Quantitative and Qualitative Composition of Bacterial Communities of Malting Barley Grain and Malt during Long-Term Storage
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Micro-Malting Procedure
2.3. Chemical Analyses
- 1.
- Barley
- (a)
- Moisture content = EBC 2010—3.2
- (b)
- Protein content = EBC 2010—3.3.1
- (c)
- Starch content = EBC 2010—3.13
- (d)
- Germination capacity = EBC 2010—3.5.2
- 2.
- Malt
- (a)
- Moisture content = EBC 2010—4.2
- (b)
- Protein content = EBC 2010—4.3.1
- (c)
- Extract content = EBC 2010—4.5.1
- (d)
- Friability = EBC 2010—4.15
2.4. Microbiological Analyses
2.5. Mass Spectrometry Identification of Isolates
2.6. Statistical Analyses
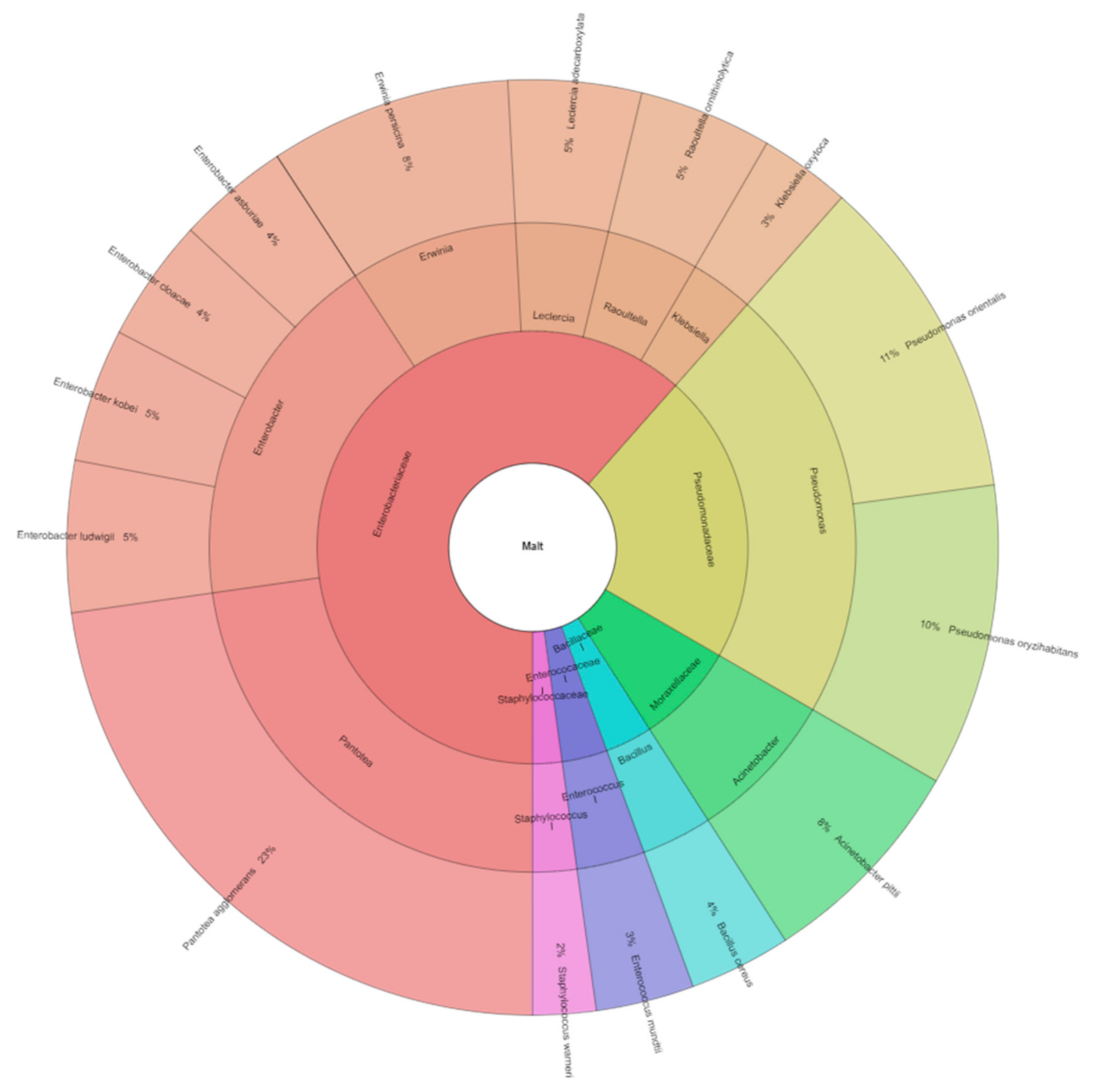
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Noots, I.; Delcour, J.A.; Michiels, C.W. From Field Barley to Malt: Detection and Specification of Microbial Activity for Quality Aspects. Crit. Rev. Microbiol. 1999, 25, 121–153. [Google Scholar] [CrossRef]
- Angelino, S.; Bol, J. Impact of microflora during storage and malting on malt properties. In Raw Materials and Sweet Wort Production, Proceedings of the Jean De Clerck Chair IV, Leuven, Belgium, 23–27 September 1990; Jean Monnet Aditorium: Leuven, Belgium, 1990; pp. 1–14. [Google Scholar]
- Flannigan, B. The microbiota of barley and malt. In Brewing Microbiology; Springer US: Boston, MA, USA, 2003; pp. 113–180. [Google Scholar]
- Laitila, A.; Sarlin, T.; Raulio, M.; Wilhelmson, A.; Kotaviita, E.; Huttunen, T.; Juvonen, R. Yeasts in malting, with special emphasis on Wickerhamomyces anomalus (synonym Pichia anomala). Antonie Van Leeuwenhoek 2011, 99, 75–84. [Google Scholar] [CrossRef]
- Doran, P.J.; Briggs, D.E. Microbes and grain germination. J. Inst. Brew. 1993, 99, 165–170. [Google Scholar] [CrossRef]
- Oliveira, P.M.; Zannini, E.; Arendt, E.K. Cereal fungal infection, mycotoxins, and lactic acid bacteria mediated bioprotection: From crop farming to cereal products. Food Microbiol. 2014, 37, 78–95. [Google Scholar] [CrossRef] [PubMed]
- Belokurova, E.; Pankina, I.; Sevastyanova, A.; Asfondyarova, I.; Katkova, N. The influence of barley weed impurities on the microbiological quality indicators. E3S Web Conf. 2020, 161, 01072. [Google Scholar] [CrossRef]
- Justé, A.; Malfliet, S.; Waud, M.; Crauwels, S.; De Cooman, L.; Aerts, G.; Marsh, T.L.; Ruyters, S.; Willems, K.; Busschaert, P.; et al. Bacterial community dynamics during industrial malting, with an emphasis on lactic acid bacteria. Food Microbiol. 2014, 39, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Bamforth, C.W. The Microbiology of Malting and Brewing. Microbiol. Mol. Biol. Rev. 2013, 77, 157–172. [Google Scholar] [CrossRef] [PubMed]
- Briggs, D.E.; McGuinness, G. Microbes on barley grains. J. Inst. Brew. 1993, 99, 249–255. [Google Scholar] [CrossRef]
- Petters, H.I.; Flannigan, B.; Austin, B. Quantitative and qualitative studies of the microflora of barley malt production. J. Appl. Bacteriol. 1988, 65, 279–297. [Google Scholar] [CrossRef]
- Luarasi, L.; Troja, R.; Pinguli, L. Microbiological safety and quality evaluation of the raw materials used in beer production. J. Hyg. Eng. Des. 2016, 16, 28–31. [Google Scholar]
- Justé, A.; Malfliet, S.; Lenaerts, M.; De Cooman, L.; Aerts, G.; Willems, K.A.; Lievens, B. Microflora during malting of barley: Overview and impact on malt quality. Brew. Sci. 2011, 64, 22–31. [Google Scholar]
- Vaughan, A.; O’Sullivan, T.; Sinderen, D. Enhancing the Microbiological Stability of Malt and Beer—A Review. J. Inst. Brew. 2005, 111, 355–371. [Google Scholar] [CrossRef]
- Dalié, D.K.D.; Deschamps, A.M.; Richard-Forget, F. Lactic acid bacteria–Potential for control of mould growth and mycotoxins: A review. Food Control 2010, 21, 370–380. [Google Scholar] [CrossRef]
- Matoulková, D.; Vontrobová, E.; Brožová, M.; Kubizniaková, P. Microbiology of brewery production–bacteria of the order Enterobacterales. Kvas. Prum. 2018, 64, 161–166. [Google Scholar] [CrossRef]
- Buňka, F.; Budinský, P.; Čechová, M.; Drienovský, V.; Pachlová, V.; Matoulková, D.; Kubáň, V.; Buňková, L. Content of biogenic amines and polyamines in beers from the Czech Republic. J. Inst. Brew. 2012, 118, 213–216. [Google Scholar] [CrossRef]
- EBC. Analysis Committee. In Analytica-EBC; Fachverlag, Hans Carl: Nürnberg, Germany, 2010; ISBN 978-3-418-00759-5. [Google Scholar]
- Kačániová, M.; Mellen, M.; Vukovic, N.L.; Kluz, M.; Puchalski, C.; Haščík, P.; Kunová, S. Combined Effect of Vacuum Packaging, Fennel and Savory Essential Oil Treatment on the Quality of Chicken Thighs. Microorganisms 2019, 7, 134. [Google Scholar] [CrossRef]
- O’Sullivan, T.F.; Walsh, Y.; O’Mahony, A.; Fitzgerald, G.F.; Sinderen, D. A Comparative Study of Malthouse and Brewhouse Microflora. J. Inst. Brew. 1999, 105, 55–61. [Google Scholar] [CrossRef]
- Priest, F.G.; Campbell, I. Brewing Microbiology; Springer: Boston, MA, USA, 2003; ISBN 978-1-4613-4858-0. [Google Scholar]
- Van Nierop, S.N.E.; Rautenbach, M.; Axcell, B.C.; Cantrell, I.C. The impact of microorganisms on barley and malt quality—A review. J. Am. Soc. Brew. Chem. 2006, 64, 69–78. [Google Scholar] [CrossRef]
- Douglas, P.E.; Flannigan, B. A microbiological evaluation of barley malt production. J. Inst. Brew. 1988, 94, 85–88. [Google Scholar] [CrossRef]
- Lewis, D. Biological mash and wort acidification. New Brew. 1998, 15, 36–45. [Google Scholar]
- Back, W. Secondary contaminations in the filling area. Brauwelt Int. 1994, 12, 326–333. [Google Scholar]
- Back, W. Biofilme in der Brauerei und Getrankeindustrie-15 Jahre Praxiserfahrung. Brauwelt 2003, 24, 766–777. [Google Scholar]
- Back, W. Colour Atlas and Hand-Book of Beverage Biology; Back, W., Ed.; Verlag Hans Carl: Nürnberg, Germany, 2005; pp. 10–112. [Google Scholar]
- Brožová, M.; Kubizniaková, P.; Matoulková, D. Brewing Microbiology–Bacteria of the Genera Bacillus, Brevibacillus and Paenibacillus and Cultivation Methods for their Detection–Part 1. Kvas. Prum. 2018, 64, 50–57. [Google Scholar] [CrossRef]
- Logan, N.A.; De Vos, P. Bacillus. In Bergey’s Manual of Systematics of Archaea and Bacteria; Whitman, W.B., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2015. [Google Scholar]
- Haakensen, M.; Ziola, B. Identification of novel horA–Harbouring bacteria capable of spoiling beer. Can. J. Microbiol. 2008, 54, 321–325. [Google Scholar] [CrossRef] [PubMed]
- Drobniewski, F.A. Bacillus cereus and related species. Clin. Microbiol. Rev. 1993, 6, 324–338. [Google Scholar] [CrossRef]
- Van Vuuren, H.J.J.; Priest, F.G. Gram-negative brewery bacteria. In Brewing Microbiology; Springer US: Boston, MA, USA, 2003; pp. 219–245. [Google Scholar]
- Blomqvist, K.; Nikkola, M.; Lehtovaara, P.; Suihko, M.L.; Airaksinen, U.; Stråby, K.B.; Knowles, J.K.; Penttilä, M.E. Characterization of the genes of the 2,3-butanediol operons from Klebsiella terrigena and Enterobacter aerogenes. J. Bacteriol. 1993, 175, 1392–1404. [Google Scholar] [CrossRef]
- Drancourt, M.; Bollet, C.; Carta, A.; Rousselier, P. Phylogenetic analyses of Klebsiella species delineate Klebsiella and Raoultella gen. nov., with description of Raoultella ornithinolytica comb. nov., Raoultella terrigena comb. nov. and Raoultella planticola comb. nov. Int. J. Syst. Evol. Microbiol. 2001, 51, 925–932. [Google Scholar] [CrossRef]

| Microorganisms | Dilution of Samples | Agar Medium | Inoculation (mL) | Temperature (°C) | Incubation Time (hours) |
|---|---|---|---|---|---|
| Total plate count | Malt 10−5, 10−6 Barley 10−3, 10−4 | PCA | 1 | 30 | 48–72 |
| Coliform | 10−3, 10−4 | VRBL | 0.1 | 37 | 24–48 |
| Sporulating | 10−1, 10−2 | PCA | 1 | 30 | 24–48 |
| LAB | 10−1, 10−2 | MRS | 0.1 | 30 | 48–72 |
| Time of Storage | Varieties | Moisture Content [%] | Protein Content [%] | Starch Content [%] | Germination Capacity [%] |
|---|---|---|---|---|---|
| 0. | Laudis | 11.63 | 11.12 | 63.03 | 95.8 |
| Kangoo | 11.72 | 11.65 | 62.88 | 98.5 | |
| Wintmalt | 11.54 | 10.57 | 64.53 | 97.0 | |
| Grain silo | |||||
| 3. months | Laudis | 12.40 | 11.15 | 63.01 | 93.5 |
| Kangoo | 12.54 | 11.78 | 62.72 | 98.3 | |
| Wintmalt | 12.74 | 10.82 | 64.14 | 99.0 | |
| 6. months | Laudis | 12.83 | 11.27 | 62.76 | 90.5 |
| Kangoo | 13.56 | 11.51 | 62.83 | 98.8 | |
| Wintmalt | 12.66 | 10.58 | 64.07 | 98.0 | |
| 9. months | Laudis | 12.90 | 11.20 | 62.85 | 90.0 |
| Kangoo | 13.82 | 11.60 | 62.75 | 98.2 | |
| Wintmalt | 12.82 | 10.70 | 64.32 | 98.0 | |
| Floor warehouse | |||||
| 3. months | Laudis | 12.64 | 11.29 | 62.78 | 92.8 |
| Kangoo | 12.65 | 11.64 | 63.11 | 98.0 | |
| Wintmalt | 12.58 | 10.94 | 63.73 | 98.3 | |
| 6. months | Laudis | 13.30 | 11.36 | 62.76 | 90.8 |
| Kangoo | 13.37 | 11.62 | 62.76 | 99.3 | |
| Wintmalt | 13.30 | 10.53 | 64.29 | 98.5 | |
| 9. months | Laudis | 13.21 | 11.27 | 62.72 | 90.2 |
| Kangoo | 13.48 | 11.68 | 63.02 | 99.0 | |
| Wintmalt | 13.07 | 10.61 | 64.15 | 98.0 | |
| Time of Storage | Varieties | TPC log cfu/g | Coli log cfu/g | LAB log cfu/g | Spor log cfu/g | Species |
|---|---|---|---|---|---|---|
| 0. | Laudis | 5.00 ± 0.04 | 3.15 ± 0.16 | 3.08 ± 0.06 | 1.26 ± 0.06 | Pantoea agglomerans |
| Kangoo | 5.23 ± 0.09 | 2.26 ± 0.02 | 2.36 ± 0.14 | 1.36 ± 0.08 | Bacillus altitudinis, Pantoea agglomerans | |
| Wintmalt | 5.84 ± 0.08 | 1.11 ± 0.04 | 1.30 ± 0.05 | 1.00 ± 0.01 | Pantoea agglomerans | |
| Granaries/storage silo | ||||||
| 3. months | Laudis | 6.04 ± 0.01 | 3.80 ± 0.09 | 1.00 ± 0.10 | 1.43 ± 0.02 | Pantoea agglomerans |
| Kangoo | 6.28 ± 0.04 | 3.60 ± 0.03 | 1.00 ± 0.08 | 2.40 ± 0.06 | Kosakonia cowanii, Bacillus altitudinis, Pantoea agglomerans | |
| Wintmalt | 6.38 ± 0.08 | 3.54 ± 0.07 | 1.00 ± 0.05 | 2.00 ± 0.01 | Pantoea agglomerans | |
| 6. months | Laudis | 7.28 ± 0.08 | 3.97 ± 0.03 | 1.00 ± 0.02 | 1.86 ± 0.10 | Pantoea agglomerans |
| Kangoo | 6.36 ± 0.07 | 4.71 ± 0.03 | 1.36 ± 0.04 | 2.15 ± 0.07 | Pantoea agglomerans | |
| Wintmalt | 6.28 ± 0.03 | 3.65 ± 0.07 | 1.00 ± 0.14 | 2.11 ± 0.12 | Pantoea agglomerans | |
| 9. months | Laudis | 6.23 ± 0.08 | 5.26 ± 0.11 | 1.30 ± 0.06 | 1.83 ± 0.10 | Pantoea agglomerans |
| Kangoo | 6.00 ± 0.02 | 5.69 ± 0.09 | 1.00 ± 0.10 | 1.77 ± 0.06 | Pantoea agglomerans | |
| Wintmalt | 5.34 ± 0.06 | 4.93 ± 0.07 | 1.11 ± 0.09 | 2.11 ± 0.09 | Pantoea agglomerans | |
| Floor warehouse | ||||||
| 3. months | Laudis | 5.92 ± 0.07 | 4.00 ± 0.06 | 1.00 ± 0.05 | 2.11 ± 0.04 | Pseudomonas poae, Ps. libanensis, Ps. synxantha, Pantoea agglomerans |
| Kangoo | 6.30 ± 0.07 | 4.18 ± 0.07 | 1.00 ± 0.05 | 2.34 ± 0.03 | Pantoea agglomerans | |
| Wintmalt | 6.15 ± 0.06 | 4.04 ± 0.07 | 1.36 ± 0.06 | 1.56 ± 0.08 | Pantoea agglomerans | |
| 6. months | Laudis | 6.23 ± 0.06 | 4.93 ± 0.07 | 1.00 ± 0.09 | 1.43 ± 0.13 | Pantoea agglomerans |
| Kangoo | 6.30 ± 0.03 | 5.08 ± 0.10 | 1.43 ± 0.06 | 1.56 ± 0.14 | Pantoea agglomerans | |
| Wintmalt | 6.32 ± 0.08 | 3.81 ± 0.05 | 1.36 ± 0.07 | 1.26 ± 0.13 | Pantoea agglomerans, Staphylococcus haemolyticus | |
| 9. months | Laudis | 6.04 ± 0.07 | 5.69 ± 0.07 | 1.00 ± 0.07 | 2.67 ± 0.06 | Pantoea agglomerans |
| Kangoo | 6.11 ± 0.06 | 5.69 ± 0.11 | 1.00 ± 0.08 | 1.00 ± 0.05 | Pantoea agglomerans | |
| Wintmalt | 6.04 ± 0.06 | 5.23 ± 0.14 | 1.00 ± 0.07 | 1.74 ± 0.09 | Pantoea agglomerans | |
| Variety/Microorganism Groups | TPC | COLI | LAB | Spor | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grain | ||||||||||||
| Laudis | 3 | 6 | 9 | 3 | 6 | 9 | 3 | 6 | 9 | 3 | 6 | 9 |
| 0 | 1.978 × 10−6 | 1.335 × 10−6 | 1.649 × 10−5 | 0.004 | 0.001 | 5.034 × 10−5 | 5.153 × 10−6 | 3.851 × 10−7 | 3.153 × 10−6 | 0.010 | 0.001 | 0.001 |
| 3 | x | 9.299 × 10−6 | 0.013 | x | 0.039 | 5.943 × 10−5 | x | 0.010 | 0.822 | x | 0.002 | 0.002 |
| 6 | x | X | 7.048 × 10−5 | x | x | 3.838 × 10−5 | x | x | 0.001 | x | x | 0.769 |
| Kangoo | 3 | 6 | 9 | 3 | 6 | 9 | 3 | 6 | 9 | 3 | 6 | 9 |
| 0 | 4.63 × 10−5 | 6.21 × 10−5 | 0.000126 | 1.82 × 10−7 | 3.82 × 10−8 | 3.78 × 10−7 | 0.000128 | 0.000291 | 0.000156 | 4.32 × 10−5 | 0.000197 | 0.001687 |
| 3 | x | 0.139483 | 0.000224 | x | 1.58 × 10−6 | 3.01 × 10−6 | x | 0,002167 | 0.96516 | x | 0.008952 | 0.000151 |
| 6 | x | x | 0.000799 | x | x | 6.74 × 10−5 | x | x | 0.003795 | x | x | 0.001856 |
| Wintmalt | 3 | 6 | 9 | 3 | 6 | 9 | 3 | 6 | 9 | 3 | 6 | 9 |
| 0 | 0.001012 | 0.000878 | 0.001049 | 8.57 × 10−7 | 6.75 × 10−7 | 1.31 × 10−7 | 0.001708 | 0.024547 | 0.033331 | 5.07 × 10−6 | 0.000102 | 2.42 × 10−5 |
| 3 | x | 0.09119 | 4.81 × 10−5 | x | 0.138831 | 1.97 × 10−5 | x | 1 | 0.131031 | x | 0.226106 | 0.126122 |
| 6 | x | x | 1.79 × 10−5 | x | x | 2.63 × 10−5 | x | x | 0.306838 | x | x | 1 |
| Floor warehouse | ||||||||||||
| Laudis | 3 | 6 | 9 | 3 | 6 | 9 | 3 | 6 | 9 | 3 | 6 | 9 |
| 0 | 3.38 × 10−5 | 7.42–06 | 2.43 × 10−5 | 0.001055 | 6.8 × 10−5 | 1.56 × 10−5 | 1.08 × 10−6 | 4.6 × 10−6 | 2.09 × 10−6 | 3.1 × 10−5 | 0.115177 | 1.03 × 10−5 |
| 3 | x | 0.003418 | 0.09487 | x | 6 × 10−5 | 4.4 × 10−6 | x | 1 | 0.948781 | x | 0.000835 | 0.00019 |
| 6 | x | x | 0.020599 | x | x | 0.000174 | x | x | 0.962091 | x | x | 0.000111 |
| Kangoo | 3 | 6 | 9 | 3 | 6 | 9 | 3 | 6 | 9 | 3 | 6 | 9 |
| 0 | 8.77 × 10−5 | 4.12 × 10−5 | 0.00015 | 9.76 × 10−7 | 1.29 × 10−6 | 7.35 × 10−7 | 1.08 × 10−6 | 4.6 × 10−6 | 2.09 × 10−6 | 3.25 × 10−5 | 0.104222 | 0.002018 |
| 3 | x | 0.945216 | 0.025161 | x | 0.000221 | 3.38 × 10−5 | x | 1 | 0.948781 | x | 0.000807 | 2.04 × 10−6 |
| 6 | x | x | 0.008236 | x | x | 0.002226 | x | x | 0.962091 | x | x | 0.003044 |
| Wintmalt | 3 | 6 | 9 | 3 | 6 | 9 | 3 | 6 | 9 | 3 | 6 | 9 |
| 0 | 0.006044 | 0.001736 | 0.028045 | 3.3 × 10−7 | 1.87 × 10−7 | 8.93 × 10−7 | 0.301257 | 0.272039 | 0.003851 | 0.000255 | 0.026285 | 0.000165 |
| 3 | x | 0.046225 | 0.089009 | x | 0.010389 | 0.000171 | x | 0.902702 | 0.002773 | x | 0.023257 | 0.061123 |
| 6 | x | x | 0.00819 | x | x | 7.11 × 10−5 | x | x | 0.002924 | x | x | 0.005776 |
| Time of Storage | Varieties | Moisture Content [%] | Protein Content [%] | Extract Content [%] | Friability [%] |
|---|---|---|---|---|---|
| 0. | Laudis | 4.82 | 10.50 | 81.19 | 83.6 |
| Kangoo | 4.70 | 11.14 | 80.24 | 85.2 | |
| Wintmalt | 5.28 | 9.9 | 80.8 | 83.8 | |
| Grain silo | |||||
| 3. months | Laudis | 3.78 | 10.69 | 82.63 | 86.1 |
| Kangoo | 3.82 | 10.93 | 82.39 | 91.6 | |
| Wintmalt | 4.37 | 10.18 | 81.44 | 85.5 | |
| 6. months | Laudis | 4.29 | 10.72 | 82.24 | 88.0 |
| Kangoo | 4.33 | 10.84 | 81.72 | 88.9 | |
| Wintmalt | 4.85 | 10.02 | 82.23 | 86.5 | |
| 9. months | Laudis | 4.15 | 10.82 | 81.97 | 87.5 |
| Kangoo | 4.50 | 10.88 | 82.01 | 89.2 | |
| Wintmalt | 4.66 | 10.05 | 82.35 | 87.2 | |
| Floor warehouse | |||||
| 3. months | Laudis | 3.96 | 10.75 | 83.12 | 87.9 |
| Kangoo | 4.02 | 10.84 | 82.07 | 91.7 | |
| Wintmalt | 4.61 | 10.39 | 82.32 | 86.9 | |
| 6. months | Laudis | 3.75 | 10.91 | 83.02 | 87.9 |
| Kangoo | 3.88 | 10.98 | 81.89 | 90.7 | |
| Wintmalt | 4.52 | 9.88 | 82.64 | 89.3 | |
| 9. months | Laudis | 3.83 | 10.75 | 82.90 | 88.5 |
| Kangoo | 3.98 | 11.02 | 81.97 | 90.5 | |
| Wintmalt | 4.72 | 9.90 | 82.53 | 89.0 | |
| Time of Storage | Varieties | TPC log cfu/g | Coli log cfu/g | LAB log cfu/g | Spor log cfu/g | Species |
|---|---|---|---|---|---|---|
| 0. | Laudis | 7.00 ± 0.01 | 4.70 ± 0.13 | 5.00 ± 0.06 | 1.36 ± 0.08 | Bacillus cereus |
| Kangoo | 6.98 ± 0.15 | 3.97 ± 0.15 | 3.56 ± 0.03 | 1.00 ± 0.11 | Enterococcus mundtii | |
| Wintmalt | 7.08 ± 0.09 | 2.70 ± 0.14 | 2.91 ± 0.14 | 1.11 ± 0.12 | Pseudomonas oryzihabitans | |
| Grain/cereal silo | ||||||
| 3. months | Laudis | 7.48 ± 0.08 | 4.97 ± 0.08 | 4.38 ± 0.09 | 2.30 ± 0.13 | Staphylococcus warneri, Enterobacter cloacae, E. ludwigii |
| Kangoo | 6.92 ± 0.07 | 3.15 ± 0.06 | 2.65 ± 0.04 | 2.51 ± 0.07 | Enterococcus mundtii | |
| Wintmalt | 7.32 ± 0.05 | 3.51 ± 0.08 | 4.11 ± 0.09 | 4.11 ± 0.05 | Pseudomonas oryzihabitans, Ps. orientalis | |
| 6. months | Laudis | 7.48 ± 0.09 | 5.99 ± 0.09 | 3.79 ± 0.09 | 2.49 ± 0.08 | Enterobacter cloacae, E. asburiae, E. kobei |
| Kangoo | 7.08 ± 0.05 | 5.34 ± 0.12 | 2.18 ± 0.07 | 2.72 ± 0.08 | Enterococcus mundtii | |
| Wintmalt | 7.48 ± 0.12 | 5.76 ± 0.12 | 3.15 ± 0.13 | 2.45 ± 0.13 | Pantoea agglomerans | |
| 9. months | Laudis | 7.73 ± 0.15 | 7.08 ± 0.06 | 4.91 ± 0.17 | 3.00 ± 0.10 | Enterobacter cloacae, E. asburiae, E. kobei |
| Kangoo | 7.08 ± 0.14 | 6.28 ± 0.14 | 2.65 ± 0.16 | 2.85 ± 0.13 | Pantoea agglomerans | |
| Wintmalt | 7.30 ± 0.09 | 7.15 ± 0.06 | 3.73 ± 0.15 | 3.11 ± 0.08 | Pantoea agglomerans | |
| Floor warehouse | ||||||
| 3. months | Laudis | 7.36 ± 0.05 | 5.28 ± 0.09 | 5.38 ± 0.14 | 2.15 ± 0.06 | Erwinia persicina |
| Kangoo | 7.20 ± 0.09 | 4.40 ± 0.14 | 3.67 ± 0.16 | 1.51 ± 0.04 | Raoultella ornithinolytica, Acinetobacter pittii, Klebsiella oxytoca | |
| Wintmalt | 7.26 ± 0.08 | 3.76 ± 0.07 | 3.67 ± 0.06 | 1.51 ± 0.06 | Pantoea agglomerans | |
| 6. months | Laudis | 7.58 ± 0.10 | 6.51 ± 0.06 | 5.08 ± 0.04 | 2.04 ± 0.14 | Pantoea agglomerans |
| Kangoo | 7.28 ± 0.07 | 5.86 ± 0.10 | 1.00 ± 0.08 | 1.36 ± 0.10 | Pantoea agglomerans | |
| Wintmalt | 7.28 ± 0.06 | 6.18 ± 0.03 | 1.00 ± 0.03 | 1.74 ± 0.04 | Pantoea agglomerans | |
| 9. months | Laudis | 8.15 ± 0.11 | 7.30 ± 0.08 | 4.58 ± 0.11 | 3.15 ± 0.03 | Pantoea agglomerans |
| Kangoo | 7.28 ± 0.15 | 6.78 ± 0.11 | 2.88 ± 0.06 | 1.76 ± 0.07 | Leclercia adecarboxylata | |
| Wintmalt | 7.56 ± 0.08 | 7.15 ± 0.01 | 2.70 ± 0.09 | 2.32 ± 0.06 | Pantoea agglomerans, Leclercia adecarboxylata | |
| Variety/Microorganism Groups | TPC | COLI | LAB | Spor | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grain | ||||||||||||
| Laudis | 3 | 6 | 9 | 3 | 6 | 9 | 3 | 6 | 9 | 3 | 6 | 9 |
| 0 | 0.000502 | 0.000682 | 0.001033 | 0.040016 | 0.000153 | 8.91 × 10−6 | 0.000514 | 4.16 × 10−5 | 0.424663 | 0.000456 | 6.66 × 10−5 | 2.42 × 10−5 |
| 3 | x | 1 | 0.061125 | x | 0.000138 | 3.62 × 10−6 | x | 0.001419 | 0.00933 | x | 0.10536 | 0.001862 |
| 6 | x | x | 0.064389 | x | x | 7.11 × 10−5 | x | x | 0.000601 | x | x | 0.002295 |
| Kangoo | 3 | 6 | 9 | 3 | 6 | 9 | 3 | 6 | 9 | 3 | 6 | 9 |
| 0 | 0.544688 | 0.329678 | 0.434449 | 0.00092 | 0.000251 | 4.29 × 10−5 | 7.86 × 10−6 | 7.16 × 10−6 | 0.000609 | 4.29 × 10−5 | 2.77 × 10−5 | 5.16 × 10−5 |
| 3 | x | 0.037567 | 0.151835 | x | 9.4 × 10−6 | 4.03 × 10−6 | x | 0.000569 | 0.973323 | x | 0.030593 | 0.018556 |
| 6 | x | x | 0,971782 | x | x | 0.000975 | x | x | 0.009395 | x | x | 0.218189 |
| Wintmalt | 3 | 6 | 9 | 3 | 6 | 9 | 3 | 6 | 9 | 3 | 6 | 9 |
| 0 | 0.016937 | 0.009634 | 0.041394 | 0.001107 | 9.85 × 10−6 | 1.13 × 10−6 | 0.000281 | 0.097998 | 0.002628 | 2.1 × 10−6 | 0.000189 | 1.48 × 10−5 |
| 3 | x | 0.09859 | 0.759835 | x | 1.11 × 10−5 | 4.06 × 10−7 | x | 0.000436 | 0.020754 | x | 3.52 × 10−5 | 4.72 × 10−5 |
| 6 | x | x | 0.105096 | x | x | 6.14 × 10−5 | x | x | 0.007229 | x | x | 0.001659 |
| Floor warehouse | ||||||||||||
| Laudis | 3 | 6 | 9 | 3 | 6 | 9 | 3 | 6 | 9 | 3 | 6 | 9 |
| 0 | 0.00027 | 0.000496 | 5.03 × 10−5 | 0.003126 | 2.54 × 10−5 | 8.38 × 10−6 | 0.011918 | 0.130162 | 0.003555 | 0.000165 | 0.001825 | 3.41 × 10−6 |
| 3 | x | 0.02561 | 0.000333 | x | 3.39 × 10−5 | 8.45 × 10−6 | x | 0.021626 | 0.001358 | x | 0.29307 | 1.44 × 10−5 |
| 6 | x | x | 0.002505 | x | x | 0.000177 | x | x | 0.001566 | x | x | 0.000183 |
| Kangoo | 3 | 6 | 9 | 3 | 6 | 9 | 3 | 6 | 9 | 3 | 6 | 9 |
| 0 | 0.017181 | 0.001915 | 0.034695 | 0.052115 | 0.000276 | 2.8 × 10−5 | 0.000171 | 5.67 × 10−8 | 1.62 × 10−6 | 0.040731 | 0.109993 | 0.00279 |
| 3 | x | 0.256915 | 0.48693 | x | 0.000128 | 1.87 × 10−5 | x | 1.01 × 10−5 | 0.001403 | x | 1 | 0.005281 |
| 6 | x | x | 0.947414 | x | x | 0.000422 | x | x | 1.73 × 10−6 | x | x | 0.007703 |
| Wintmalt | 3 | 6 | 9 | 3 | 6 | 9 | 3 | 6 | 9 | 3 | 6 | 9 |
| 0 | 0.003925 | 0.00112 | 0.000239 | 0.000394 | 4.58 × 10−5 | 1.35 × 10−5 | 1.01 × 10−5 | 3.62 × 10−8 | 2.62 × 10−6 | 0.053766 | 0.001849 | 7.66 × 10−5 |
| 3 | x | 0.73302 | 0.008581 | x | 7.94 × 10−7 | 1.16 × 10−6 | x | 2.77 × 10−7 | 9.53 × 10−5 | x | 0.005367 | 7.6 × 10−5 |
| 6 | x | x | 0.007432 | x | x | 9.2 × 10−5 | x | x | 5.33 × 10−6 | x | x | 0.000158 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Felšöciová, S.; Kowalczewski, P.Ł.; Krajčovič, T.; Dráb, Š.; Kačániová, M. Quantitative and Qualitative Composition of Bacterial Communities of Malting Barley Grain and Malt during Long-Term Storage. Agronomy 2020, 10, 1301. https://doi.org/10.3390/agronomy10091301
Felšöciová S, Kowalczewski PŁ, Krajčovič T, Dráb Š, Kačániová M. Quantitative and Qualitative Composition of Bacterial Communities of Malting Barley Grain and Malt during Long-Term Storage. Agronomy. 2020; 10(9):1301. https://doi.org/10.3390/agronomy10091301
Chicago/Turabian StyleFelšöciová, Soňa, Przemysław Łukasz Kowalczewski, Tomáš Krajčovič, Štefan Dráb, and Miroslava Kačániová. 2020. "Quantitative and Qualitative Composition of Bacterial Communities of Malting Barley Grain and Malt during Long-Term Storage" Agronomy 10, no. 9: 1301. https://doi.org/10.3390/agronomy10091301
APA StyleFelšöciová, S., Kowalczewski, P. Ł., Krajčovič, T., Dráb, Š., & Kačániová, M. (2020). Quantitative and Qualitative Composition of Bacterial Communities of Malting Barley Grain and Malt during Long-Term Storage. Agronomy, 10(9), 1301. https://doi.org/10.3390/agronomy10091301







