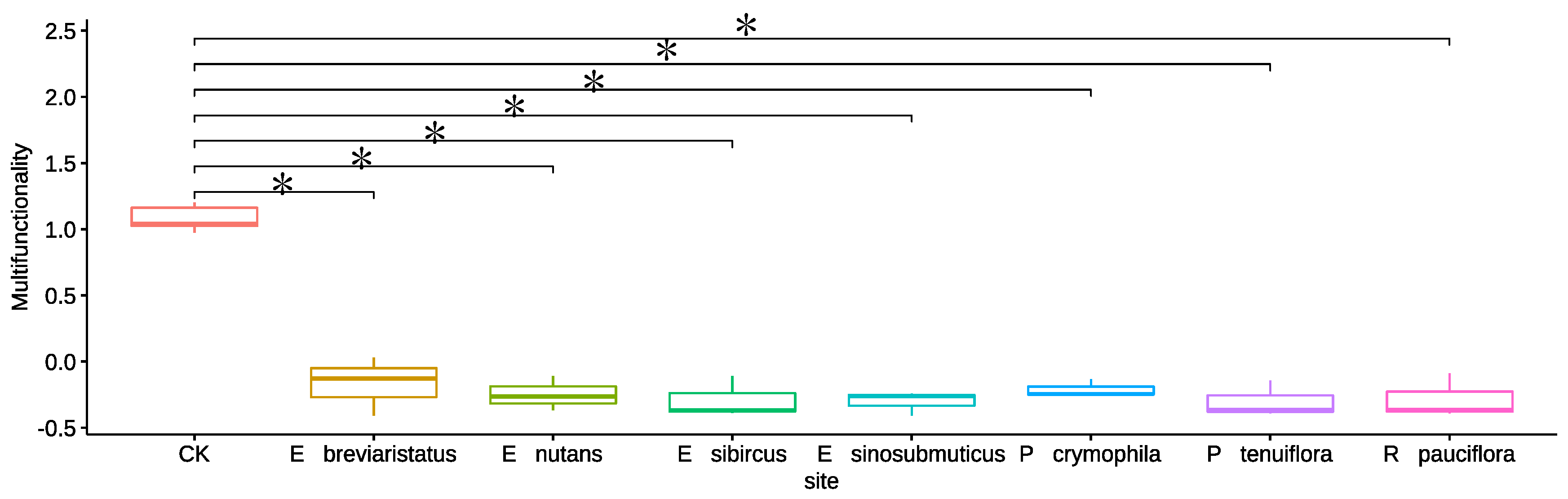
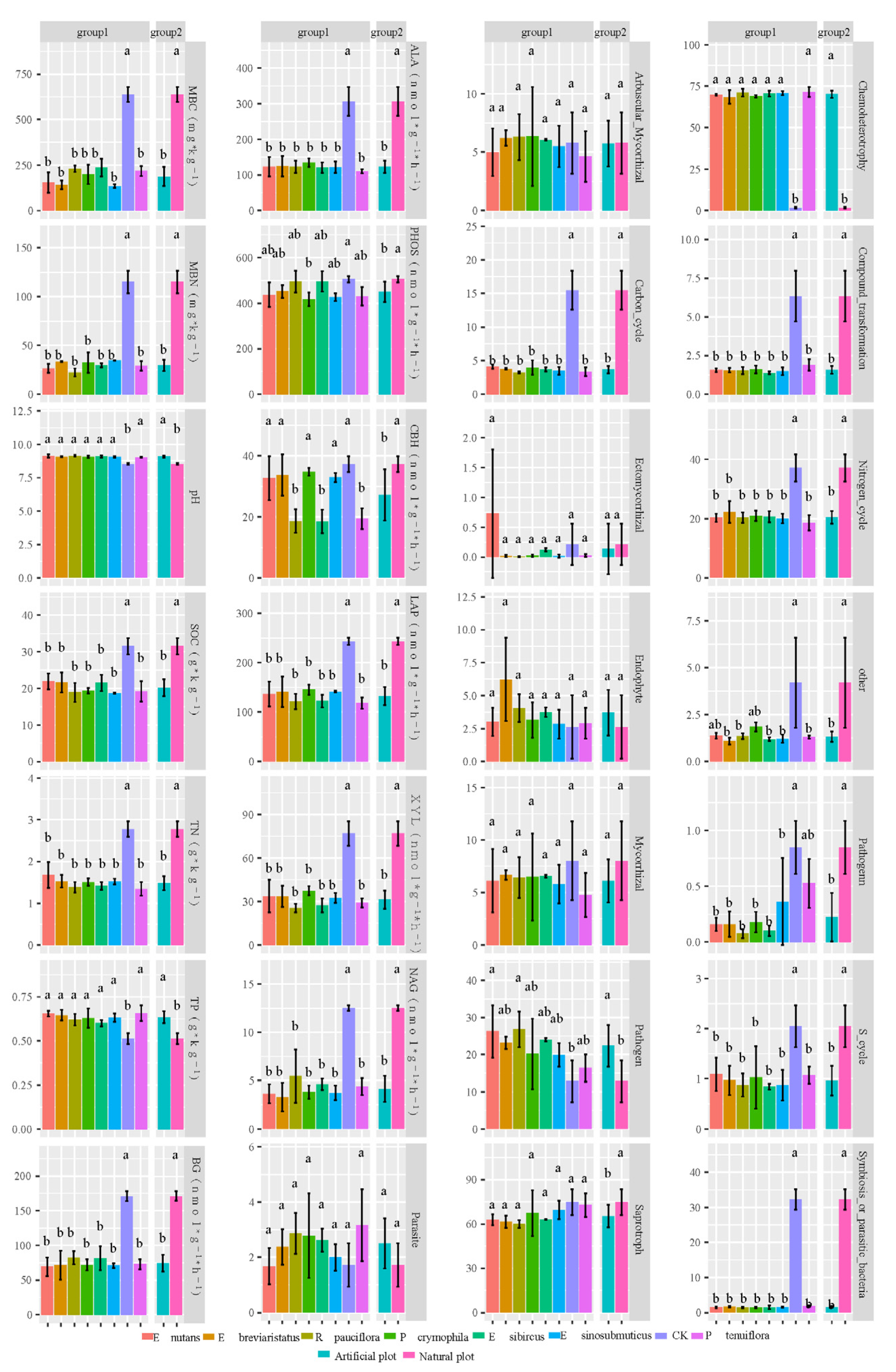
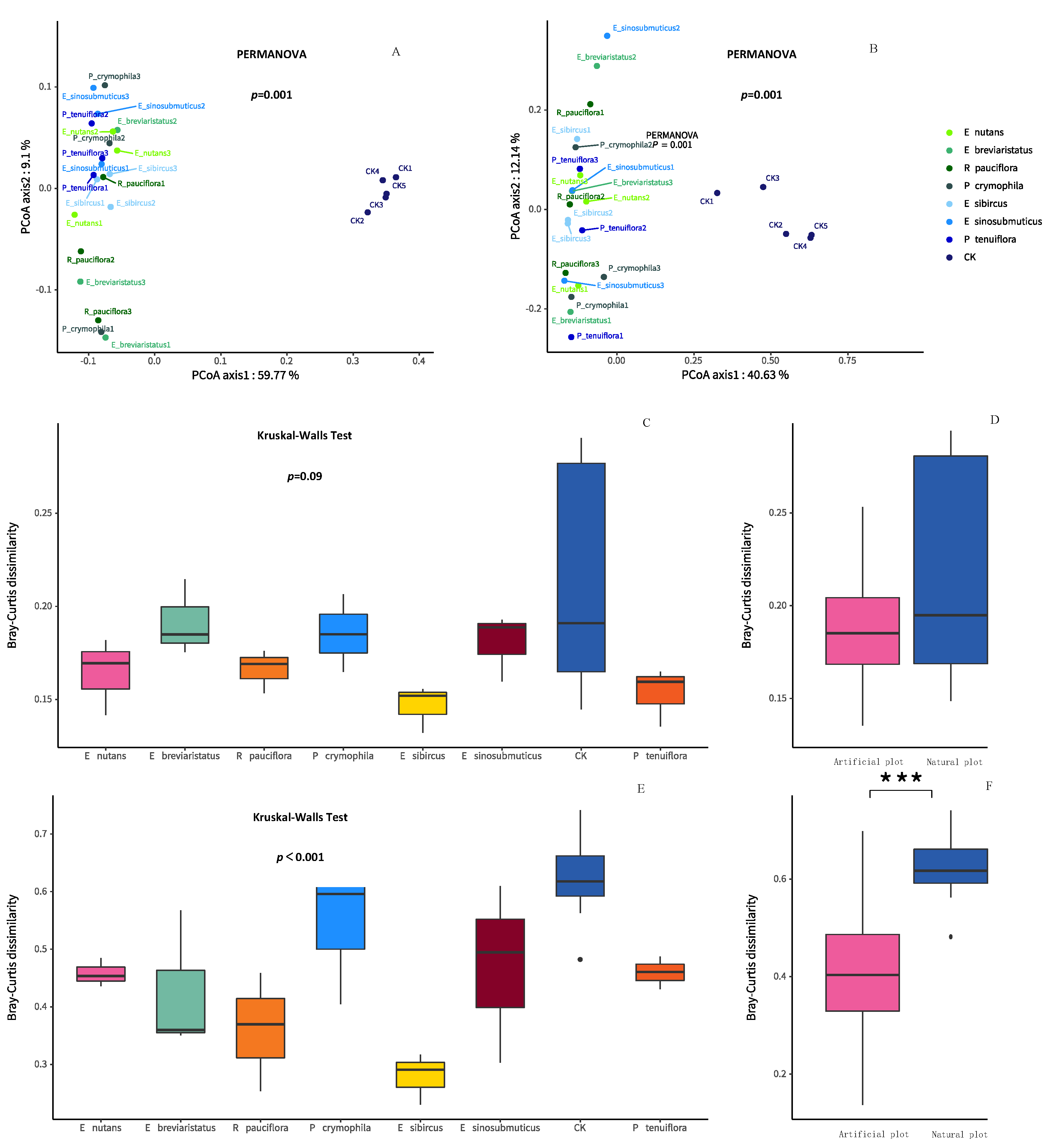
3.1. Impacts of Artificial Grasslands on Soil EMF, Single Functions, Abiotic Factors and Microbial Diversity of the Temperate Steppe
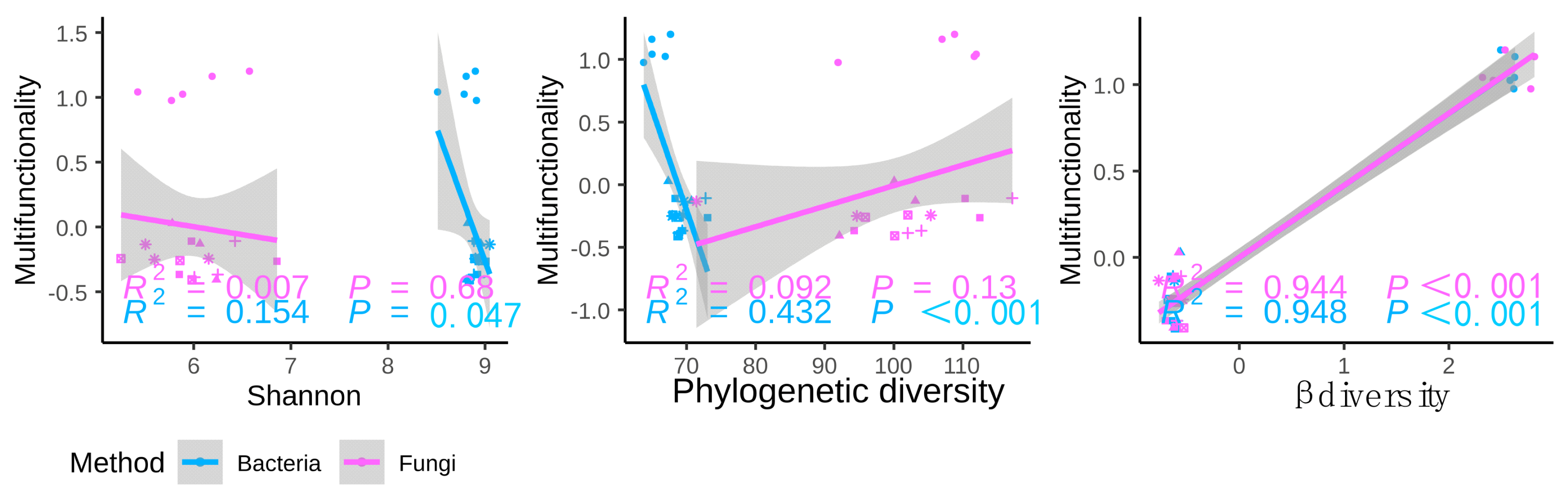
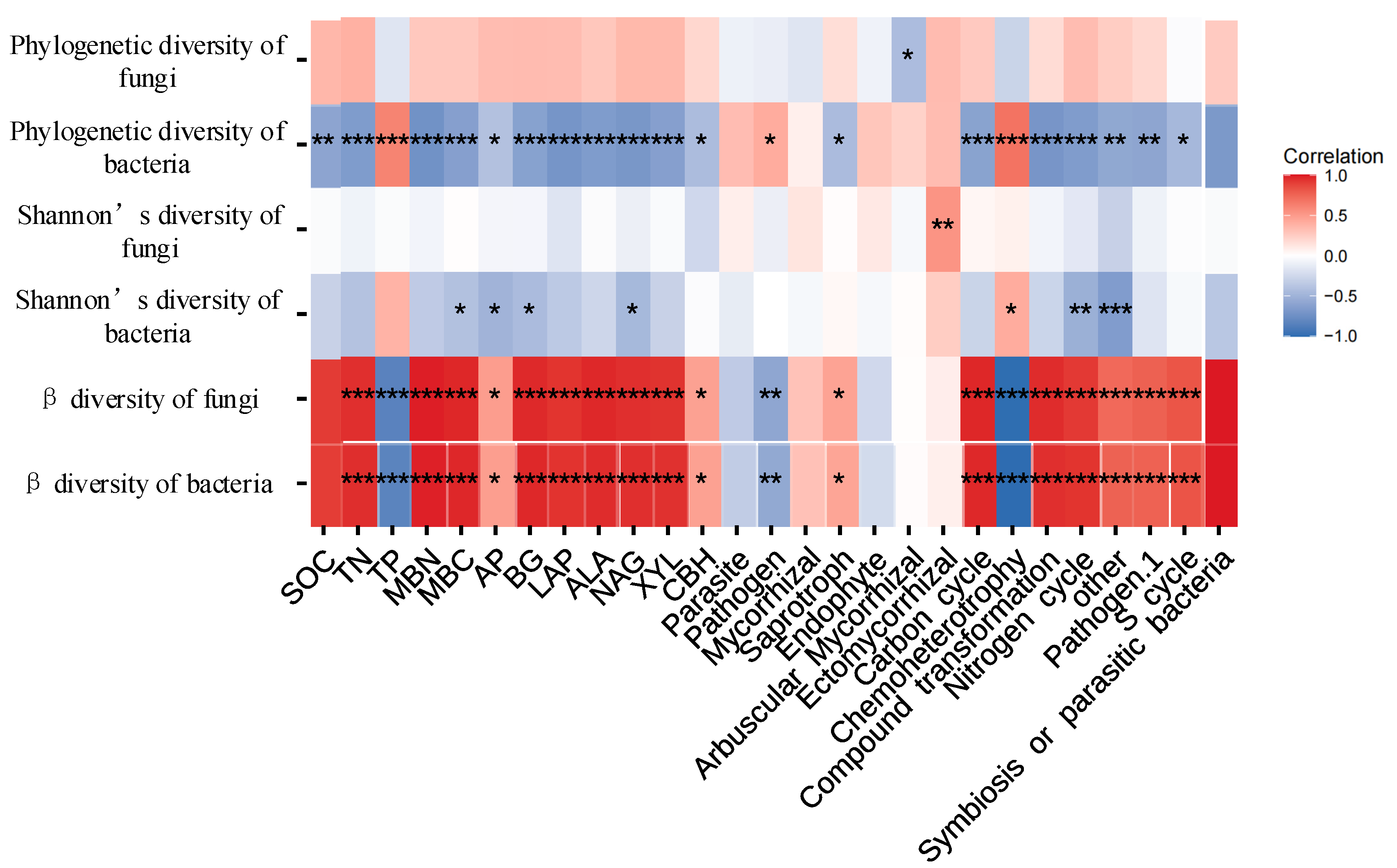
3.2. Relationships between Soil Microbial Diversity and Ecosystem Functionality
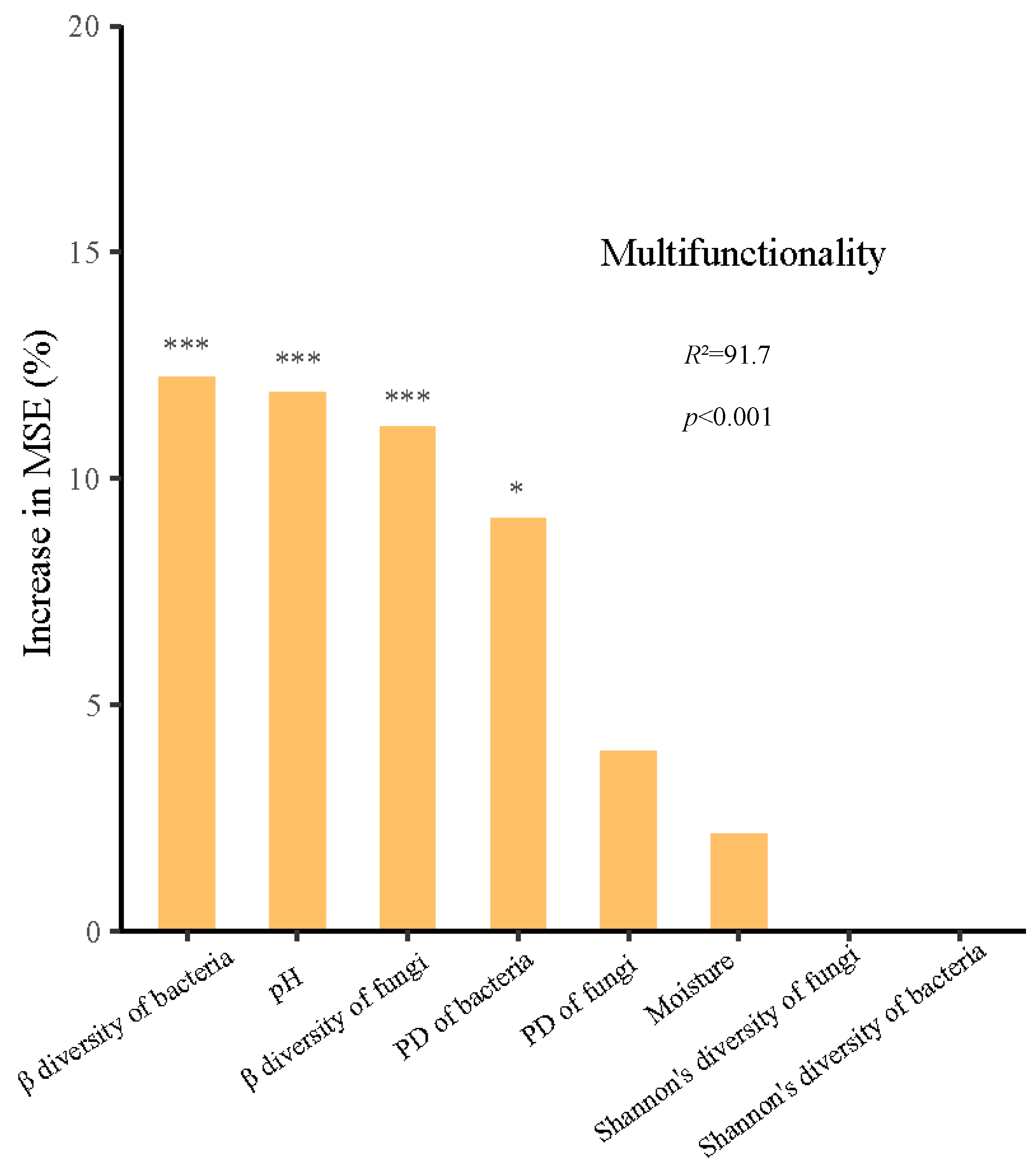
3.3. Possible Drivers of Soil EMF
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dong, S.; Sherman, R. Enhancing the resilience of coupled human and natural systems of alpine rangelands on the qinghai-tibetan plateau. Rangel. J. 2015, 37, i–iii. [Google Scholar] [CrossRef] [Green Version]
- Wei, D.; Liu, Y.; Wang, Y.; Wang, Y. Three-year study of co2 efflux and ch4/n2o fluxes at an alpine steppe site on the central tibetan plateau and their responses to simulated n deposition. Geoderma 2014, 232, 88–96. [Google Scholar] [CrossRef]
- Bai, Y.; Yu, Z.; Yang, Q.; Wan, H.; Huang, J.; Ji, B.; Li, A. Mechanisms regulating the productivity and stability of artificial grasslands in china: Issues, progress, and prospects. Chin. Sci. Bull. 2018, 63, 511–520. [Google Scholar]
- Yang, R.; Wu, P.; Wei, X. Effects of the transformation from natural alpine grassland to artificial oat grassland on the soil nematode communities. Acta Ecol. Sin. 2020, 40, 4903–4920. [Google Scholar]
- Wu, W.; Zhang, L.; Huang, X.; Yang, X.; Xue, L.; Liu, Y. Difference in soil microbial diversity in artificial grasslands of the northwest plateau of sichuan province. Acta Prataculturae Sin. 2019, 28, 29–41. [Google Scholar]
- Qiu, Y.; Wu, P.; Wei, X. Differences among three artificial grasslands in dynamics and community diversity of soil microarthropods. Acta Prataculturae Sin. 2019, 29, 21–32. [Google Scholar]
- Maestre, F.T.; Quero, J.L.; Gotelli, N.J.; Escudero, A.; Ochoa, V.; Delgado-Baquerizo, M.; García-Gómez, M.; Bowker, M.A.; Soliveres, S.; Escolar, C. Plant species richness and ecosystem multifunctionality in global drylands. Science 2012, 335, 214–218. [Google Scholar] [CrossRef] [Green Version]
- Li, K.; Zhang, H.; Li, X.; Wang, C.; Zhang, J.; Jiang, R.; Feng, G.; Liu, X.; Zuo, Y.; Yuan, H. Field management practices drive ecosystem multifunctionality in a smallholder-dominated agricultural system. Agric. Ecosyst. Environ. 2021, 313, 107389. [Google Scholar] [CrossRef]
- Zheng, Q.; Hu, Y.; Zhang, S.; Noll, L.; Böckle, T.; Dietrich, M.; Herbold, C.W.; Eichorst, S.A.; Woebken, D.; Richter, A. Soil multifunctionality is affected by the soil environment and by microbial community composition and diversity. Soil Biol. Biochem. 2019, 136, 107521. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Xu, T.; Cheng, J.; Wei, G.; Lin, Y. Above-and belowground biodiversity drives soil multifunctionality along a long-term grassland restoration chronosequence. Sci. Total Environ. 2021, 772, 145010. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Zhang, Q.; Buyantuev, A.; Liu, Q.; Niu, J. Plant functional β diversity is an important mediator of effects of aridity on soil multifunctionality. Sci. Total Environ. 2020, 726, 138529. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.-L.; Ding, J.; Zhu, D.; Hu, H.-W.; Delgado-Baquerizo, M.; Ma, Y.-B.; He, J.-Z.; Zhu, Y.-G. Rare microbial taxa as the major drivers of ecosystem multifunctionality in long-term fertilized soils. Soil Biol. Biochem. 2020, 141, 107686. [Google Scholar] [CrossRef]
- Delgado-Baquerizo, M.; Trivedi, P.; Trivedi, C.; Eldridge, D.J.; Reich, P.B.; Jeffries, T.C.; Singh, B.K. Microbial richness and composition independently drive soil multifunctionality. Funct. Ecol. 2017, 31, 2330–2343. [Google Scholar] [CrossRef] [Green Version]
- Luo, G.; Rensing, C.; Chen, H.; Liu, M.; Wang, M.; Guo, S.; Ling, N.; Shen, Q. Deciphering the associations between soil microbial diversity and ecosystem multifunctionality driven by long-term fertilization management. Funct. Ecol. 2018, 32, 1103–1116. [Google Scholar] [CrossRef]
- Yang, Y.; Cheng, H.; Gao, H.; An, S. Response and driving factors of soil microbial diversity related to global nitrogen addition. Land Degrad. Dev. 2020, 31, 190–204. [Google Scholar] [CrossRef]
- Muñoz-Arenas, L.C.; Fusaro, C.; Hernández-Guzmán, M.; Dendooven, L.; Estrada-Torres, A.; Navarro-Noya, Y.E. Soil microbial diversity drops with land-use change in a high mountain temperate forest: A metagenomics survey. Environ. Microbiol. Rep. 2020, 12, 185–194. [Google Scholar] [CrossRef]
- Yousfi, S.; Marín, J.; Parra, L.; Lloret, J.; Mauri, P.V. A rhizogenic biostimulant effect on soil fertility and roots growth of turfgrass. Agronomy 2021, 11, 573. [Google Scholar] [CrossRef]
- Du, J.; Li, Y.; Ur-Rehman, S.; Mukhtar, I.; Yin, Z.; Dong, H.; Wang, H.; Zhang, X.; Gao, Z.; Zhao, X.; et al. Synergistically promoting plant health by harnessing synthetic microbial communities and prebiotics. iScience 2021, 24, 102918. [Google Scholar] [CrossRef]
- Narwani, A.; Matthews, B.; Fox, J.; Venail, P. Using phylogenetics in community assembly and ecosystem functioning research. Funct. Ecol. 2015, 29, 589–591. [Google Scholar] [CrossRef] [Green Version]
- Flynn, D.F.; Mirotchnick, N.; Jain, M.; Palmer, M.I.; Naeem, S. Functional and phylogenetic diversity as predictors of biodiversity–ecosystem-function relationships. Ecology 2011, 92, 1573–1581. [Google Scholar] [CrossRef] [Green Version]
- Srivastava, D.S.; Cadotte, M.W.; MacDonald, A.A.M.; Marushia, R.G.; Mirotchnick, N. Phylogenetic diversity and the functioning of ecosystems. Ecol. Lett. 2012, 15, 637–648. [Google Scholar] [CrossRef] [PubMed]
- Mori, A.S.; Isbell, F.; Seidl, R. B-diversity, community assembly, and ecosystem functioning. Trends Ecol. Evol. 2018, 33, 549–564. [Google Scholar] [CrossRef]
- Xu, Y.; Dong, S.; Gao, X.; Yang, M.; Li, S.; Shen, H.; Xiao, J.; Han, Y.; Zhang, J.; Li, Y. Aboveground community composition and soil moisture play determining roles in restoring ecosystem multifunctionality of alpine steppe on qinghai-tibetan plateau. Agric. Ecosyst. Environ. 2021, 305, 107163. [Google Scholar] [CrossRef]
- Winfree, R.; Fox, J.W.; Williams, N.M.; Reilly, J.R.; Cariveau, D.P. Abundance of common species, not species richness, drives delivery of a real-world ecosystem service. Ecol. Lett. 2015, 18, 626–635. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Wang, W.; Zhou, H.; Mao, X.; Liu, Y. Dynamic of the soil soluble nitrogen and productivity of artificial pasture on the qingzang plateau. Chin. J. Plant. Ecol. 2021, 45, 562–572. [Google Scholar] [CrossRef]
- Chen, W.; Zhou, H.; Wu, Y.; Wang, J.; Zhao, Z.; Li, Y.; Qiao, L.; Chen, K.; Liu, G.; Xue, S. Direct and indirect influences of long-term fertilization on microbial carbon and nitrogen cycles in an alpine grassland. Soil Biol. Biochem. 2020, 149, 107922. [Google Scholar] [CrossRef]
- Nelson, D.; Sommers, L.E. Total carbon, organic carbon, and organic matter. Methods Soil Anal. Part 2 Chem. Microbiol. Prop. 1983, 9, 539–579. [Google Scholar]
- Bremner, J.; Mulvaney, C. Nitrogen-Total 1. Methods of Soil Analysis. Part 2. Chemical and Microbiological Properties, (Methodsofsoilan2); American Society of Agronomy, Soil Science Society of America: Madison, WI, USA, 1982. [Google Scholar]
- Olsen, S. Anion resin extractable phosphorus. Methods Soil Anal. 1982, 2, 423–424. [Google Scholar]
- German, D.P.; Weintraub, M.N.; Grandy, A.S.; Lauber, C.L.; Rinkes, Z.L.; Allison, S.D. Optimization of hydrolytic and oxidative enzyme methods for ecosystem studies. Soil Biol. Biochem. 2011, 43, 1387–1397. [Google Scholar] [CrossRef]
- Wang, J.; Wang, X.; Liu, G.; Zhang, C.; Wang, G. Bacterial richness is negatively related to potential soil multifunctionality in a degraded alpine meadow. Ecol. Indic. 2021, 121, 106996. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Mills, D.A.; Caporaso, J.G. Quality-filtering vastly improves diversity estimates from illumina amplicon sequencing. Nat. Methods 2013, 10, 57–59. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. Uchime improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Yu, L.; Zhang, J.; Zhang, X.; Xue, Y.; Liu, J.; Zou, X. Characterization of the core microbiome in tobacco leaves during aging. MicrobiologyOpen 2020, 9, e984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. Funguild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Kembel, S.W.; Cowan, P.D.; Helmus, M.R.; Cornwell, W.K.; Morlon, H.; Ackerly, D.D.; Blomberg, S.P.; Webb, C.O. Picante: R tools for integrating phylogenies and ecology. Bioinformatics 2010, 26, 1463–1464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Byrnes, J.E.; Gamfeldt, L.; Isbell, F.; Lefcheck, J.S.; Griffin, J.N.; Hector, A.; Cardinale, B.J.; Hooper, D.U.; Dee, L.E.; Emmett Duffy, J. Investigating the relationship between biodiversity and ecosystem multifunctionality: Challenges and solutions. Methods Ecol. Evol. 2014, 5, 111–124. [Google Scholar] [CrossRef] [Green Version]
- Hothorn, T.; Bretz, F.; Westfall, P. Simultaneous inference in general parametric models. Biom. J. J. Math. Methods Biosci. 2008, 50, 346–363. [Google Scholar] [CrossRef] [Green Version]
- Dixon, P. Vegan, a package of r functions for community ecology. J. Veg. Sci. 2003, 14, 927–930. [Google Scholar] [CrossRef]
- Liaw, A.; Wiener, M. Classification and regression by randomforest. R News 2002, 2, 18–22. [Google Scholar]
- Gómez-Rubio, V. Ggplot2-elegant graphics for data analysis. J. Stat. Softw. 2017, 77, 1–3. [Google Scholar] [CrossRef] [Green Version]
- Guan, H.; Fan, J.; Li, Y. The impact of different introduced artificial grassland species combinations on community biomass and species diversity in temperate steppe of the qinghai-tibetan plateau. Acta Prataculturae Sin. 2019, 28, 192–201. [Google Scholar]
- Varallyay, G. Role of soil multifunctionality in sustainable development. Soil Water Res. 2010, 5, 102–107. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Ji, C.; Ma, W.; Wang, S.; Wang, S.; Han, W.; Mohammat, A.; Robinson, D.; Smith, P. Significant soil acidification across northern china’s grasslands during 1980s–2000s. Glob. Chang. Biol. 2012, 18, 2292–2300. [Google Scholar] [CrossRef]
- Treseder, K.K. Nitrogen additions and microbial biomass: A meta-analysis of ecosystem studies. Ecol. Lett. 2008, 11, 1111–1120. [Google Scholar] [CrossRef] [Green Version]
- Jiao, S.; Peng, Z.; Qi, J.; Gao, J.; Wei, G. Linking bacterial-fungal relationships to microbial diversity and soil nutrient cycling. Msystems 2021, 6, e01052-01020. [Google Scholar] [CrossRef]
- Chen, W.; Zhou, H.; Wu, Y.; Li, Y.; Qiao, L.; Wang, J.; Zhai, J.; Song, Y.; Zhao, Z.; Zhang, Z. Plant-mediated effects of long-term warming on soil microorganisms on the qinghai-tibet plateau. CATENA 2021, 204, 105391. [Google Scholar] [CrossRef]
- Wang, H.; Bu, L.; Tian, J.; Hu, Y.; Song, F.; Chen, C.; Zhang, Y.; Wei, G. Particular microbial clades rather than total microbial diversity best predict the vertical profile variation in soil multifunctionality in desert ecosystems. Land Degrad. Dev. 2021, 32, 2157–2168. [Google Scholar] [CrossRef]
- Cadotte, M.W. Experimental evidence that evolutionarily diverse assemblages result in higher productivity. Proc. Natl. Acad. Sci. USA 2013, 110, 8996. [Google Scholar] [CrossRef] [Green Version]
- Venail, P.; Gross, K.; Oakley, T.H.; Narwani, A.; Allan, E.; Flombaum, P.; Isbell, F.; Joshi, J.; Reich, P.B.; Tilman, D.; et al. Species richness, but not phylogenetic diversity, influences community biomass production and temporal stability in a re-examination of 16 grassland biodiversity studies. Funct. Ecol. 2015, 29, 615–626. [Google Scholar] [CrossRef] [Green Version]
- Becker, J.; Eisenhauer, N.; Scheu, S.; Jousset, A. Increasing antagonistic interactions cause bacterial communities to collapse at high diversity. Ecol. Lett. 2012, 15, 468–474. [Google Scholar] [CrossRef]
- Louca, S.; Polz, M.F.; Mazel, F.; Albright, M.B.; Huber, J.A.; O’Connor, M.I.; Ackermann, M.; Hahn, A.S.; Srivastava, D.S.; Crowe, S.A. Function and functional redundancy in microbial systems. Nat. Ecol. Evol. 2018, 2, 936–943. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, K.; Zhou, H.; Lu, B.; Wu, Y.; Wang, J.; Zhao, Z.; Li, Y.; Wang, M.; Zhang, Y.; Chen, W.; et al. Single-Species Artificial Grasslands Decrease Soil Multifunctionality in a Temperate Steppe on the Qinghai–Tibet Plateau. Agronomy 2021, 11, 2092. https://doi.org/10.3390/agronomy11112092
Chen K, Zhou H, Lu B, Wu Y, Wang J, Zhao Z, Li Y, Wang M, Zhang Y, Chen W, et al. Single-Species Artificial Grasslands Decrease Soil Multifunctionality in a Temperate Steppe on the Qinghai–Tibet Plateau. Agronomy. 2021; 11(11):2092. https://doi.org/10.3390/agronomy11112092
Chicago/Turabian StyleChen, Kelu, Huakun Zhou, Bingbing Lu, Yang Wu, Jie Wang, Ziwen Zhao, Yuanze Li, Mei Wang, Yue Zhang, Wenjing Chen, and et al. 2021. "Single-Species Artificial Grasslands Decrease Soil Multifunctionality in a Temperate Steppe on the Qinghai–Tibet Plateau" Agronomy 11, no. 11: 2092. https://doi.org/10.3390/agronomy11112092
APA StyleChen, K., Zhou, H., Lu, B., Wu, Y., Wang, J., Zhao, Z., Li, Y., Wang, M., Zhang, Y., Chen, W., Liu, G., & Xue, S. (2021). Single-Species Artificial Grasslands Decrease Soil Multifunctionality in a Temperate Steppe on the Qinghai–Tibet Plateau. Agronomy, 11(11), 2092. https://doi.org/10.3390/agronomy11112092





