Analysis of Genetic Factors Defining Head Blight Resistance in an Old Hungarian Wheat Variety-Based Mapping Population
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Artificial Inoculation
2.3. Statistical Analyses
2.4. Molecular Methods
2.4.1. DNA Extraction
2.4.2. DNA-Based Markers (SSR, AFLP, SNP)
2.4.3. QTL Identification
3. Results
3.1. Review of Type II Resistance of the Offspring Lines Originating from the ’BKT9086-95/Mv Magvas’ Experimental Cross under Greenhouse Conditions
3.2. Review of Type II Resistance of the Offspring Lines ’BKT9086-95/Mv Magvas’ under Field Conditions
3.3. Molecular Tests in the ’BKT9086-95/Mv Magvas’ Offspring Population
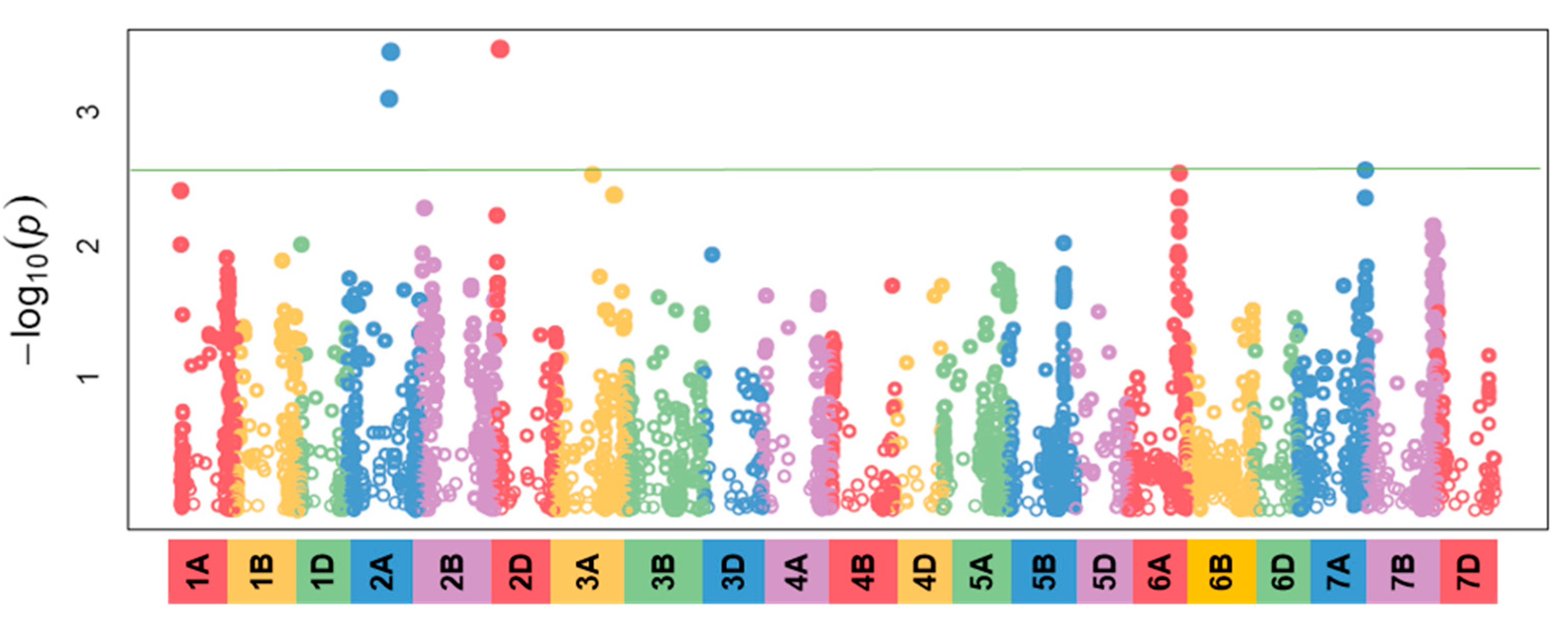
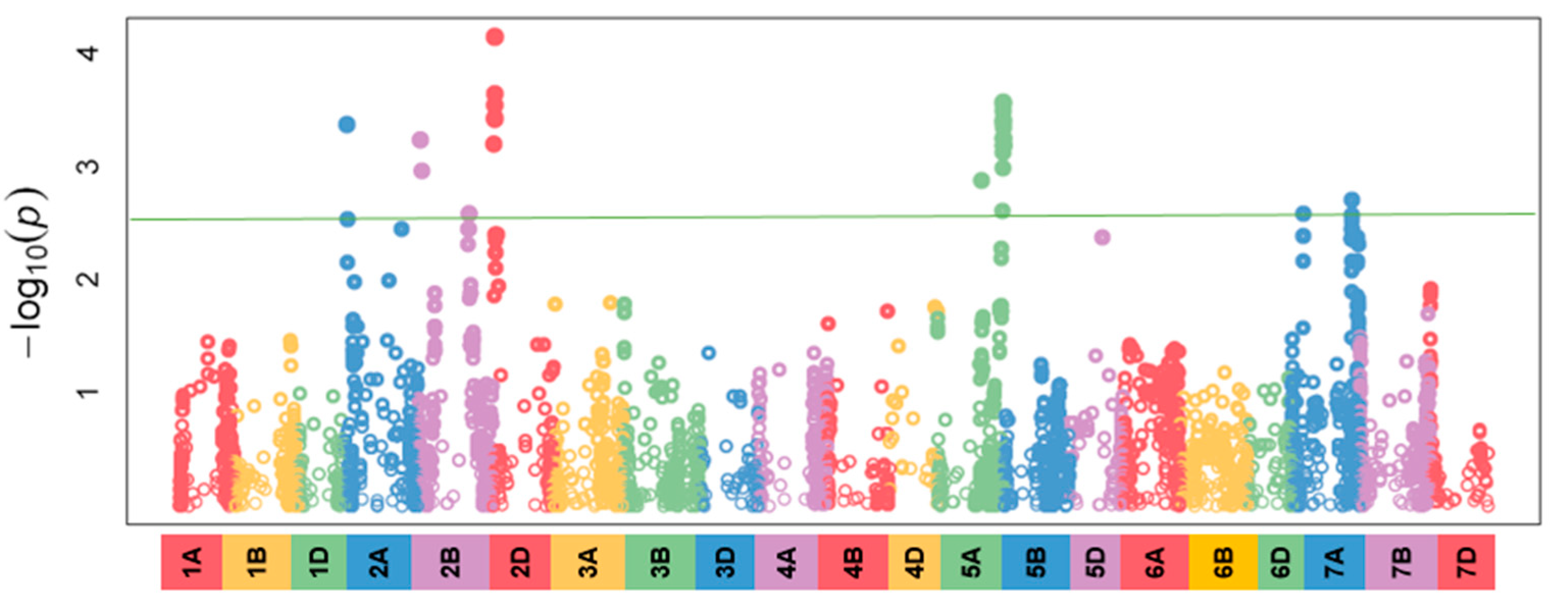
3.4. Identification of the Genetic Background Related To Infection Severity
3.5. Identification of the Genetic Background In Correlation With the Size of the Area Under the Disease Progress Curve
3.6. Comparison of the Genetic Regions Related To the Infection Percentage and the AUDPC Values
3.7. Linkage of Resistance to Head Blight and Other Phenotype Properties
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Windels, C.E. Economic and Social Impacts of Fusarium Head Blight: Changing Farms and Rural Communities in the Northern Great Plains. Phytopathology 2000, 90, 17–21. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Halder, J.; White, R.; Hughes, D.; Ye, Z.; Wang, C.; Xu, R.; Gan, B.; Fitt, B.D. Climate change increases risk of fusarium ear blight on wheat in central China. Ann. Appl. Boil. 2014, 164, 384–395. [Google Scholar] [CrossRef]
- Vaughan, M.; Backhouse, D.; Del Ponte, E.M. Climate change impacts on the ecology of Fusarium graminearum species complex and susceptibility of wheat to Fusarium head blight: A review. World Mycotoxin J. 2016, 9, 685–700. [Google Scholar] [CrossRef]
- Manzini, M.; Rodriguez-Estrada, M.T.; Meca, G.; Mañes, J. Reduction of beauvericin and enniatins bioaccessibility by prebiotic compounds, evaluated in static and dynamic simulated gastrointestinal digestion. Food Control. 2015, 47, 203–211. [Google Scholar] [CrossRef]
- Tibola, C.S.; Fernandes, J.; Guarienti, E.M. Effect of cleaning, sorting and milling processes in wheat mycotoxin content. Food Control. 2016, 60, 174–179. [Google Scholar] [CrossRef]
- Keller, M.D.; Bergstrom, G.C.; Shields, E.J. The aerobiology of Fusarium graminearum. Aerobiol. 2013, 30, 123–136. [Google Scholar] [CrossRef]
- Salgado, J.D.; Madden, L.V.; Paul, P.A. Efficacy and Economics of Integrating In-Field and Harvesting Strategies to Manage Fusarium Head Blight of Wheat. Plant Dis. 2014, 98, 1407–1421. [Google Scholar] [CrossRef]
- Buerstmayr, H.; Ban, T.; Anderson, J.A. QTL mapping and marker-assisted selection forFusariumhead blight resistance in wheat: A review. Plant Breed. 2009, 128, 1–26. [Google Scholar] [CrossRef]
- Buerstmayr, M.; Steiner, B.; Buerstmayr, H. Breeding for Fusarium head blight resistanxe in wheat – Progress and challenges. Plant Breed. 2020, 139, 429–454. [Google Scholar] [CrossRef]
- Bai, G.; Kolb, F.L.; Shaner, G.; Domier, L.L. Amplified Fragment Length Polymorphism Markers Linked to a Major Quantitative Trait Locus Controlling Scab Resistance in Wheat. Phytopathology 1999, 89, 343–348. [Google Scholar] [CrossRef]
- Steiner, B.; Lemmens, M.; Griesser, M.; Scholz, U.; Schondelmaier, J.; Buerstmayr, H. Molecular mapping of resistance to Fusarium head blight in the spring wheat cultivar Frontana. Theor. Appl. Genet. 2004, 109, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Mardi, M.; Pazouki, L.; Delavar, H.; Kazemi, M.B.; Ghareyazie, B.; Steiner, B.; Nolz, R.; Lemmens, M.; Buerstmayr, H. QTL analysis of resistance to Fusarium head blight in wheat using a ’Frontana’-derived population. Plant Breed. 2006, 125, 313–317. [Google Scholar] [CrossRef]
- Miedaner, T.; Würschum, T.; Maurer, H.P.; Korzun, V.; Ebmeyer, E.; Reif, J.C. Association mapping for Fusarium head blight resistance in European soft winter wheat. Mol. Breed. 2010, 28, 647–655. [Google Scholar] [CrossRef]
- Mirdita, V.; Liu, G.; Zhao, Y.; Miedaner, T.; Longin, C.F.H.; Gowda, M.; Mette, M.F.; Reif, J.C. Genetic architecture is more complex for resistance to Septoria tritici blotch than to Fusarium head blight in Central European winter wheat. BMC Genom. 2015, 16, 430. [Google Scholar] [CrossRef][Green Version]
- Jin, F.; Zhang, D.; Bockus, W.; Baenziger, P.; Carver, B.; Bai, G. Fusarium Head Blight Resistance in U.S. Winter Wheat Cultivars and Elite Breeding Lines. Crop. Sci. 2013, 53, 2006–2013. [Google Scholar] [CrossRef]
- Zhou, Z.; Hao, Y.; Mergoum, M.; Bai, G.; Humphreys, G.; Cloutier, S.; Xia, X.; He, Z. Breeding wheat for sesistance to Fusarium head blight in the Global North: China, USA and Canada. Crop J. 2019, 7, 730–738. [Google Scholar] [CrossRef]
- Bradshaw, D.; Gans, C.; Jones, P.; Rizzuto, G.; Steiner, N.; Mitton, W.; Ng, J.; Koester, R.; Hartzman, R.; Hurley, C. Novel HLA-A locus alleles including A*01012, A*0306, A*0308, A*2616, A*2617, A*3009, A*3206, A*3403, A*3602 and A*6604. Tissue Antigens 2002, 59, 325–327. [Google Scholar] [CrossRef]
- Schroeder, H.W.; Christensen, J.J. Factors affecting resistance of wheat to scab caused by Gibberella zeae. Phytopathology 1963, 53, 831–838. [Google Scholar]
- Mesterházy, Á. Types and components of resistance to Fusarium head blight of wheat. Plant Breed. 1995, 114, 377–386. [Google Scholar] [CrossRef]
- Buerstmayr, M.; Huber, K.; Heckmann, J.; Steiner, B.; Nelson, J.C.; Buerstmayr, H. Mapping of QTL for Fusarium head blight resistance and morphological and developmental traits in three backcross populations derived from Triticum dicoccum × Triticum durum. Theor. Appl. Genet. 2012, 125, 1751–1765. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhao, Y.; Rodemann, B.; Plieske, J.; Kollers, S.; Korzun, V.; Ebmeyer, E.; Argillier, O.; Hinze, M.; Ling, J.; et al. Potential and limits to unravel the genetic architecture and predict the variation of Fusarium head blight resistance in European winter wheat (Triticum aestivum L.). Heredity 2014, 114, 318–326. [Google Scholar] [CrossRef] [PubMed]
- Mesterhazy, A.; Toth, B.; Varga, M.; Bartók, T.; Szabó-Hevér, Á.; Farády, L.; Lehoczki-Krsjak, S. Role of Fungicides, Application of Nozzle Types, and the Resistance Level of Wheat Varieties in the Control of Fusarium Head Blight and Deoxynivalenol. Toxins 2011, 3, 1453–1483. [Google Scholar] [CrossRef] [PubMed]
- Snape, J.W.; Riggs, T.J. Genetical consequences of single seed descent in the breeding of self-pollinating crops. Heredity 1975, 35, 211–219. [Google Scholar] [CrossRef]
- Juhász, A.; Larroque, O.R.; Tamas, L.; Hsam, S.L.K.; Zeller, F.J.; Békés, F.; Bedő, Z. Bánkúti 1201—An old Hungarian wheat variety with special storage protein composition. Theor. Appl. Genet. 2003, 107, 697–704. [Google Scholar] [CrossRef]
- Földi, M.; Drexler, D. Organic field trials to promote Hungarian organic cereal production–Testing wheat varieties. Acta Fytotech. et Zootech. 2015, 18, 94–97. [Google Scholar] [CrossRef][Green Version]
- Nirenberg, H. Untersuchungen über die morphologische und biologische Differenzierung in der Fusarium-Section Liseola. In Mitteilungen der Biol. Bundesanstalt für Land- und Forstwirtschaft; Arno Brynda GmbH: Berlin, Germany, 1976; pp. 1–117. [Google Scholar]
- Bal, G.H.; Shaner, G. Scab of Wheat: Prospects For Control. Plant Dis. 1994, 78, 760. [Google Scholar] [CrossRef]
- Snijders, C.H.A.; Van Eeuwijk, F.A. Genotype × strain interactions for resistance to Fusarium head blight caused by Fusarium culmorum in winter wheat. Theor. Appl. Genet. 1991, 81, 239–244. [Google Scholar] [CrossRef]
- Stack, R.W. A comparison of the inoculum potential of ascospores and conidia of Gibberella zeae. Can. J. Plant Pathol. 1989, 11, 137–142. [Google Scholar] [CrossRef]
- Dill-Macky, R. Inoculation methods and evaluation of Fusarium head blight resistance in wheat. In Fusarium Head Blight of Wheat and Barley; Leonard, K.J., Bushnell, W.R., Eds.; APS Press: St. Paul, MN, USA, 2003; pp. 184–210. [Google Scholar]
- Tischner, T.; Kőszegi, B.; Veisz, O. Climatic programmes used in the Martonvásár phytotron most frequently in recent years. Acta Agron. Hungarica 1997, 45, 85–104. [Google Scholar]
- Lancashire, P.D.; Bleiholder, H.; Boom, T.V.D.; Langelüddeke, P.; Stauss, R.; Weber, E.; Witzenberger, A. A uniform decimal code for growth stages of crops and weeds. Ann. Appl. Biol. 1991, 119, 561–601. [Google Scholar] [CrossRef]
- Madden, L.V.; Hughes, G.; Bosch, F.V.D. The Study of Plant Disease Epidemics; Scientific Societies; The Am. Phytopathol. Soc.: St. Paul, MN, USA, 2017; p. 421. [Google Scholar]
- The R Project for Statistical Computing. Available online: http://www.r-project.org/ (accessed on 13 February 2012).
- Søren Højsgaard, A.; Halekoh, U.; Søren Højsgaard, M. Package “doBy”. Available online: https://cran.r-project.org/web/packages/doBy/doBy.pdf. (accessed on 2 November 2018).
- Peterson, B.G. Package “PerformanceAnalytics”. Available online: https://cran.r-project.org/web/packages/PerformanceAnalytics/PerformanceAnalytics.pdf. (accessed on 2 November 2018).
- Yang, Z.; Gilbert, J.; Somers, D.; Fedak, G.; Procunier, J.; McKenzie, I. Marker Assisted Selection of Fusarium Head Blight Resistance Genes in Two Doubled Haploid Populations of Wheat. Mol. Breed. 2003, 12, 309–317. [Google Scholar] [CrossRef]
- Wen, W.; He, Z.; Gao, F.; Liu, J.; Jin, H.; Zhai, S.; Qu, Y.; Xia, X. A High-Density Consensus Map of Common Wheat Integrating Four Mapping Populations Scanned by the 90K SNP Array. Front. Plant Sci. 2017, 8, 8. [Google Scholar] [CrossRef] [PubMed]
- Somers, D.J.; Isaac, P.; Edwards, K. A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 2004, 109, 1105–1114. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Liu, X.; Wang, J.; Li, M.; Wang, Q.; Tian, F.; Su, Z.; Pan, Y.; Liu, D.; Lipka, A.E.; et al. GAPIT Version 2: An Enhanced Integrated Tool for Genomic Association and Prediction. Plant Genome 2016, 9, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Rasheed, A.; Hao, Y.; Xia, X.; Khan, A.; Xu, Y.; Varshney, R.K.; He, Z. Crop Breeding Chips and Genotyping Platforms: Progress, Challenges, and Perspectives. Mol. Plant 2017, 10, 1047–1064. [Google Scholar] [CrossRef]
- Arruda, M.P.; Brown, P.J.; Lipka, A.E.; Krill, A.M.; Thurber, C.; Kolb, F.L. Genomic Selection for Predicting Fusarium Head Blight Resistance in a Wheat Breeding Program. Plant Genome 2015, 8, 1–12. [Google Scholar] [CrossRef]
- Arruda, M.P.; Brown, P.; Brown-Guedira, G.; Krill, A.M.; Thurber, C.; Merrill, K.R.; Foresman, B.J.; Kolb, F.L. Genome-Wide Association Mapping of Fusarium Head Blight Resistance in Wheat using Genotyping-by-Sequencing. Plant Genome 2016, 9. [Google Scholar] [CrossRef]
- Zeng, A.; Chen, P.; Korth, K.; Hancock, F.; Pereira, A.; Brye, K.; Wu, C.; Shi, A. Genome-wide association study (GWAS) of salt tolerance in worldwide soybean germplasm lines. Mol. Breed. 2017, 37, 85. [Google Scholar] [CrossRef]
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting Linear Mixed-Effects Models Using lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- Parikka, P.; Hakala, K.; Tiilikkala, K. Expected shifts in Fusarium species’ composition on cereal grain in Northern Europe due to climatic change. Food Addit. Contam. Part A 2012, 29, 1543–1555. [Google Scholar] [CrossRef]
- László, E.; Puskás, K.; Vida, G.; Bedö, Z.; Veisz, O. Study of Fusarium head blight resistance in old Hungarian wheat cultivars. Cereal Res. Commun. 2007, 35, 717–720. [Google Scholar] [CrossRef]
- Lu, Q.; Szabó-Hevér, Á.; Bjørnstad, Å.; Lillemo, M.; Semagn, K.; Mesterházy, Á.; Ji, F.; Shi, J.; Skinnes, H. Two Major Resistance Quantitative Trait Loci are Required to Counteract the Increased Susceptibility to Fusarium Head Blight of the Rht-D1b Dwarfing Gene in Wheat. Crop. Sci. 2011, 51, 2430–2438. [Google Scholar] [CrossRef]
- Mao, S.-L.; Wei, Y.-M.; Cao, W.; Lan, X.-J.; Yu, M.; Chen, Z.-M.; Chen, G.-Y.; Zheng, Y.-L. Confirmation of the relationship between plant height and Fusarium head blight resistance in wheat (Triticum aestivum L.) by QTL meta-analysis. Euphytica 2010, 174, 343–356. [Google Scholar] [CrossRef]
- McIntosh, R.A.; Dubcovsky, J.; Rogers, W.J.; Morris, C.; Xia, X.C. Catalogue of gene symbols for wheat. National BioResource Project (NBRP): KOMUGI-Integrated Wheat Science Database: Genome / Development. Available online: https://shigen.nig.ac.jp/wheat/komugi/genes/symbolClassList.jsp (accessed on 29 November 2018).
- Waldron, B.L.; Moreno-Sevilla, B.; Anderson, J.A.; Stack, R.W.; Frohberg, R.C. RFLP Mapping of QTL for Fusarium Head Blight Resistance in Wheat. Crop. Sci. 1999, 39, 805–811. [Google Scholar] [CrossRef]
- Paillard, S.; Schnurbusch, T.; Tiwari, R.; Messmer, M.; Winzeler, M.; Keller, B.; Schachermayr, G. QTL analysis of resistance to Fusarium head blight in Swiss winter wheat (Triticum aestivum L.). Theor. Appl. Genet. 2004, 109, 323–332. [Google Scholar] [CrossRef]
- Somers, D.J.; Fedak, G.; Clarke, J.; Cao, W. Mapping of FHB resistance QTLs in tetraploid wheat. Genome 2006, 49, 1586–1593. [Google Scholar] [CrossRef]
- Gervais, L.; Dedryver, F.; Morlais, J.-Y.; Bodusseau, V.; Negre, S.; Bilous, M.; Groos, C.; Trottet, M. Mapping of quantitative trait loci for field resistance to Fusarium head blight in an European winter wheat. Theor. Appl. Genet. 2002, 106, 961–970. [Google Scholar] [CrossRef]
- Shen, X.; Ittu, M.; Ohm, H. Quantitative Trait Loci Conditioning Resistance to Fusarium Head Blight in Wheat Line F201R. Crop. Sci. 2003, 43, 850–857. [Google Scholar] [CrossRef]
- Kumar, S.; Stack, R.W.; Friesen, T.L.; Faris, J.D. Identification of a Novel Fusarium Head Blight Resistance Quantitative Trait Locus on Chromosome 7A in Tetraploid Wheat. Phytopathology 2007, 97, 592–597. [Google Scholar] [CrossRef]
- Buerstmayr, H.; Lemmens, M.; Hartl, L.; Doldi, L.; Steiner, B.; Stierschneider, M.; Ruckenbauer, P. Molecular mapping of QTLs for Fusarium head blight resistance in spring wheat. I. Resistance to fungal spread (Type II resistance). Theor. Appl. Genet. 2002, 104, 84–91. [Google Scholar] [CrossRef]
- Buerstmayr, H.; Steiner, B.; Hartl, L.; Griesser, M.; Angerer, N.; Lengauer, D.; Miedaner, T.; Schneider, B.; Lemmens, M. Molecular mapping of QTLs for Fusarium head blight resistance in spring wheat. II. Resistance to fungal penetration and spread. Theor. Appl. Genet. 2003, 107, 503–508. [Google Scholar] [CrossRef] [PubMed]




| Head Infection (%) | |||||
|---|---|---|---|---|---|
| Parents | Lines (n = 173) | ||||
| ’BKT9086-95′ | ’Mv Magvas’ | Average | Range | Standard Deviation | |
| 2007 | 32.25 | 95.24 | 52.95 | 4.09–95.91 | 24.75 |
| 2008 | 12.00 | 94.43 | 24.17 | 5.15–96.45 | 17.99 |
| 2009 | 38.06 | 97.52 | 65.20 | 3.85–100.00 | 25.44 |
| 21 DAI | ||||
| SST | Df | F-value | Pr > F | |
| Genotype | 448,802 | 174 | 4.001 | 1.45 × 10 −10 *** |
| Year | 5751 | 1 | 2.461 | 0.035 * |
| Genotype × Year | 123,863 | 174 | 0.552 | 1.000 |
| Residual value | 1,026,146 | 796 | ||
| AUDPC | ||||
| SST | Df | F-value | Pr > F | |
| Genotype | 51,294,274 | 174 | 2.361 | 1.2 × 10 −15 *** |
| Year | 50,592 | 1 | 0.405 | 0.525 |
| Genotype × Year | 16,902,477 | 174 | 0.778 | 0.979 |
| Residual value | 99,403,034 | 796 | ||
| Head Infection (%) | ||||||
|---|---|---|---|---|---|---|
| Parents | Lines (n = 221) | |||||
| Isolate | ’BKT 9086-95′ | ’Mv Magvas’ | Average | Range | Dev. | |
| 2006 | Fc | 6.67 | 100.0 | 30.90 | 4.35–76.90 | 18.14 |
| Fg | 8.51 | 92.86 | 25.27 | 4.35–92.56 | 14.14 | |
| 2009 | Fg | 26.19 | 100.0 | 47.10 | 7.01–91.67 | 17.49 |
| 2011 | Fc | 25.53 | 72.75 | 53.16 | 8.95–100.00 | 18.83 |
| Fg | 23.99 | 85.99 | 65.95 | 9.93–100.00 | 20.62 | |
| Ave. | Fc | 16.10 | 86.38 | 42.03 | 4.37–100.00 | 18.22 |
| Fg | 19.56 | 92.95 | 46.10 | 9.93–100.00 | 20.74 | |
| Main average | 18.18 | 89.67 | 44.48 | 11.19–90.83 | 12.99 | |
| PH | 125.0 | 85.00 | 115.6 | 85.00–142.50 | 11.74 | |
| Heading | 19.0 | 21.00 | 21.60 | 14–30 | 2.51 | |
| Ear comp. | 1.67 | 2.50 | 2.10 | 1.53–2.78 | 0.28 | |
| 21 DAI | ||||
| SST | Df | F-value | Pr > F | |
| Genotype | 609,534 | 220 | 2.9057 | 2.2 × 10−16 *** |
| Year | 55,986 | 2 | 5.4406 | 6.829 × 10 −7 *** |
| Date of infection | 37,636 | 12 | 3.3491 | 0.019742 * |
| Plant height | 5095 | 16 | 3.7365 | 7.247 × 10 −5 *** |
| Ear compactness | 12,525 | 25 | 3.3436 | 0.009695 ** |
| Genotype × Year | 190,936 | 192 | 1.0619 | 0.272554 |
| Residual value | 685,810 | 2868 | ||
| AUDPC | ||||
| SST | Df | F-value | Pr > F | |
| Genotype | 39,177,695 | 220 | 2.8577 | 2 × 10−16 *** |
| Year | 1,875,384 | 2 | 1.9151 | 0.01534 * |
| Date of infection | 1,610,250 | 12 | 2.1925 | 0.0994 * |
| Plant height | 28,482 | 16 | 0.4654 | 0.49518 |
| Ear compactness | 385,892 | 25 | 1.5763 | 0.17782 |
| Genotype×Year | 12,957,508 | 192 | 1.1027 | 0.16585 |
| Residual value | 75,532,613 | 2868 | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Varga-László, E.; Puskás, K.; Varga, B.; Farkas, Z.; Veisz, O.; Vida, G. Analysis of Genetic Factors Defining Head Blight Resistance in an Old Hungarian Wheat Variety-Based Mapping Population. Agronomy 2020, 10, 1128. https://doi.org/10.3390/agronomy10081128
Varga-László E, Puskás K, Varga B, Farkas Z, Veisz O, Vida G. Analysis of Genetic Factors Defining Head Blight Resistance in an Old Hungarian Wheat Variety-Based Mapping Population. Agronomy. 2020; 10(8):1128. https://doi.org/10.3390/agronomy10081128
Chicago/Turabian StyleVarga-László, Emese, Katalin Puskás, Balázs Varga, Zsuzsanna Farkas, Ottó Veisz, and Gyula Vida. 2020. "Analysis of Genetic Factors Defining Head Blight Resistance in an Old Hungarian Wheat Variety-Based Mapping Population" Agronomy 10, no. 8: 1128. https://doi.org/10.3390/agronomy10081128
APA StyleVarga-László, E., Puskás, K., Varga, B., Farkas, Z., Veisz, O., & Vida, G. (2020). Analysis of Genetic Factors Defining Head Blight Resistance in an Old Hungarian Wheat Variety-Based Mapping Population. Agronomy, 10(8), 1128. https://doi.org/10.3390/agronomy10081128






