Evaluation of Selection Methods for Resistance to a Specialist Insect Pest of Squash (Cucurbita pepo)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plants
2.2. Acalymma vittatum Assays
2.3. Trichoplusia ni Assays
2.4. Founder Phenotypic Evaluation
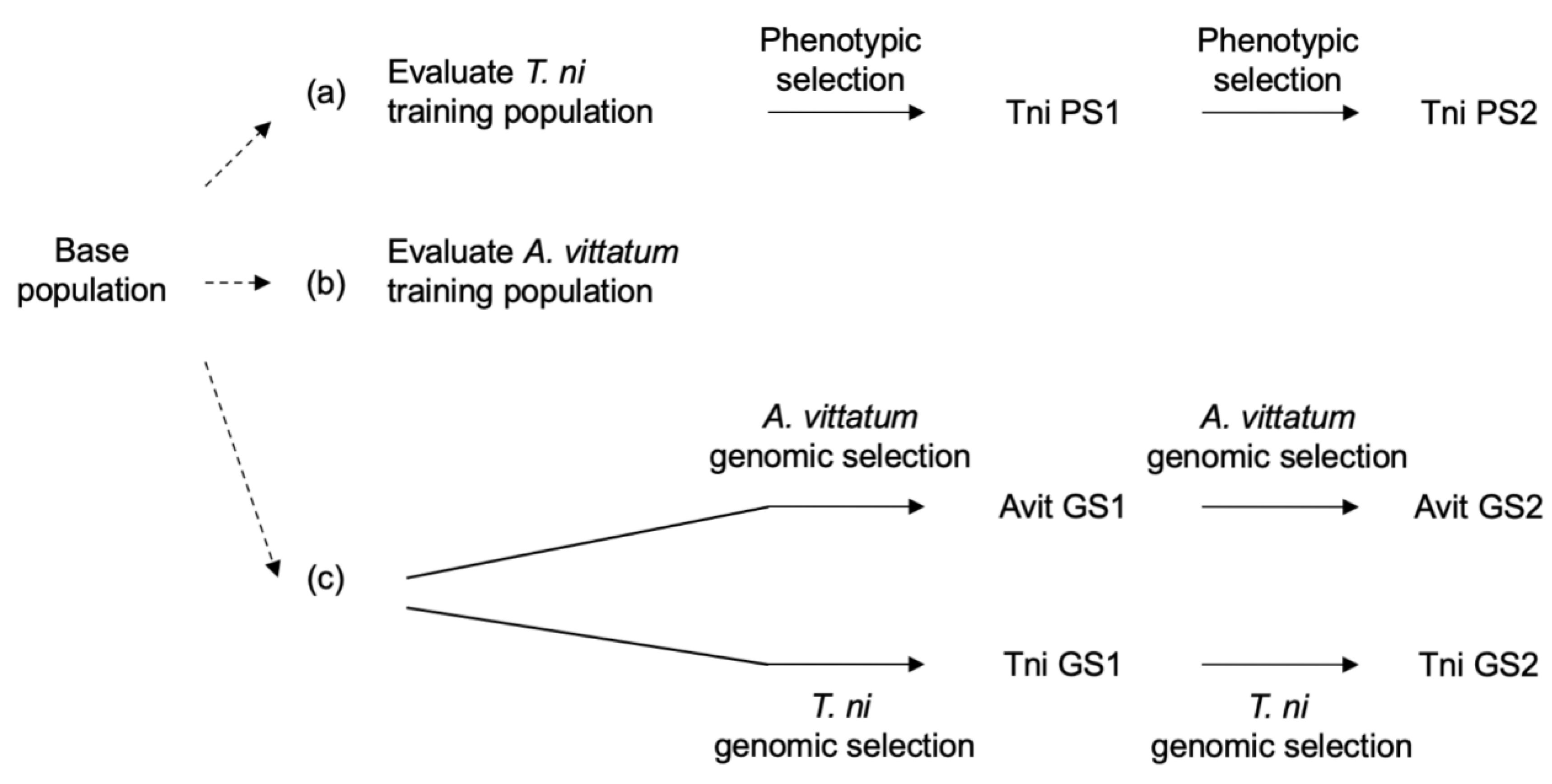
2.5. Indirect Selection Using Resistance to Trichoplusia ni
2.6. Genomic Selection Training Population Evaluation
2.6.1. Resistance to T. ni
2.6.2. Resistance to A. vittatum
2.7. Genotyping and SNP Calling–Training Populations and Genomic Selection
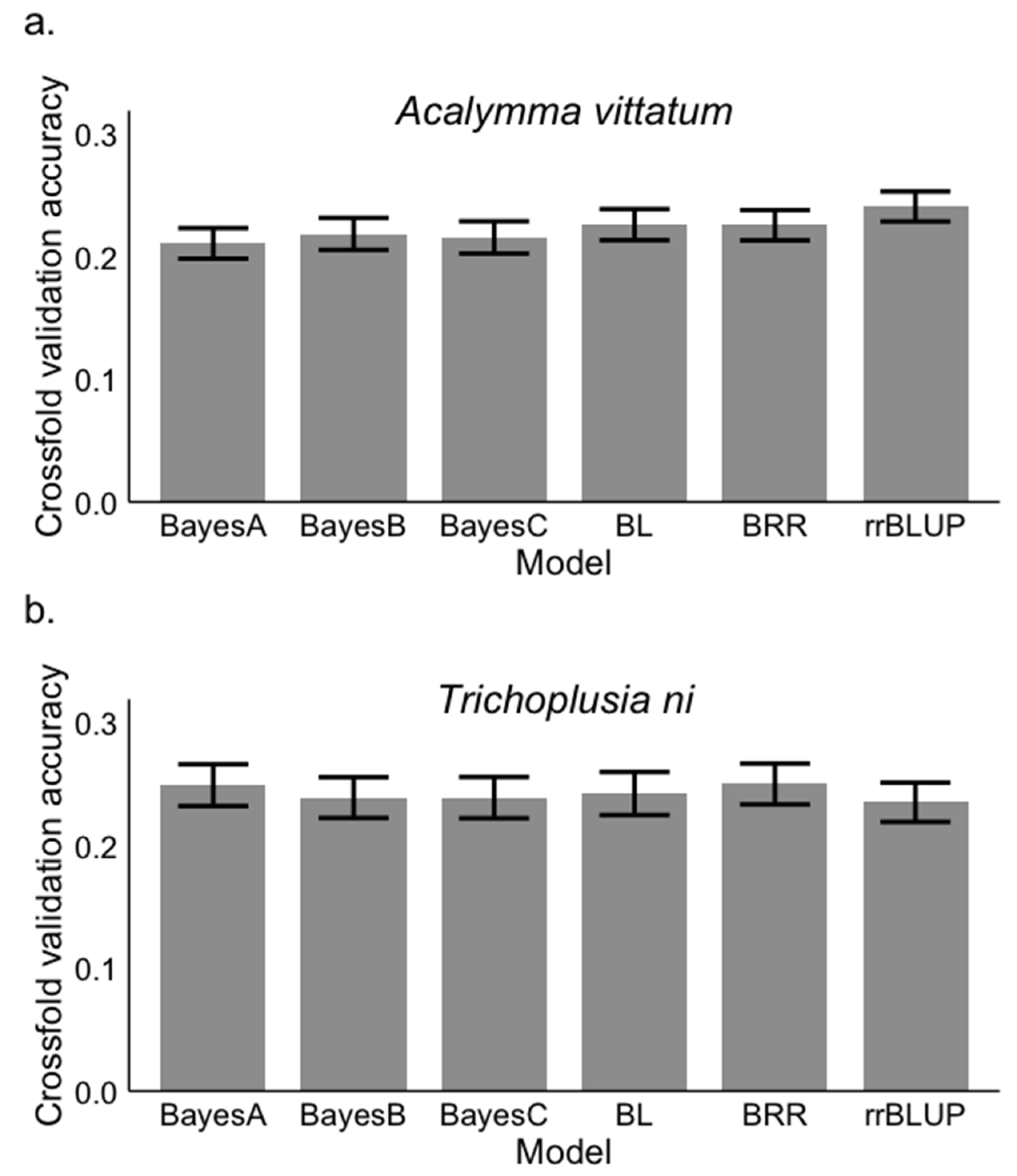
2.8. Genomic Selection
2.9. Realized Gains from Selection
2.9.1. Resistance to Acalymma vittatum
2.9.2. Resistance to T. ni
2.10. Statistics
3. Results
3.1. Phenotypic Extremes in Population Founders
3.2. Indirect Selection Approach
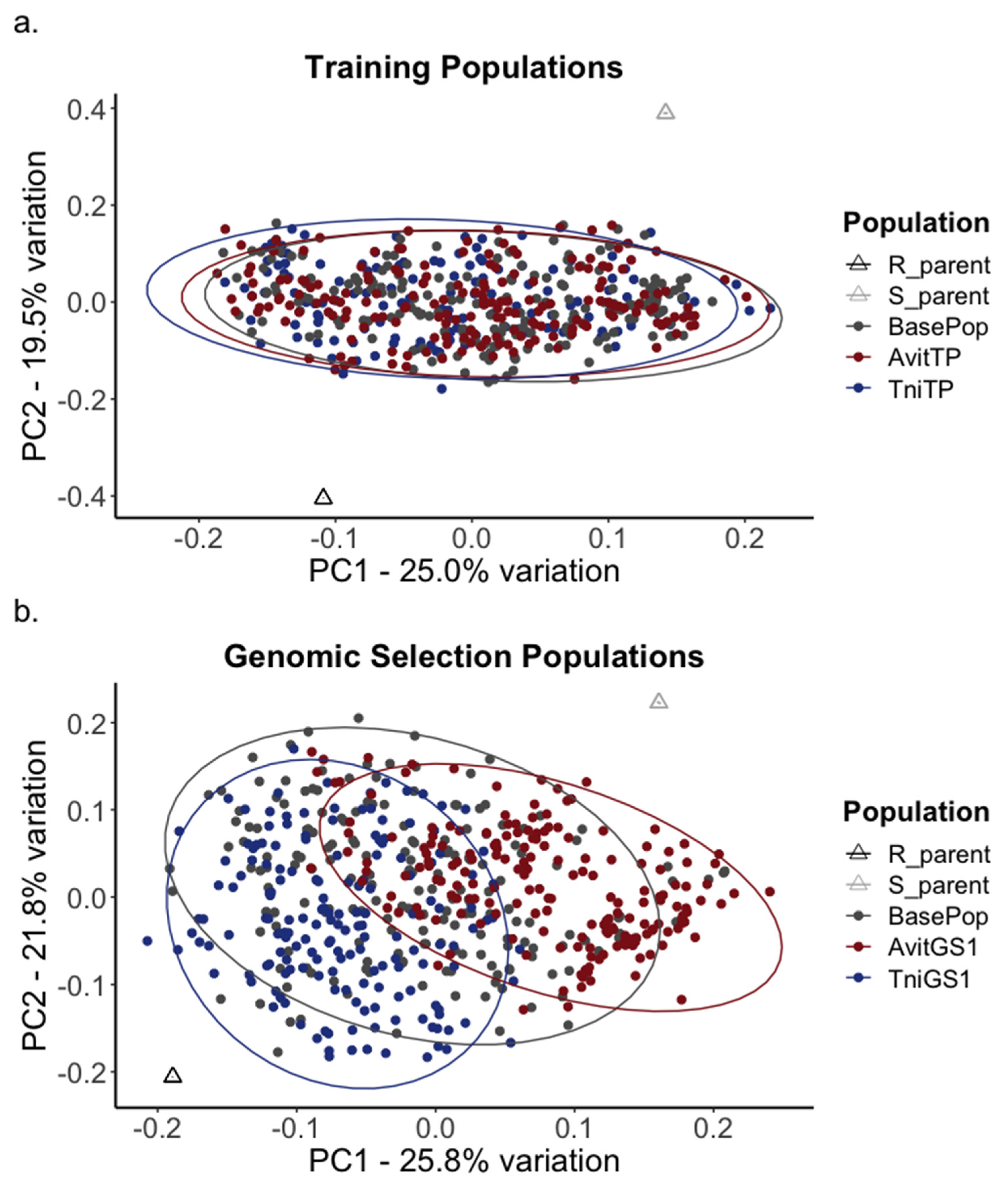
3.3. Genomic Selection Approach
3.4. Realized Gains for Resistance to A. vittatum and T. ni
4. Discussion
4.1. Indirect Selection Approaches
4.2. Genomic Selection Approaches
4.3. Insights into Mechanisms of Resistance to A. vittatum and Future Breeding Directions
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mitchell, C.; Brennan, R.M.; Graham, J.; Karley, A.J. Plant Defense against Herbivorous Pests: Exploiting Resistance and Tolerance Traits for Sustainable Crop Protection. Front. Plant Sci. 2016, 7, 1–8. [Google Scholar] [CrossRef]
- Smith, C.M.; Clement, S.L. Molecular bases of plant resistance to arthropods. Annu. Rev. Entomol. 2012, 57, 309–328. [Google Scholar] [CrossRef]
- Stuart, J. Insect effectors and gene-for-gene interactions with host plants. Curr. Opin. Insect Sci. 2015, 9, 56–61. [Google Scholar] [CrossRef]
- Kliebenstein, D.J. Quantitative Genetics and Genomics of Plant Resistance to Insects. Annu. Plant Rev. Online 2017, 47, 235–262. [Google Scholar] [CrossRef]
- Lush, J. Animal Breeding; Collegiate Press: Ames, IA, USA, 1937. [Google Scholar]
- Cobb, J.N.; DeClerck, G.; Greenberg, A.; Clark, R.; McCouch, S. Next-generation phenotyping: Requirements and strategies for enhancing our understanding of genotype-phenotype relationships and its relevance to crop improvement. Theor. Appl. Genet. 2013, 126, 867–887. [Google Scholar] [CrossRef]
- Widstrom, N.W.; Snook, M.E. Recurrent selection for maysin, a compound in maize silks, antibiotic to earworm. Plant Breed. 2001, 120, 357–359. [Google Scholar] [CrossRef]
- Ben-Mahmoud, S.; Smeda, J.R.; Chappell, T.M.; Stafford-Banks, C.; Kaplinsky, C.H.; Anderson, T.; Mutschler, M.A.; Kennedy, G.G.; Ullman, D.E. Acylsugar amount and fatty acid profile differentially suppress oviposition by western flower thrips, frankliniella occidentalis, on tomato and interspecific hybrid flowers. PLoS ONE 2018, 13, e0201583. [Google Scholar] [CrossRef] [PubMed]
- Moore, V.M.; Tracy, W.F. Recurrent Full-sib Family Selection for Husk Extension in Sweet Corn. J. Am. Soc. Hortic. Sci. 2019, 144, 63–69. [Google Scholar] [CrossRef]
- Berzonsky, W.A.; Ding, H.; Haley, S.D.; Harris, M.O.; Lamb, R.J.; McKenzie, R.I.; Ohm, H.W.; Patterson, F.L.; Peairs, F.B.; Porter, D.R.; et al. Breeding Wheat for Resistance to Insects. In Plant Breeding Reviews; Janick, J., Ed.; John Wiley & Sons: Hoboken, NJ, USA, 2003; Volume 22, pp. 221–230. ISBN 0-471-21541-4. [Google Scholar]
- Falconer, D.S.; MacKay, T.F. Correlated Characters. In Introduction to Quantitative Genetics; Longmans Green: Harlow, UK, 1996; pp. 312–334. [Google Scholar]
- Eberhart, S.A. Factors effecting efficiencies of breeding methods. African Soils 1970, 15, 655–680. [Google Scholar]
- Cobb, J.N.; Juma, R.U.; Biswas, P.S.; Arbelaez, J.D.; Rutkoski, J.; Atlin, G.; Hagen, T.; Quinn, M.; Ng, E.H. Enhancing the rate of genetic gain in public-sector plant breeding programs: Lessons from the breeder’s equation. Theor. Appl. Genet. 2019, 132, 627–645. [Google Scholar] [CrossRef] [PubMed]
- Heffner, E.L.; Sorrells, M.E.; Jannink, J.L. Genomic selection for crop improvement. Crop Sci. 2009, 49, 1–12. [Google Scholar] [CrossRef]
- Heslot, N.; Jannink, J.-L.; Sorrells, M.E. Perspectives for Genomic Selection Applications and Research in Plants. Crop Sci. 2015, 55, 1. [Google Scholar] [CrossRef]
- Bhat, J.A.; Ali, S.; Salgotra, R.K.; Mir, Z.A.; Dutta, S.; Jadon, V.; Tyagi, A.; Mushtaq, M.; Jain, N.; Singh, P.K.; et al. Genomic selection in the era of next generation sequencing for complex traits in plant breeding. Front. Genet. 2016, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Crossa, J.; Pérez-Rodríguez, P.; Cuevas, J.; Montesinos-López, O.; Jarquín, D.; de los Campos, G.; Burgueño, J.; González-Camacho, J.M.; Pérez-Elizalde, S.; Beyene, Y.; et al. Genomic Selection in Plant Breeding: Methods, Models, and Perspectives. Trends Plant Sci. 2017, 22, 961–975. [Google Scholar] [CrossRef] [PubMed]
- Lenz, P.R.N.; Nadeau, S.; Mottet, M.; Perron, M.; Isabel, N.; Beaulieu, J.; Bousquet, J. Multi-trait genomic selection for weevil resistance, growth, and wood quality in Norway spruce. Evol. Appl. 2019, 76–94. [Google Scholar] [CrossRef]
- Metcalf, R.L.; Metcalf, E.R. Diabroticite rootworm beetles. In Plant kairomones in Insect Ecology and Control; Miller, T.A., van Emden, H.F., Eds.; Chapman & Hall: New York, NY, USA, 1992; pp. 64–108. [Google Scholar]
- Rand, R.V. Transmission and control of bacterial wilt of cucurbits. J. Agric. Res. 1916, 6, 417–434. [Google Scholar]
- Gergerich, R.C.; Scott, H.A.; Fulton, J.P. Evaluation of Diabrotica beetles as vectors of plant viruses. In Methods for the Study of Pest Diabrotica; Krysan, J.L., Miller, T.A., Eds.; Springer: New York, NY, USA, 1986; pp. 227–250. [Google Scholar]
- Hoffmann, M.P.; Robinson, R.W.; Kyle, M.M.; Kirkwyland, J.J. Defoliation and infestation of Cucurbita pepo genotypes by Diabroticite beetles. HortScience 1996, 31, 439–442. [Google Scholar] [CrossRef]
- Brzozowski, L.J.; Leckie, B.M.; Gardner, J.; Hoffmann, M.P.; Mazourek, M. Cucurbita pepo subspecies delineates striped cucumber beetle (Acalymma vittatum) preference. Hortic. Res. 2016, 3, 16028. [Google Scholar] [CrossRef]
- Hoffmann, M.P.; Ayyappath, R.; Gardner, J. Effect of striped cucumber beetle (Coleoptera: Chrysomelidae) foliar feeding on pumpkin yield. J. Entomol. Sci. 2003, 38, 439–448. [Google Scholar] [CrossRef]
- Smyth, R.R.; Hoffmann, M.P. A male-produced aggregation pheromone facilitating Acalymma vittatum [F.] (Coleoptera: Chrysomelidae) early-season host plant colonization. J. Insect Behav. 2003, 16, 347–359. [Google Scholar] [CrossRef]
- Metcalf, R.L.; Metcalf, R.A.; Rhodes, A.M. Cucurbitacins as kairomones for diabroticite beetles. Proc. Natl. Acad. Sci. USA 1980, 77, 3769–3772. [Google Scholar] [CrossRef] [PubMed]
- Metcalf, R.L.; Rhodes, A.M.; Metcalf, R.A.; Ferguson, J.; Metcalf, E.R.; Lu, P. Cucurbitacin contents and Diabroticite (Coleoptera: Chrysomelidae) feeding upon Cucurbita ssp. Environ. Entomol. 1982, 11, 931–937. [Google Scholar] [CrossRef]
- Brzozowski, L.J.; Mazourek, M.; Agrawal, A.A. Mechanisms of Resistance to Insect Herbivores in Isolated Breeding Lineages of Cucurbita pepo. J. Chem. Ecol. 2019, 45, 313–325. [Google Scholar] [CrossRef] [PubMed]
- Theis, N.; Barber, N.A.; Gillespie, S.D.; Hazzard, R.V.; Adler, L.S. Attracting mutualists and antagonists: Plant trait variation explains the distribution of specialist floral herbivores and pollinators on crops and wild gourds. Am. J. Bot. 2014, 101, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, D.W.S.; Greene, G.L. Suppression and Management of Cabbage Looper Populations. USDA Tech. Bull. 1984, 1684, 1–13. [Google Scholar]
- Hernandez, C.; Wyatt, L.E.; Mazourek, M. Genomic Prediction and Selection for Fruit Traits in Winter Squash. Submitted.
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Garrick, D.J.; Taylor, J.F.; Fernando, R.L. Deregressing estimated breeding values and weighting information for genomic regression analyses. Genet. Sel. Evol. 2009, 41, 1–8. [Google Scholar] [CrossRef]
- Montero-Pau, J.; Blanca, J.; Bombarely, A.; Ziarsolo, P.; Esteras, C.; Martí-Gómez, C.; Ferriol, M.; Gómez, P.; Jamilena, M.; Mueller, L.; et al. De novo assembly of the zucchini genome reveals a whole-genome duplication associated with the origin of the Cucurbita genus. Plant Biotechnol. J. 2018, 16, 1161–1171. [Google Scholar] [CrossRef]
- Glaubitz, J.C.; Casstevens, T.M.; Lu, F.; Harriman, J.; Elshire, R.J.; Sun, Q.; Buckler, E.S. TASSEL-GBS: A high capacity genotyping by sequencing analysis pipeline. PLoS ONE 2014, 9, e90346. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Fragoso, C.A.; Heffelfinger, C.; Zhao, H.; Dellaporta, S.L. Imputing genotypes in biallelic populations from low-coverage sequence data. Genetics 2016, 202, 487–495. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mohammadi, M.; Tiede, T.; Smith, K.P. PopVar: A Genome-Wide Procedure for Predicting Genetic Variance and Correlated Response in Biparental Breeding Populations. Crop Sci. 2015, 55, 2068–2077. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2016. [Google Scholar]
- Bates, D.; Maechler, M.; Bolker, B.; Walker, S. Fitting Linear Mixed-Effects Models Using lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- Clark, S.A.; van der Werf, J. Genomic Best Linear Unbiased Prediction (gBLUP) for the Estimation of Genomic Breeding Values. In Genome-Wide Association Studies and Genomic Prediction. Methods in Molecular Biology (Methods and Protocols); Gondro, C., van der Werf, J., Hayes, B., Eds.; Humana Press: Totowa, NJ, USA, 2013. [Google Scholar]
- Li, R.; Tsaih, S.W.; Shockley, K.; Stylianou, I.M.; Wergedal, J.; Paigen, B.; Churchill, G.A. Structural model analysis of multiple quantitative traits. PLoS Genet. 2006, 2, 1046–1057. [Google Scholar] [CrossRef] [PubMed]
- Byrne, P.F.; McMullen, M.D.; Wiseman, B.R.; Snook, M.E.; Musket, T.A.; Theuri, J.M.; Widstrom, N.W.; Coe, E.H. Maize silk maysin concentration and corn earworm antibiosis: QTLS and genetic mechanisms. Crop Sci. 1998, 38, 461–471. [Google Scholar] [CrossRef]
- Lee, E.A.; Byrne, P.F.; McMullen, M.D.; Snook, M.E.; Wiseman, B.R.; Widstrom, N.W.; Coe, E.H. Genetic mechanisms underlying apimaysin and maysin synthesis and corn earworm antibiosis in maize (Zea mays L.). Genetics 1998, 149, 1997–2006. [Google Scholar] [PubMed]
- Howe, G.A.; Jander, G. Plant Immunity to Insect Herbivores. Annu. Rev. Plant Biol. 2008, 59, 41–66. [Google Scholar] [CrossRef]
- Ali, J.G.; Agrawal, A.A. Specialist versus generalist insect herbivores and plant defense. Trends Plant Sci. 2012, 17, 293–302. [Google Scholar] [CrossRef] [PubMed]
- Bernardo, R.; Yu, J. Prospects for genomewide selection for quantitative traits in maize. Crop Sci. 2007, 47, 1082–1090. [Google Scholar] [CrossRef]
- Voss-Fels, K.P.; Cooper, M.; Hayes, B.J. Accelerating crop genetic gains with genomic selection. Theor. Appl. Genet. 2019, 132, 669–686. [Google Scholar] [CrossRef] [PubMed]
- Asoro, F.G.; Newell, M.A.; Beavis, W.D.; Scott, M.P.; Tinker, N.A.; Jannink, J.L. Genomic, marker-assisted, and pedigree-BLUP selection methods for β-glucan concentration in elite oat. Crop Sci. 2013, 53, 1894–1906. [Google Scholar] [CrossRef]
- Rutkoski, J.; Singh, R.P.; Huerta-Espino, J.; Bhavani, S.; Poland, J.; Jannink, J.L.; Sorrells, M.E. Genetic Gain from Phenotypic and Genomic Selection for Quantitative Resistance to Stem Rust of Wheat. Plant Genome 2015, 8, 1–10. [Google Scholar] [CrossRef]
- Mattson, W.J. Herbivory in Relation to Plant Nitrogen Content. Annu. Rev. Ecol. Syst. 1980, 11, 119–161. [Google Scholar] [CrossRef]
- Fernandez, P.; Hilker, M. Host plant location by Chrysomelidae. Basic Appl. Ecol. 2007, 8, 97–116. [Google Scholar] [CrossRef]
| Generation | As % Less than Mid-Parent | As % Less than High Parent | H2 | ||
|---|---|---|---|---|---|
| µ0 | µselected | µ0 | µselected | ||
| Generation 1 (Base → TniPS1) | 1.04% | 27.60% | 23.69% | 44.18% | 0.228 |
| Generation 2 (TniPS1 → TniPS2) | 18.81% | 59.40% | 46.25% | 73.12% | 0.289 |
| Training Population | Phenotype | H2 |
|---|---|---|
| Acalymma vittatum | Estimated percent leaf damage (%) | 0.066 |
| Measured percent leaf damage (%) | 0.369 | |
| Log(Measured leaf damage) | 0.314 | |
| Measured total damage (cm2) | 0 | |
| Trichoplusia ni | Larvae mass | 0.228 |
| Experiment | Population | Damage (%) | SE | Statistics | ||
|---|---|---|---|---|---|---|
| June 2019 | Base | 12.05 | ± | 2.90 | a | F4,386 = 1.008 p = 0.403 |
| TniPS1 | 14.71 | ± | 2.90 | a | ||
| TniPS2 | 14.79 | ± | 2.90 | a | ||
| TniGS1 | 11.85 | ± | 2.90 | a | ||
| AvitGS1 | 14.35 | ± | 2.90 | a | ||
| August 2019 | Base | 30.94 | ± | 5.07 | b | F4,388 = 9.496 p < 0.001 |
| TniGS1 | 17.32 | ± | 5.07 | a | ||
| TniGS2 | 32.60 | ± | 5.06 | b | ||
| AvitGS1 | 14.98 | ± | 5.06 | a | ||
| AvitGS2 | 22.98 | ± | 5.06 | ab |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brzozowski, L.J.; Mazourek, M. Evaluation of Selection Methods for Resistance to a Specialist Insect Pest of Squash (Cucurbita pepo). Agronomy 2020, 10, 847. https://doi.org/10.3390/agronomy10060847
Brzozowski LJ, Mazourek M. Evaluation of Selection Methods for Resistance to a Specialist Insect Pest of Squash (Cucurbita pepo). Agronomy. 2020; 10(6):847. https://doi.org/10.3390/agronomy10060847
Chicago/Turabian StyleBrzozowski, Lauren J., and Michael Mazourek. 2020. "Evaluation of Selection Methods for Resistance to a Specialist Insect Pest of Squash (Cucurbita pepo)" Agronomy 10, no. 6: 847. https://doi.org/10.3390/agronomy10060847
APA StyleBrzozowski, L. J., & Mazourek, M. (2020). Evaluation of Selection Methods for Resistance to a Specialist Insect Pest of Squash (Cucurbita pepo). Agronomy, 10(6), 847. https://doi.org/10.3390/agronomy10060847





