Genetic Diversity and Molecular Characterization of Worldwide Prairie Grass (Bromus catharticus Vahl) Accessions Using SRAP Markers
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Samples and DNA Extraction
2.2. SRAP Analysis
2.3. Data Analysis
3. Results
3.1. SRAP Genetic Diversity and Polymorphism in the Germplasm Collection
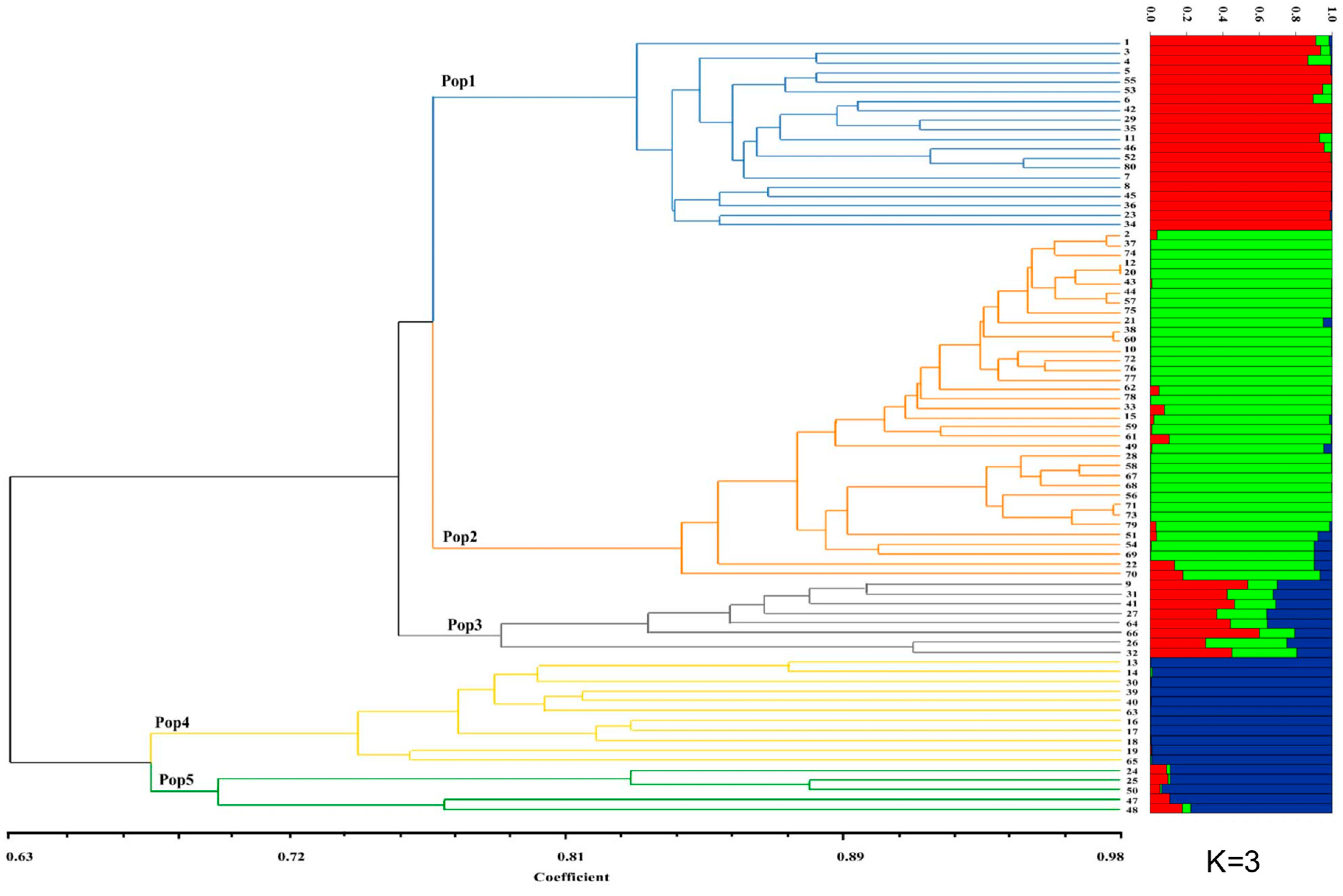
3.2. Hierarchal Clustering, Pcoa, Neighbor-Net and Population Structure Analysis
3.3. Genetic Structure of Inferred Clustering Groups with UPGMA and the Geographic Groups
4. Discussion
4.1. Genetic Polymorphisms and Discriminating Capacity of the SRAP Primers
4.2. Genetic Relatedness and Population Structure
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kole, C. Wild Crop Relatives: Genomic and Breeding Resources, Millets and Grasses; Springer: Berlin/Heidelberg, Germany, 2011; pp. 15–30. [Google Scholar]
- Nizam, I.; Gulcu, R.; Tuna, G.S.; Tuna, M. Determination of nuclear DNA content and ploidy of some Bromus l. germplasm by flow cytometery. Pak. J. Bot. 2020, 52, 909–913. [Google Scholar] [CrossRef]
- Abbott, L.; Filippini, S.; Delfino, H.; Pistorale, S. Stability analysis of forage production in Bromus catharticus (prairie grass) using three methodologies. Int. J. Agric. Nat. Resour. 2012, 39, 331–338. [Google Scholar] [CrossRef]
- Muzafar, I.; Khuroo, A.; Mehraj, G.; Hamid, M.; Rashid, I. Bromus catharticus Vahl (Poaceae): A new plant record for Kashmir Himalaya, India. Check List 2016, 12, 1–5. [Google Scholar] [CrossRef]
- Aulicino, M.B.; Arturi, M.J. Phenotypic diversity in Argentinian populations of Bromus catharticus (Poaceae). Genetic and environmental components of quantitative traits. N. Z. J. Bot. 2002, 40, 223–234. [Google Scholar] [CrossRef]
- Arturi, M.J.; Aulicino, M.B. Regional variation in Argentinean populations of Bromus catharticus (Poaceae) as measured by morphological divergence associated with environmental conditions. In Anales del Jardín Botánico de Madrid; CSIC-Real Jardín Botánico: Madrid, Spain, 2008; Volume 65, pp. 135–147. [Google Scholar]
- Rosso, B.; Pagano, E.; Rimieri, P.; Ríos, R. Characteristics of Bromus cartharticus Vahl (Poaceae) natural populations collected in the central area of Argentina. Sci. Agric. 2009, 66, 276–279. [Google Scholar] [CrossRef]
- Wolff, R.; Abbott, L.; Pistorale, S. Reproductive behavior of Bromus catharticus Vahl. (Cebadilla criolla) in natural and cultivated populations. J. Genet. Breed. 1996, 50, 121–128. [Google Scholar]
- Ma, X.; Zhou, C.J.; Zhang, C.L.; Sun, M.; Guo, Z.H.; Wang, X.L.; Zhang, J.B. Patterna of Morphological Variation and Agronomic Traits in A Worldwide Sample of Prairie Grass Germplasm. Acta Agrestia Ainica 2015, 23, 1048–1056. [Google Scholar]
- Massa, A.N.; Larson, S.R.; Jensen, K.B.; Hole, D.J. AFLP variation in Bromus section Ceratochloa germplasm of Patagonia. Crop Sci. 2000, 41, 136–144. [Google Scholar] [CrossRef]
- Puecher, D.I.; Robredo, C.G.; Rios, R.D.; Rimieri, P. Genetic variability measures among Bromus catharticus Vahl. populations and cultivars with RAPD and AFLP markers. Euphytica 2001, 121, 229–236. [Google Scholar] [CrossRef]
- Sun, M.; Dong, Z.X.; Yang, J.; Wu, W.D.; Zhang, C.; Zhang, J.; Zhao, J.; Xiong, Y.; Jia, S.; Ma, X. Transcriptomic resources for prairie grass (Bromus catharticus): Expressed transcripts, tissue-specific genes, and identification and validation of EST-SSR markers. BMC Plant Biol. 2021, 21, 264. [Google Scholar] [CrossRef]
- Cuyeu, R.; Pagano, E.; Rosso, B.; Soto, G.; Ayub, N.D. The genetic diversity of wild rescuegrass is associated with precipitation levels. J. Genet. 2014, 94, 1–6. [Google Scholar] [CrossRef][Green Version]
- Li, G.; Quiros, C.F. Sequence-related amplified polymorphism (SRAP), a new marker system based on a simple PCR reaction: Its application to mapping and gene tagging in Brassica. Theor. Appl. Genet. 2001, 103, 455–461. [Google Scholar] [CrossRef]
- Robarts, D.W.H.; Wolfe, A.D. Sequence-related amplified polymorphism (SRAP) markers: A potential resource for studies in plant molecular biology1. Appl. Plant Sci. 2014, 2, 1400017. [Google Scholar] [CrossRef]
- Yu, Q.; Liu, Q.; Xiong, Y.; Xiong, Y.; Dong, Z.; Yang, J.; Liu, W.; Ma, X.; Bai, S. Genetic diversity and population divergence of a rare, endemic grass (Elymus breviaristatus) in the southeastern Qinghai-Tibetan plateau. Sustainability 2019, 11, 5863. [Google Scholar] [CrossRef]
- Luo, Y.; Zhang, X.; Xu, J.; Zheng, Y.; Pu, S.; Duan, Z.; Li, Z.; Liu, G.; Chen, J.; Wang, Z. Phenotypic and molecular marker analysis uncovers the genetic diversity of the grass Stenotaphrum secundatum. BMC Genet. 2020, 21, 1–12. [Google Scholar] [CrossRef]
- Wu, F.; Chen, J.; Wang, J.; Wang, X.; Lu, Y.; Ning, Y.; Li, Y. Intra-population genetic diversity of Buchloe dactyloides (Nutt.) Engelm (buffalograss) determined using morphological traits and sequence-related amplified polymorphism markers. 3 Biotech 2019, 9, 1–8. [Google Scholar] [CrossRef]
- Zeng, B.; Zhang, X.Q.; Lan, Y.; Yang, W.Y. Evaluation of genetic diversity and relationships in orchardgrass (Dactylis glomerata L.) germplasm based on SRAP markers. Can. J. Plant Sci. 2008, 88, 53–60. [Google Scholar] [CrossRef]
- Zheng, Y.; Xu, S.; Liu, J.; Zhao, Y.; Liu, J. Genetic diversity and population structure of Chinese natural bermudagrass [Cynodon dactylon (L.) Pers.] germplasm based on SRAP markers. PLoS ONE 2017, 12, e0177508. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, N.C.; Hauvermale, A.L.; Dhingra, A.; Burke, I.C. Population structure and genetic diversity of Bromus tectorum within the small grain production region of the pacific northwest. Ecol. Evol. 2017, 7, 8316–8328. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.B.; Coulman, B.E.; Ferdinandez, Y.; Cayouette, J.; Peterson, P.M. Genetic diversity of fringed brome (Bromus ciliatus) as determined by amplified fragment length polymorphism. Can. J. Bot. 2005, 83, 1322–1328. [Google Scholar] [CrossRef]
- Gu, X.Y.; Guo, Z.; Ma, X.; Bai, S.; Zhang, X.; Zhang, C.; Chen, S.; Peng, Y.; Yan, Y.; Huang, L.; et al. Population genetic variability and structure of Elymus breviaristatus (Poaceae: Triticeae) endemic to Qinghai–Tibetan Plateau inferred from SSR markers. Biochem. Syst. Ecol. 2015, 58, 247–256. [Google Scholar] [CrossRef]
- Powell, W.; Morgante, M.; Andre, C.; Hanafey, M.; Vogel, J.; Tingey, S.; Rafalski, A. The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol. Breed. 1996, 2, 225–238. [Google Scholar] [CrossRef]
- Prevost, A.; Wilkinson, M.J. A new system of comparing PCR primers applied to ISSR fingerprinting of potato cultivars. Appl. Genet. 1999, 98, 107–112. [Google Scholar] [CrossRef]
- Amiryousefi, A.; Hyvonen, J.; Poczai, P. iMEC: Online Marker Efficiency Calculator. Appl. Plant Sci. 2018, 6, e1159. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Zeller, K.A.; Creech, T.G.; Millette, K.L.; Crowhurst, R.S.; Long, R.A.; Wagner, H.H.; Balkenhol, N.; Landguth, E.L. Using simulations to evaluate Mantel-based methods for assessing landscape resistance to gene flow. Ecol. Evol. 2016, 6, 4115–4128. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.X.; Feng, J.; Fu, W.J.; Li, X.Y.; Li, B. Deciphering of microbial community and antibiotic resistance genes in activated sludge reactors under high selective pressure of different antibiotics. Water Res. 2019, 151, 388–402. [Google Scholar] [CrossRef] [PubMed]
- Huson, D.H.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Jakobsson, M.; Rosenberg, N.A. CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 2007, 23, 1801–1806. [Google Scholar] [CrossRef]
- Ma, M.; Wang, T.; Lu, B. Assessment of genetic diversity in Amomum tsao-ko Crevost & Lemarie, an important medicine food homologous crop from Southwest China using SRAP and ISSR markers. Genet. Resour. Crop. Evol. 2021, 20, 1–13. [Google Scholar]
- Hassan, R.; Waheed, M.Q.; Shokat, S.; Rehman-Arif, M.A.; Tariq, R.; Arif, M.; Arif, A. Estimation of genomic diversity using sequence related amplified polymorphism (SRAP) markers in a mini core collection of wheat germplasm from Pakistan. Cereal Res. Commun. 2020, 48, 33–40. [Google Scholar] [CrossRef]
- Gao, S.; Cong, R.; Gao, L.; Zhu, Y.; Meng, Y.; Zhou, Y. Genetic diversity analysis of phenotypic character and SRAP molecular markers in 45 tree peony cultivars. Rev. Bras. Botânica 2020, 43, 291–302. [Google Scholar] [CrossRef]
- Li, R.F.; Ding, H.; Wang, C.; Lu, L.; Zhang, X. Genetic Diversity of Clover by SRAP. Agric. Biotechnol. 2019, 8, 6–8. [Google Scholar]
- Bolc, P.; Łapiński, B.; Podyma, W. Genetic Diversity and Population Structure of Algerian Endemic Plant Species Avena macrostachya Bal. ex Cross. et Durieu. Agronomy 2020, 10, 1984. [Google Scholar] [CrossRef]
- Xie, X.M.; Zhou, F.; Zhang, X.Q.; Zhang, J.M. Genetic variability and relationship between MT-1 elephant grass and closely related cultivars assessed by SRAP markers. J. Genet. 2009, 88, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Nybom, H.; Bartish, I.V. Effects of life history traits and sampling strategies on genetic diversity estimates obtained with RAPD markers in plants. Perspect. Plant Ecol. Evol. Syst. 2000, 3/2, 93–114. [Google Scholar] [CrossRef]
- Varshney, R.K.; Chabane, K.; Hendre, P.S.; Aggarwal, R.K.; Graner, A. Comparative assessment of EST-SSR, EST-SNP and AFLP markers for evaluation of genetic diversity and conservation of genetic resources using wild, cultivated and elite barleys. Plant Sci. 2007, 173, 638–649. [Google Scholar] [CrossRef]
- Sun, X.; Xie, Y.; Bi, Y.; Liu, J.; Amombo, E.; Hu, T.; Fu, J. Comparative study of diversity based on heat tolerant-related morpho-physiological traits and molecular markers in tall fescue accessions. Sci. Rep. 2015, 5, 18213. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
| Primer Group | TNB | NPB | PBB | DP | H | PIC | RP | MI |
|---|---|---|---|---|---|---|---|---|
| Me2 + em3 | 7 | 6 | 85.7% | 0.607 | 0.468 | 0.425 | 4.325 | 2.550 |
| Me3 + em2 | 6 | 3 | 50.0% | 0.913 | 0.417 | 0.210 | 0.775 | 0.630 |
| Me4 + em1 | 10 | 7 | 70.0% | 0.766 | 0.499 | 0.337 | 3.175 | 2.359 |
| Me4 + em8 | 11 | 11 | 100.0% | 0.854 | 0.472 | 0.350 | 5.350 | 3.850 |
| Me4 + em16 | 11 | 9 | 81.8% | 0.779 | 0.498 | 0.376 | 4.925 | 3.384 |
| Me4 + em20 | 9 | 9 | 100.0% | 0.952 | 0.343 | 0.229 | 2.750 | 2.061 |
| Me5 + em1 | 9 | 8 | 88.9% | 0.772 | 0.499 | 0.328 | 3.750 | 2.624 |
| Me6 + em6 | 12 | 8 | 66.7% | 0.891 | 0.443 | 0.394 | 5.100 | 3.152 |
| Me7 + em7 | 10 | 10 | 100.0% | 0.867 | 0.464 | 0.349 | 5.450 | 3.490 |
| Me7 + em15 | 6 | 4 | 66.7% | 0.532 | 0.432 | 0.218 | 1.025 | 0.872 |
| Me7 + em19 | 10 | 8 | 80.0% | 0.772 | 0.499 | 0.262 | 2.700 | 2.096 |
| Me8 + em3 | 3 | 3 | 100.0% | 0.451 | 0.383 | 0.348 | 1.550 | 1.044 |
| Me8 + em7 | 9 | 7 | 77.8% | 0.655 | 0.485 | 0.344 | 3.575 | 2.408 |
| Me8 + em16 | 5 | 2 | 40.0% | 0.202 | 0.190 | 0.184 | 0.425 | 0.368 |
| Me9 + em1 | 13 | 7 | 53.8% | 0.766 | 0.499 | 0.227 | 1.925 | 1.589 |
| Me9 + em20 | 14 | 13 | 92.9% | 0.629 | 0.476 | 0.331 | 5.850 | 4.303 |
| Me10 + em9 | 15 | 11 | 73.3% | 0.866 | 0.465 | 0.281 | 4.375 | 3.091 |
| Me10 + em16 | 8 | 6 | 75.0% | 0.528 | 0.430 | 0.399 | 3.450 | 2.394 |
| Me11 + em3 | 11 | 10 | 90.9% | 0.812 | 0.491 | 0.354 | 5.375 | 3.540 |
| Me11 + em6 | 13 | 10 | 76.9% | 0.613 | 0.470 | 0.355 | 5.500 | 3.550 |
| Me11 + em8 | 10 | 7 | 70.0% | 0.818 | 0.489 | 0.276 | 2.575 | 1.932 |
| Me12 + em10 | 7 | 6 | 85.7% | 0.775 | 0.490 | 0.311 | 2.550 | 1.866 |
| Me12 + em17 | 10 | 7 | 70.0% | 0.907 | 0.424 | 0.274 | 2.825 | 1.918 |
| Me13 + em9 | 7 | 6 | 85.7% | 0.505 | 0.417 | 0.367 | 3.400 | 2.202 |
| Me13 + em18 | 13 | 8 | 61.5% | 0.758 | 0.500 | 0.342 | 3.725 | 2.736 |
| Me13 + em19 | 12 | 8 | 66.7% | 0.59 | 0.460 | 0.299 | 3.400 | 2.392 |
| Me14 + em3 | 10 | 8 | 80.0% | 0.677 | 0.491 | 0.328 | 3.900 | 2.624 |
| Me14 + em12 | 12 | 10 | 83.3% | 0.777 | 0.498 | 0.343 | 5.200 | 3.430 |
| Me15 + em6 | 15 | 8 | 53.3% | 0.848 | 0.476 | 0.359 | 4.550 | 2.872 |
| Me15 + em13 | 6 | 5 | 83.3% | 0.751 | 0.500 | 0.287 | 2.050 | 1.435 |
| Me16 + em6 | 8 | 4 | 50.0% | 0.784 | 0.498 | 0.362 | 2.175 | 1.448 |
| Me17 + em7 | 9 | 7 | 77.8% | 0.655 | 0.485 | 0.319 | 3.225 | 2.233 |
| Me17 + em13 | 9 | 8 | 88.9% | 0.766 | 0.500 | 0.249 | 2.450 | 1.992 |
| Me17 + em16 | 10 | 5 | 50.0% | 0.468 | 0.394 | 0.347 | 2.700 | 1.735 |
| Me18 + em1 | 8 | 4 | 50.0% | 0.913 | 0.417 | 0.331 | 2.075 | 1.324 |
| Me18 + em2 | 9 | 6 | 66.7% | 0.703 | 0.496 | 0.321 | 3.050 | 1.926 |
| Me18 + em4 | 9 | 5 | 55.6% | 0.782 | 0.498 | 0.383 | 2.975 | 1.915 |
| Me18 + em8 | 10 | 7 | 70.0% | 0.768 | 0.499 | 0.336 | 3.450 | 2.352 |
| Me18 + em10 | 11 | 9 | 81.8% | 0.68 | 0.491 | 0.327 | 4.575 | 2.943 |
| Me18 + em17 | 7 | 6 | 85.7% | 0.684 | 0.492 | 0.307 | 2.350 | 1.842 |
| Me18 + em18 | 13 | 11 | 84.6% | 0.782 | 0.498 | 0.363 | 5.825 | 3.993 |
| Me18 + em19 | 9 | 8 | 88.9% | 0.574 | 0.453 | 0.288 | 3.250 | 2.304 |
| Me19 + em1 | 11 | 9 | 81.8% | 0.754 | 0.500 | 0.265 | 3.075 | 2.385 |
| Me19 + em14 | 14 | 11 | 78.6% | 0.695 | 0.495 | 0.303 | 4.300 | 3.333 |
| Me20 + em6 | 11 | 9 | 81.8% | 0.749 | 0.500 | 0.300 | 3.675 | 2.700 |
| Me20 + em10 | 12 | 9 | 75.0% | 0.850 | 0.475 | 0.279 | 3.625 | 2.511 |
| Me20 + em20 | 6 | 2 | 33.3% | 0.836 | 0.482 | 0.324 | 0.875 | 0.648 |
| sum | 460 | 345 | ||||||
| mean | 9.8 | 7.3 | 75% | 0.753 | 0.465 | 0.317 | 3.387 | 2.314 |
| Geographical Origin | N | Na | Ne | I | He | uHe |
|---|---|---|---|---|---|---|
| South America | 22 | 1.936 | 1.556 | 0.484 | 0.323 | 0.330 |
| North America | 14 | 1.928 | 1.585 | 0.498 | 0.335 | 0.347 |
| Africa | 13 | 1.887 | 1.558 | 0.476 | 0.319 | 0.332 |
| Europe | 8 | 1.594 | 1.476 | 0.403 | 0.273 | 0.309 |
| Oceania | 8 | 1.728 | 1.507 | 0.431 | 0.289 | 0.291 |
| Asia | 15 | 1.472 | 1.325 | 0.295 | 0.194 | 0.200 |
| Mean | 13.333 | 1.757 | 1.501 | 0.431 | 0.289 | 0.302 |
| Source of Variation | df | SS | MS | Est.Var. | Fst | PMV (%) |
|---|---|---|---|---|---|---|
| Among geo-groups | 5 | 434.307 | 86.861 | 2.548 | 0.045 | 5% |
| Within geo-groups | 74 | 3977.906 | 53.755 | 53.755 | 95% | |
| Total | 79 | 4412.213 | 56.303 | 100% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yi, L.; Dong, Z.; Lei, Y.; Zhao, J.; Xiong, Y.; Yang, J.; Xiong, Y.; Gou, W.; Ma, X. Genetic Diversity and Molecular Characterization of Worldwide Prairie Grass (Bromus catharticus Vahl) Accessions Using SRAP Markers. Agronomy 2021, 11, 2054. https://doi.org/10.3390/agronomy11102054
Yi L, Dong Z, Lei Y, Zhao J, Xiong Y, Yang J, Xiong Y, Gou W, Ma X. Genetic Diversity and Molecular Characterization of Worldwide Prairie Grass (Bromus catharticus Vahl) Accessions Using SRAP Markers. Agronomy. 2021; 11(10):2054. https://doi.org/10.3390/agronomy11102054
Chicago/Turabian StyleYi, Limei, Zhixiao Dong, Yu Lei, Junming Zhao, Yanli Xiong, Jian Yang, Yi Xiong, Wenlong Gou, and Xiao Ma. 2021. "Genetic Diversity and Molecular Characterization of Worldwide Prairie Grass (Bromus catharticus Vahl) Accessions Using SRAP Markers" Agronomy 11, no. 10: 2054. https://doi.org/10.3390/agronomy11102054
APA StyleYi, L., Dong, Z., Lei, Y., Zhao, J., Xiong, Y., Yang, J., Xiong, Y., Gou, W., & Ma, X. (2021). Genetic Diversity and Molecular Characterization of Worldwide Prairie Grass (Bromus catharticus Vahl) Accessions Using SRAP Markers. Agronomy, 11(10), 2054. https://doi.org/10.3390/agronomy11102054






