Abstract
Currently, the global agricultural system is focused on a limited number of crop species, thereby presenting a threat to food security and supply, especially with predicted global climate change conditions. The importance of ‘underutilized’ crop species in meeting the world’s demand for food has been duly recognized by research communities, governments and policy makers worldwide. The development of underutilized crops, with their vast genetic resources and beneficial traits, may be a useful step towards solving food security challenges by offering a multifaceted agricultural system that includes additional important food resources. Bambara groundnut is among the beneficial underutilized crop species that may have a positive impact on global food security through organized and well-coordinated multidimensional breeding programs. The excessive degrees of allelic difference in Bambara groundnut germplasm could be exploited in breeding activities to develop new varieties. It is important to match recognized breeding objectives with documented diversity in order to significantly improve breeding. This review assesses the genetic diversity of Bambara groundnut, as well as important factors involved in realizing and harnessing the potential of this crop.
1. Introduction
There is increasing concern over the state of food and nutritional security in the world due to the over-reliance on major crop species as the main source of food and nutrition [1]. The global population is projected to reach about 9.7 billion by 2050 and the harvested yield from the major food crops may be insufficient to meet the food demand of the projected population [2,3]. Similarly, the threat of global climate degradation on food security has become evident, for example, in fluctuating temperatures, prolonged droughts, soil degradation, salinity and flooding, as well as an increase in disease and pest conditions that considerably disrupt the growth and performance of the major crop species. Therefore, it is imperative to explore the available plant genetic diversity resources to boost food production and supply as a means of reducing the overreliance on major food crops and to promote global food security [4,5]. A more diversified system of agriculture that can provide a solution to the aforementioned problems could be achieved through the incorporation and utilization of underutilized crop species as the chief source of nutrition. This will prove beneficial in achieving global food security [6].
The term ‘Underutilized’ can be used to describe the abandonment of a species by indigenous and international research communities. Underutilized species are generally categorized as crops with little relevance on the global level. These crops, however, play a significant role in terms of climate change and food security, while improving the living standards of low-income families and consumers in developing nations [7,8]. Underutilized crop species, with essential nutrients, can fit into several niches in food production systems and can better adapt to low-input systems in resource-poor regions of the world [9,10]. Presently, these species are either harvested in the wild for cultivation as future or domesticated crops; however, the underutilized crops are mostly cultivated and valued at subsistence or regional levels. In all cases, there is a dearth of research on the production of these underutilized crop species. Despite their neglect, underutilized crops have gained considerable attention from the media in recent times [11], due to their prospects in addressing several UN Sustainable Development [12], and in poor nations of Africa [13] and the Latin America [14]. Additionally, the increasing interest on healthy food alternatives by Western consumers have aided the recent popularity of underutilized crop species [15]. While limited scientific research has been conducted on underutilized crop species, a small number of these species have been adequately researched, mainly due to the commitment of institutions and researchers in the developing world, as well as technical and financial aid from the developed world [16]. Considering their numerous dietary values and tolerance to harsh environmental conditions, some of these underutilized crop species, such as Bambara groundnut, are regarded as crops for the future. Additionally, these crops are highly fortified, with essential nutrients that are excellent in promoting health, and have the ability to combat malnutrition and other related diseases. However, these important species usually fall short of essential, commercial valued crops, such as high yielding varieties that may attract growers with extensive resources and inputs required to achieve the improved yield performance of the crops [17,18]. Incorporating these underutilized crop species, specifically Bambara groundnut, in diversifying the food chain can have a positive impact by serving as a general tool for improving human nutrition. Bambara groundnut is highly nutritious, making it relevant in the nutritional formulation of people that cannot afford expensive protein sources, especially animal-based protein [19]. The seed of Bambara groundnut contains 61–69% carbohydrate, protein 17–27%, fiber 3.3–6.4%, ash 3.1–4.4%, and fat 3.6–7.4%, thus making it a valuable dietary source [20,21]. It also contains 95.5–99.0 mg Ca, 5.1–9.0 mg K, and 2.9–10.6 mg Na per 100 g, with a substantial amount of Zinc (20.98 ± 1.07 mg/100g), which may have a significant impact on preventing prostate cancer in men [22]. The freshly harvested pods, as well as the dry seeds, are processed and consumed in different forms (Figure 1). Freshly harvested pods are boiled or grilled and consumed as snacks [23]. The seeds have a higher content (80%) of high-quality amino acids, such as arginine, leucine, valine, methionine, and lysine, as compared to cowpea, soybeans and groundnut (64%, 74% and 65%, respectively), and may potentially complement the deficient essential amino acid content of foods [22].

Figure 1.
Some examples of processed food products made from Bambara groundnut.
Bambara groundnut (Vigna subterranea (L.) Verdc.) is one of the essential but forgotten and underutilized annual leguminous crops belonging to the Fabaceae family, which is indigenous to the tropical African region. This crop is commonly cultivated in Central and West African regions, mainly for its nutritional benefits and high tolerance to drought stress, unlike other essential legumes [24]. Generally, the plant looks similar to peanuts, with compound trifoliate. The pods contain one-to-two seeds that are born underground just like peanuts. Being a legume, it provides a benefit to other crops due to its ability to fix atmospheric nitrogen to the soil. It is also resistant to pests and diseases and can grow on poorly drained soils where many other crops cannot thrive [25]. It is widely accepted that Africa is the geographical origin of Bambara groundnut [26,27,28]. Despite the agreement on Africa as the geographical origin of Bambara, the precise area of its domestication has been widely speculated [29]. The most abundant genetic resources exist between the corridor of Nigeria and Cameroon, which is believed to be its origin of dispersal [30]. Goli [31] reported the distribution of wild Bambara types from Jos Plateau and Yola Adamawa in Nigeria to Garoua in Cameroon. It was thought that Bambara groundnut was first introduced to East Africa and Madagascar and then subsequently to South and South-East Asia during the slave trade era. In a study by Takahashi et al. [2], it was stated that landraces of the Thai’s Bambara were from both West Africa (Nigeria) and East Africa, thus implying that this crop may have been introduced to Thailand on several occasions. Beyond its cultural importance, one of the major reasons why Bambara groundnut is still cultivated by local farmers is due to its characteristic high yield under drought conditions [32]. However, this crop remains cultivated as landraces comprising selected inbred lines based on agro-ecology. Hence, this review provides an overview of the impact and constraints to Bambara groundnut production, and the major achievements recorded to date in Bambara groundnut research with regards to its agronomy, breeding, and improvement. Through retrospective assessments, this review ultimately intended to present the highlights of the improvement prospects of this crop for future directions.
2. Constraints to Bambara Groundnut Use
Although Bambara groundnut has a long history, its cultivation is still from the local landraces rather than established varieties with a specific breeding purpose suited to a particular agro-ecological region or system of production as stated by many researchres [33,34,35]. However, until recently, the Bambara breeding system has not been fully understood as no true variety was produced. Like other indigenous African crops, Bambara groundnut has almost been ignored by researchers and funding bodies due to its position as a poor man’s crop, cultivated for sustenance instead of cash [36]. While it is hard to precisely state which characteristics defined a crop as being underutilized, the underutilization of crops is mostly connected to the following constraints:
- (1)
- The traditional or cultural heritage of areas where the crop originated;
- (2)
- Inadequate knowledge and documentation on the crop’s morphology and cultivation;
- (3)
- Adaptation to particular agroecological zones;
- (4)
- Lack of genetic improvement and access to laboratory equipment and facilities [37].
Unlike in major crops such as wheat, rice, maize and potato, Bambara groundnut has been neglected and underutilized due to the perception by the industry, international research bodies and the scientific community that this crop lacks economic importance [38,39]. This underutilization has also resulted in a lack of genetic improvement, lower crop yield and quality. One of the reasons why Bambara groundnut remained neglected and underutilized may be attributed to the introduction of Groundnut from Brazil to West Africa by the Portuguese, which may have replaced traditional Bambara groundnut seeds because Groundnut contains a reasonable quantity of lipid and can be cultivated as an oilseed crop. Therefore, a subsistence food crop of the indigenous population is replaced with a cash crop with export potentials. Another important reason for Bambara groundnut’s underutilization is its hard-to-cook characteristic, as well as the inadequate processing methods for this crop [40]. Hard-to-cook for Bambara groundnut is characterized by the prolonged cooking time (3–4 h) required to achieve suitable softening in cooking [41], and the hardness of the seed coat makes it extremely difficult to achieve better dehulling [42].
Consequently, a lack of functional value chains, which ensures the delivery of this valuable crop from the farmers to the final consumers as a processed and refined product, is among the constraints (bottleneck) facilitating the underutilization of this crop [43]. Additionally, there is limited information and knowledge on germplasm collection, biological aspects such as photoperiodic sensitivity, reproductive biology and pod-filling that affects the time/range of planting, geographic location and yield stability. The problem of mechanization and the unavailability of machinery for post-harvest operations like dehulling have also contributed to its underutilization [41]. However, despite the enumerated beneficial characteristics of Bambara groundnut in the introductory stage of this review, the low yield has been identified as another key constraint to the improvement of this crucial crop. Farmers have reported low yields of Bambara groundnut in sub-Saharan Africa, thus necessitating the need for research on breeding and the improvement of varieties with better agronomic practices aimed at yield improvement. In future breeding attempts, it is critical to consider the local practices, suitability and close involvement of farmer groups to enable them to identify suitable lines for selection to meet their immediate requirements so that the identified traits will be back-crossed to the local lines. Massawe et al. [44] and Hillocks et al. [43] emphasized the significance of this method from a socio-economic point of view.
3. Genetic Diversity in Bambara Groundnut
The assessment of available genetic diversity is fundamental in the improvement of Bambara groundnut, which is mostly restricted to small scale traditional farming systems in which they have been commonly cultivated from the existing landraces [45]. Landraces are more phenotypically and genotypically diverse compared to pure lines and are excellent sources of genetic variation for breeding [46]. Cultivated landraces were developed from the wild progenitor (Vigna subterranea var. spontanea) [47]. Bambara groundnut is grown from landraces in all the major growing regions, particularly in sub-Saharan Africa, and its yield can be unstable and unpredictable across different geographical regions. While being adapted to their current environment, landraces may not contain the optimal combination of traits [44].
Globally, up to 6145 Bambara groundnut landraces/accessions are conserved ex-situ and these collections are kept in trust by international or regional gene banks, which are comprised of several countries (Table 1). Genetic variability, which could be beneficial for the improvement of the genetic performance of any crop species [48], is largely preserved in the form of landraces [48]. A significant quantity of genetic diversity has been maintained in the landraces of Bambara groundnut under low input systems of farming [47]. Traditional farmers of Bambara groundnut depend on the prevailing diversity among the cultivated landraces and this has enhanced the maintenance of on-farm genetic diversity in its conservation [49]. Ex-situ conservation of Bambara groundnut landraces is necessary for the crop’s future genetic improvement programs. However, landraces are problematic when it comes to understanding the genetic background of traits of interest for crop improvement because they are a mixture of numerous genotypes (Figure 2), which may bring about confusion between genotypic and environmental effects [50]. These genetic resources are the basis for present and future food security [51]. Genetic diversity within lines and populations is central to breeding and germplasm conservation programs [52]. As such, it is pertinent to know the genetic diversity among breeding materials to avoid the risks related to increased uniformity in elite germplasm, and to ensure long-term selection gain as a cross between the limited number of elite lines that put them at risk of losing their genetic diversity [53].

Table 1.
Bambara groundnut landraces/accessions and wild-type genotypes held by international institutions across some selected countries.
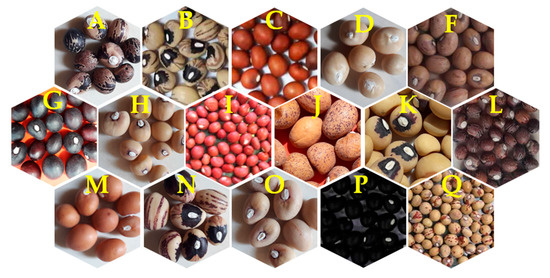
Figure 2.
Seed morphological genetic diversity among different landraces of Bambara groundnut (Note: A = Bidi, B = Maiki, C = Bidilalle, D = Karu, F = Maizane, G = Doii, H = Ex-Sokoto, I = Jatau, J = Bidiyashi, K = Hawayenzaki, L = Zabuwa, M = Yar-Gombe, N = Maibargo, O = Giwa, P-Dunabaki, Q = Cancaraki).
The landmass of a country is usually made up of predominantly different soil types [56] and the variation in soil types may contribute to the diversity of Bambara groundnut. In addition to autogamy and farmer preferences, geographic and ecological isolation fosters agro-morphological diversity among local populations of Bambara groundnut for their adjustment and adaptation to their respective growing areas [57]. The constant influence of these evolutionary factors results in the adaptive characteristics of the accessions of each population [48,58]. Knowledge of the genetic potentials of Bambara groundnut is managed by local farmers in the different agro-ecological zones, which are considered limited, and as a result, the crop has been poorly exploited [31]. Furthermore, in-depth information on the constraints and difficulties faced by an individual underutilized crop, like Bambara groundnut, may perhaps be translated to the general improvement of other underutilized species. Generally, the results demonstrated that different local populations of Bambara groundnut have several genotypes with the potential capacity to produce and adapted to changing environmental conditions [59]. These genotypes could avail options of plant breeders in regards to improving the adaptation and yield of Bambara groundnut in different agro-ecological zones, in consideration of climate change. To achieve a well-coordinated plant breeding and improvement program, it is necessary to embark on an excellent germplasm collection of superior qualities, where scientists and breeders can utilize the information to classify and select parental genotypes for crop improvement schemes [60,61]. Genotypes collected from a country or different locations in a country may be similar or may have common ancestry with diverse native terms [44]. For example, Bonny et al. [62] conducted a study on variability, based on agro-morphological characters among and within the population of four agro-ecological regions of the species in Côte d’Ivoire. Results from their study indicated a significant phenotypic variation among all the traits studied and in the overall population. Consequently, their findings proposed that at every agro-ecological region, Bambara groundnut farmers hold a significant genetic diversity on their farms. Taking into account the results of individual and overall population, mean, dispersion and the coefficient of variation obtained for every character were significantly greater than those obtained in a similar study by Ntundu et al. [63] who worked on 100 accessions in Tanzania and at IITA in Nigeria by Goli [28] who used 1384 accessions collected across Africa.
Bambara groundnut has become a model drought-tolerant crop due to its ability to grow in a wide range of agro-ecological zones with varying soil conditions and the ability to produce a significant amount of yield under moderate or extreme drought stress. This tolerance to a wide range of environmental stress conditions makes Bambara groundnut an important crop for cultivation in an arid environment and equally as a future crop in regions where projections of climate change indicate a likely occurrence of drought and/or intermittent rainfall patterns [35]. Furthermore, the crop has adapted to different environments, ranging from high daytime temperatures and extreme low night-time temperatures in arid areas of Botswana, to the more humid and far milder climate of Indonesia. Thus, their ability to grow under terminal soil and climate conditions cannot be underestimated. For instance, Mabhaudhi et al. [24] predicted that Bambara groundnut yield and water productivity can increase by 37.5% and 33%, respectively, under the predicted climate change scenario. Similarly, Mabhaudhi et al. [64] established that a suitable environment for the production of Bambara groundnut under climate change would increase, thereby ascertaining its resilience to change in the climate.
Identification of important traits through agro-morphological characterization remains the first step in the collection of available genetic germplasm resources [45]. This procedure is inexpensive, direct and easy to practice; therefore, it is among the standard procedures for assessing genetic variability in many species, particularly for under-researched crop species like Bambara groundnut [65,66], and this procedure is employed for legume characterization using qualitative and quantitative characters. Additionally, Hoque et al. [67] employed a similar process to achieve significant agro-morphological diversity in characterizing rice genotypes. Additionally, this procedure is used to select morphological traits that can be positively correlated with grain yield and can allow plant breeders to make a decision on traits of choice for selection among the evaluated species. According to Eckert et al. [68], agro-morphological divergence in a population is primarily due to environmental influences. Thus, this spatial structuration of agro-morphological diversity could be the result of the influence of variations in climatic factors, especially rainfall, temperature and probably day length. Several studies have shown that temperature [69], humidity [70] and day length [71] have variable effects on the vegetative, phonological and reproductive development of Bambara groundnut. This spatial structuration could also be accentuated by divergent selection and continuous inbreeding because of the species autogamy [72]. Wet climates usually entice farmers to grow accessions of the spreading type that are well suited to long cropping seasons, while in dry climates, farmers would prefer to grow the bunch type due to the short cropping season. Previous research has shown that a significant agro-morphological variation exists among many local genotypes of Bambara groundnut [56]. Bambara groundnut demonstrates a considerable amount of genotypic variability for various phenotypic, physiological, agronomic and seed traits [55]. Studies focused on variation in inter and intra-landrace morphological descriptors (both as sovereign research and as an extensive classification of germplasm) in Bambara groundnut has been comprehensively reported [34,73,74]. Bambara groundnut standard descriptors (Table 2) were used for the phenotypic classification exercise [74,75]. The International Institute of Tropical Agriculture (IITA) in Nigeria evaluated and characterized approximately 1400 accessions of Bambara groundnut using 28 characters (Table 2), and it was discovered that there was significant agronomic and morphological diversity among the accessions, both in terms of qualitative and quantitative characters, and recommended their confirmation through molecular markers [63].

Table 2.
Select documented qualitative morphological variants in Bambara groundnut germplasm.
.
Ntundu et al. [31] and Bonny et al. [62] discovered certain vegetative characters that had prominence in principal component analysis and can be used to distinguish between landraces of Bambara groundnut. Using isozyme markers, traits such as pod weight, seed weight, number of petioles, internode length, leaflet width, and leaflet length were recognized as significant traits to distinguish between domesticated and wild species of Bambara groundnuts [81]. Furthermore, other researchers who worked on crops such as Mungbean, Black gram, rice [82,83] and wheat have successfully used several numerical taxonomic procedures to classify and measure genetic diversity patterns among the collected germplasms, using morphological and agronomic traits to classify the traits responsible for qualities such as yield in a similar manner to Bambara groundnut [84] and soybean [85]. To date, there is no report on the relative characterization of larger Bambara groundnut landraces with regards to the photoperiodic response, pigmentation on banner and wings, and stem hairiness in quantitative traits analysis of this plant. The lack of reporting on the photoperiodic reaction in Bambara groundnut landraces is due to the nonexistence of the protocol for high throughput phenotyping. In many ecological settings, such a phenotyping protocol requires a highly controlled environment, particularly around tropical zones where day length variation is partial. Nonetheless, research on Bambara groundnut focusing on the mechanism of the photo-thermal reaction have been documented by many researchers [86,87,88,89,90]. Aliyu and Massawe [81] used petiole pigmentation in their investigation of the photo-thermal reaction; however, this was not among the listed standard descriptors for Bambara groundnut.
Molecular genetic diversity analyses have aided breeding conclusions and germplasm conservation in crop species [91,92]. Over the past twenty years, various molecular markers for analyzing genetic diversity, specifically designed for Bambara groundnut, were documented to either oppose or validate the reports on phenotypic descriptors (Table 3; [93]. Numerous studies have discussed ways that molecular markers can: offer information and knowledge on genetic diversity in Bambara groundnut [93], determine heterozygosity for the purification of seed [94], link germplasm to specific features such as the geographic region [45], function in quality regulation and can be useful in realizing breeding objectives [77]. Similarly, various studies have been conducted to explore the genetic mechanism of significant traits, such as leaf appearance, stress tolerance [95,96] and can provide a basis for the comparison of crops, with and without complete genome sequences, to pave the way for the translation of positional data into underutilized crops [97]. The use of isozymes among 79 domesticated accessions of 21 wild type populations, to study their genetic diversity, was reported by Pasquet et al. [98], the use Random Amplified Polymorphic DNA RAPD was documented by Rungnoi et al. [99], and Massawe et al. [100] and Ntundu et al. [74] used amplified Fragment Length Polymorphism AFLP on 16 landraces, with cluster analysis that categorized the landraces into three clusters, and principally based their location or geographical origin. The low cost of genotyping per sample has allowed for the development of high throughput genotyping for several species of legume [101]. For example, a 60 k cowpea iSelect Consortium Array to screen 51,128 Single Nucleotide Polymorphism SNPs [100] and a flexible throughput Competitive Allele-Specific Polymerase chain reaction PCR (KASPar) assay for 2005 SNPs in chickpea (Cicer arietinum) [102]. Lately, the University of Nottingham was credited with the development of molecular mapping populations that can aid in the marker-assisted selection of additional improved lines for daylength insensitivity [35].

Table 3.
Some reported molecular marker analyses of genetic diversity in Bambara groundnut.
4. Research to Date
While the yield of Bambara groundnut is somewhat unpredictable, Bambara groundnut collected from the Republic of Benin showed a high degree of morphological variability [34]. Thus, accessions from West Africa were expected to have higher genetic variability, assuming a single region of domestication. However, Aliyu et al. [59] suggested the possibility of the Southern/Eastern African region being a secondary center of domestication or diversity for the crop, based on a detailed analysis of the genetic population structure. This placed the West African and the Southern/Eastern African accessions into two distinct clusters [47]. Seed yields (t ha−1) vary in African countries between locations and landraces (0.5–3 t ha−1) with potential yields of 3–4 t ha−1 [106]. On average, a yield of 0.85 t ha−1 is achievable in Bambara groundnut [50]; however, the crop has been reported to have a genetic potential to produce up to 4000 kg ha−1 [106]. Reports by Somta et al. [94] and Molosiwa et al. [76] advocated Bambara groundnut being individual plants that are extremely inbred. Nevertheless, farmer landraces are quite a mixture of (mostly related) inbred lines and molecular evaluation, and this has shown that this condition persists even amongst the released varieties [107]. An average of 1.3% heterozygosity was reported by Somta et al. [94], with the exception of ten accessions from Burkina Faso and three accessions from Guinea, which had higher heterozygosity (2.99% and 4.99%, respectively), signifying a greater chance of outcrossing in their history. Equally, Molosiwa [65] reported 12 SSR markers used on 123 accessions, which indicated that the inbreeding coefficient ranges between 0.9 and 1.00, with average heterozygosity observed at 0.01; therefore, single seed descent (SSD) line advancement can be the first step in the development of near homozygous pre-breeding Bambara groundnut materials. Based on the inbreeding level observed, a single seed from one plant may represent a variety that can be used to produce pure lines, known as Single Plant Decent (SPD).
This report has shown that new areas of Bambara Groundnut production include, amongst others, Thailand, Indonesia and Malaysia [59]. One of the remarkable attributes of this crop is its ability to flourish in several agro-ecologies, thus enabling its wider coverage and acceptability as an alternative food source in regions where it is cultivated and beyond [108]. Several studies showed the effect of soil variation [109,110] and ecological factors, such as soil moisture stress [111] temperature [68] and day length [112] on vegetative development, phenology and the yield of Bambara groundnut. Considering a probable interaction effect (genotype × environment), the structure of the agro-morphological diversity of local accessions, based on the geographical origin, would be the expression of adaptive characters linked to each collection zone. Milla et al. [113] reported that natural and artificial selection continually contributes to the shape of the adaptation of organisms in different environments. Ouedraogo et al. [114] studied the phenotypic variability of Bambara groundnut accessions from northern Burkina Faso, and reported that the plant canopy, number of pods per plant, seed width, seed length, seed weight per plant and a 100 seed weight were positively correlated with seed yield per plant. However, a negative correlation was observed between days to 50% flowering and yield plant−1, meaning that longer vegetative growth could likely reduce the yield of Bambara groundnut, where more vegetative yields lead to lower seed yields. A limitation of this study is that the landraces were only collected from one region [115]. Similarly, studies on phenology in the southern part of Ghana, where the rainfall pattern is bimodal, indicated that Bambara groundnut can be cultivated twice a year [116,117]. It was also discovered that sowing time affects the yield of Bambara groundnut. Higher yields were recorded during minor rainfall seasons, as compared to the main rainy season [118]. Mabhaudhi and Modi [119] also discovered that there were higher yields in the transition agro-ecological zones where rainfall is low and temperatures are higher compared to the forest zones. Up to 4 t ha−1 of pod yields were obtained in some varieties at the transition agro-ecological region in Ghana. The finding also reported that cultivating Bambara groundnut at the right time in the forest zone could give relatively higher yields. Another important attribute of Bambara groundnut is the availability of genotypes with varying maturity periods, ranging from 3 to 6 months, which makes it suitable for production and harvesting all year round, while minimizing yield loss [120].
In the past three decades, several researchers made efforts to describe the mechanisms of drought tolerance in Bambara groundnut [121,122,123]. Bambara groundnut can retain leaf turgor pressure through the reduction in leaf area, regulation of stomatal operation and osmotic adjustment [124]. This implies that the plant can potentially maintain turgor at 2 megapascal, which is much less than groundnut at 1.2–1.6. Nevertheless, variations exist in the level of drought resistance for landraces and their center of origin, the intensity and speed of drought stress and the stage of phenological effect [125]. For instance, in a study conducted by Berchie et al. [119] that evaluated several landraces of Bambara groundnut for drought resistance and heat tolerance, the researchers found that when the irrigation system was stopped at 30 days after planting, some landraces of Bambara were able to withstand drought stress up to 120 days. A landrace that was collected from Burkina Faso, a Sahelian country, was identified to be the most resistant to drought stress, based on the performance recorded among all the landraces studied, and it had the highest plant biomass with a few pods produced when others could not produce any. Despite drought generally decreasing the yield of several crops, including Bambara groundnut, a reasonable yield of 1.7 t ha−1 was produced from dried pods of Bambara groundnut [48].
Agro-morphological characterization is the first step in the collection of genetic materials and the identification of desirable characters of interest [72]. Preliminary research has shown a significant agro-morphological variability among some local accessions of Bambara groundnut [126]. Goli et al. [118] characterized 1384 from of over 2000 accessions kept at IITA, and found significant genetic variation in their growth habits and leaf shapes. Similar reports were made by Ntundu et al. [62] on the morphological diversity of Bambara groundnut landraces in Tanzania, and they observed variation among Bambara groundnut landraces, which showed that 63% of them were semi-bunch, 30% bunch and 7% spreading. Ofori et al. [127] characterized Bambara groundnut landraces and observed variations in the primary leaf color of emerging seedlings to be 29% green and 71% purple. They observed that 89% of leaves were oval in shape, while 5.5% each were lanceolate and round. The number of days to flowering, pod length, and pod width showed low coefficients of variability when compared to the number of leaves per plant, canopy spread and petiole length. Shelling percentage and shell thickness varies from 11.7% to 50.6% and 0.2 to 0.9 mm, respectively. In general, Ntundu et al. [62] and Onwubiko et al. [127] found that there was sufficient variation to breed Bambara groundnut. Olukolu et al. [45] showed that variability among yield parameters may be related to variations among leaf shape, stem length, pod and seed production. The researchers further stated that, in a principal component analysis of nine characters, a similarity of 40% was recorded from five groups, which correspond to 58 different morpho-types out of the 70 accessions, accounting for 82% of these accessions. The Pod color was observed to be 57% yellowish, 37% brownish and 6% reddish-brown, while pod textures included smooth (14%), little grooved (77%), and much grooved (9%) [45]. Qualitative traits were found to be significantly variable [71], thus indicating the importance of phenotypic markers for Bambara groundnut in genetic studies and improvement. Olukolu et al. [45] proposed the integration of qualitative and quantitative traits with molecular characterization in germplasm studies for pre-breeding. Maréchal et al. [128] found that variations exist between Bambara groundnut and other species of the genus Vigna, such as cowpea (Vigna unguiculata [L.] Walp), to which Bambara groundnut belongs. Bambara groundnut differs from cowpea in that the latter bears its long pods above the ground on long robust stems. Distinctive morphological features of Bambara groundnut have been described by several authors [31,73,129,130]. Morphological descriptions used by previous studies were based on criteria defined by The International Plant Genetic Resources Institute IPGRI [74]. The morphological differences observed between different groups of phenotypic diversity suggest that accessions are maintained under very different evolutionary processes in their respective agroecosystems. Agroecosystems are likely to exert variable selective pressures [131] and anthropogenic pressures on genotypes [93]. Farmers’ management of seeds, such as selective sorting, crop recycling and agricultural practices, lead to selection, which is expressed as the maintenance or creation of significant phenotypic diversity [93] or disappearance of some morphotypes.
Initially, genetic diversity of some Bambara groundnut landraces have been evaluated using amplified fragment length polymorphism (AFLP) and random amplified polymorphic DNA (RAPD) markers [100,106]. Additionally, Basu et al. [132] developed ten SSR markers for Bambara groundnut from a genomic library of enriched-microsatellite. Higher throughput genotyping was achieved by the development of dominant DArT, which enabled the hybridization system-based array-genotyping, comprising four key subpopulations identified out of 40 genotypes collected from nine countries, as indicated by 554 DArT markers [45,99]. However, despite the feasibility of transferring SSR markers to closely related species, only 20 amplifiable SSRs out of 178 from Vigna unguiculata, Vigna angularis and Vigna radiata were identified as informative among 240 accessions of Bambara groundnut [2,94]. The discovery of the next-generation sequencing aided the optimization of additional 74 SSR markers from a leaf transcriptome [61,77]. The DArTseq platform on the bases of Illumina next-generation sequencing produces both co-dominant SNP markers (variation contain within the sequence tags of markers) and dominant silico DArT markers [133,134,135,136].
While genome-wide association studies have not been used in Bambara groundnut to unravel the projection and variation sources linked to traits of agronomic relevance, the reports on genetic assets are generated either among or throughout species. In a breeding program of heterosis hybrid in pigeon pea (Cajanuscajan), a significant number of developed SSRs are available to differentiate the maintainer of cytoplasmic male sterility and fertility restorer lines, as well as to evaluate the genetic purity of hybrid lines [137]. AFLP and SSR molecular markers were both used in attempts to cross-breed Bambara groundnut to evaluate the true hybrid parents before proceeding to the advanced populations and recombinant inbred lines. Evaluation of heterogeneity in Bambara groundnut has also been achieved using SSR in commercial cultivars before individual plant selections for the entire genome sequencing [99]. Genetic linkage maps comprising simple sequence repeats (SSR), single nucleotide polymorphism (SNP) and diversity arrays technology (DArT) markers using four populations of Bambara groundnut have been developed (F2 generation and beyond), comprised of domesticated and wild types [96,98]. The purpose of the development is to create segregating populations for several traits, such as growth pattern, photoperiodic sensitivity, drought tolerance and disease resistance to the development of improved varieties.
The major molecular marker analysis of polymorphism in Bambara groundnut consists of 20 SSRs and 201 DArT Array markers, assessed on 363 sole accessions from diverse local varieties [76]. The results indicate a high degree of both intra and inter-allelic diversity and grouping of genotypes based on the recognized geographical sites of origin that have been described [79]. For example, the Central African accession and the East African accession were separated into individual groups and each group clustered to the West Africa genotypes [98]. The results of the structure analysis indicated that there are two major clusters and all the landraces fitted into the first subpopulation, while only thirteen landraces belonged to the second subpopulation [101]. Recently, studies were conducted on the genetic link of Bambara groundnut in relation to low temperatures [138], water stress [3] and other related legumes [6]. Rungnoi et al. [100] noted that all 363 landraces of Bambara groundnut studied might have similar population structures, while stating that West Africa (comprising Nigeria/Cameroon region) remains the crop’s location of diversity/domestication. The researchers further claimed that the crop could have been introduced to Southeast Asia (Thailand) from West Africa through East Africa. Generally, the West-Central African genotypes and the South-Eastern African genotypes are always clustered separately throughout the molecular analysis [76]. In Bambara groundnut, studies on comparative genomics analysis between leaf transcriptome in Bambara groundnut and other crop species have been evaluated to enable the detection of suitable cross-species orthologues and the crop model genes [94,97,139]. The outcome revealed that soybean scored a similar maximum similarity transcript sequence to that of Bambara groundnut, and among the other species (Vigna radiate, Riciniscommunis, Populastrichocarpa, Arabidopsis lyrata, Vitisvinifera, and Medicagotruncatula) examined in the study and may be used in the form of a gene model in Bambara groundnut gene expression profiling [97]. The DNA of Bambara groundnut has been hybridized to Arabidopsis ATH1 and Medicago truncatula Affymetrix Gene Marks for high and low stems, due to the lack of availability of Affymetrix Gene marks in Bambara groundnut [97]. The approach of cross species microarray, linked with genetics and genomics in Bambara groundnut, was carried out using the gene marks array of soybean.
5. Future Developments
Advancements in molecular biology, crop genomics, transcriptomics and bioinformatics offer the opportunity to study and understand aspects of plant biology in a better-unified system, while easing the transfer of information from major to minor species, such as Bambara groundnut [140]. The robust practice of the conventional system of breeding is necessary for successful genomic research on neglected and underutilized crop species like Bambara groundnut [141]. Exploiting the benefit of translation and biotechnological tools in research, moving from major crops to underutilized species, can offer desirable results in breeding methods by using two major procedures, which are the transformation of technologies, next-generation sequencing and translation of the real genetic-character data set from associated species on location bases or network exploration [76]. Due to Bambara groundnut’s relevance in providing useful information on the species, the use of comparative genomics is gaining wider acceptance, especially in crop species that have genomes that are yet to be sequenced, as compared to identified species [142]. Rapid improvement in crop genomics has enabled in-depth structural and functional assessments of the genes contained in several biological procedures within major species and other crop plants. Consequently, the use of bioinformatics tools in comparative genomics can offer the opportunity for transferring significant information from the major crops to the Bambara groundnut. Additionally, underutilized crops can provide better models for character and character analysis, which is lacking in major crops [143,144]. For instance, if drought tolerance alleles in Bambara groundnut can be traced, the fundamental physiological mechanism and responsible genes could be utilized through marker-assisted selection to screen for alleles among the species of interest or the character can be transferred by direct gene transfer to major crop species. An experiment was conducted using XSpecies microarray to detect and identify specific genes and gene modules related to low temperature responses in Bambara groundnut; 375 and 659 variably expressed genes were identified (p < 0.01) at 23 °C and 18 °C sub-optimal temperatures, respectively. Validation of 52 from the 100 diversity expressed genes was carried out using next-generation sequencing (NGS) technology created from the same population used to generate cross-species microarray information. More than 50% similarity was observed from the result between next-generation sequencing technology and the XSpecies Microarray approach. The gene modules identified can be used to breed varieties of Bambara groundnut for tolerance to low-temperature stress [137]. Polymorphisms among individuals for gene expression analysis can be identified through these approaches. Additionally, mutation can be seen to speed up the generation markers generation for specific characters in underutilized crop species [145].
6. Conclusions
Developing underutilized crops with an abundance of genetic resources and potentially advantageous traits can be the solution to a more diversified and robust agricultural system with additional food sources needed to address food and nutritional security. There is a need for translating knowledge, skills and edge cutting tools, developed for a well-researched crop species, to underutilized crops. Prioritizing a marker-assisted program of breeding is essential; however, establishing in-depth knowledge of Bambara groundnut to certify that breeding approaches are based on demand and are acceptable in precise agro-ecological zones is equally important. Besides, in the genetic improvement of the crop based on the farmers demand, there is a need to focus on basic agricultural mechanization systems to developed novel and composite system of agriculture for the onward delivery of better and improved food security in the future. A concerted effort is being carried out by the International Institute of Tropical Agriculture (IITA) Nigeria, Crop For the Future (CFF) and the University of Nottingham Malaysia, to tap into their diverse network. In this effort, a total of 420 accessions from indigenous farmers, breeder seeds and gene banks are being characterized and are being both phenotypically and genotypically characterized. The background information of each of those accessions will be made readily available and can be easily be accessed by researchers and producers, thus helping to improve breeding activities. The combination of genomics-assisted breeding, high-throughput approaches, which are progressively becoming cheaper, and phenotyping, which is becoming popular and more accessible, would be better established in underutilized crop species. For climate change, genomic research in underutilized crops is highly valued, due to their inherent resistance to biotic and abiotic stress conditions. Accurately narrowing down the molecular and physiological genetics of such essential traits will not only improve the genetic breeding of underutilized crop species, but also the entire crop breeding, the adaptation to climate change mitigation, and improvement in income generation and food security.
Author Contributions
I.M., M.Y.R., and Y.O. drafted the manuscript, while the proofreading, editing, and finishing were carried out by S.I.R., M.H.N., A.R.H., I.M., F.A., S.C.C., B.S.H., I.S.A., J.H. and I.W.A. All authors offered suggestions on various drafts of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
The authors are grateful to the Ministry of Education Malaysia, for adequate research Higher Institution Centres of Excellence (HICoE) funding to conduct research on improvement of crop varieties for adaption to biotic and abiotic stress.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Godfray, H.C.J.; Garnett, T. Food security and sustainable intensification food security and sustainable intensification. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 6–11. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Somta, P.; Muto, C.; Iseki, K.; Naito, K.; Pandiyan, M.; Natesan, S.; Tomooka, N. Novel genetic resources in the genus vigna unveiled from gene bank accessions. PLoS ONE 2016, 11, e0147568. [Google Scholar] [CrossRef]
- Khan, F.; Chai, H.H.; Ajmera, I.; Hodgman, C.; Mayes, S.; Lu, C. A Transcriptomic comparison of two bambara groundnut landraces under dehydration stress. Genes (Basel) 2017, 8, 121. [Google Scholar] [CrossRef] [PubMed]
- Mayes, S.; Massawe, F.J.; Alderson, P.G.; Roberts, J.A.; Azam-Ali, S.N.; Hermann, M. The potential for underutilized crops to improve security of food production. J. Exp. Bot. 2012, 63, 1075–1079. [Google Scholar] [CrossRef] [PubMed]
- Chanyalew, S.; Ferede, S.; Damte, T.; Fikre, T.; Genet, Y.; Kebede, W. Significance and prospects of an orphan crop tef. Planta 2019. [Google Scholar] [CrossRef]
- Ho, W.K.; Chai, H.H.; Kendabie, P.; Ahmad, N.S.; Jani, J.; Massawe, F.; Kilian, A.; Mayes, S. Integrating genetic maps in bambara groundnut [Vigna subterranea (L.) Verdc.] and their syntenic relationships among closely related legumes. BMC Genom. 2017, 18, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Glenn, K.C.; Alsop, B.; Bell, E.; Goley, M.; Jenkinson, J.; Liu, B.; Martin, C.; Parrott, W.; Souder, C.; Sparks, O.; et al. Bringing new plant varieties to market: Plant breeding and selection practices advance beneficial characteristics while minimizing unintended changes. Crop Sci. 2017, 57, 2906–2921. [Google Scholar] [CrossRef]
- Mabhaudhi, T.; Grace, V.; Chimonyo, P.; Hlahla, S.; Massawe, F. Prospects of orphan crops in climate change. Planta 2019, 250, 695–708. [Google Scholar] [CrossRef] [PubMed]
- Gruber, K. The Living Library. Nature 2017, 544, S8–S10. [Google Scholar] [CrossRef]
- AOCC. The African Orphan Crops Consortium. 2019. Available online: http://africanorphancrops.org/ (accessed on 10 January 2019).
- EconomistEconomist. No crop left behind: Improving the plants that Africans eat and breeders neglect. Economist 2017. Available online: http://www.economist.com/news/science-and-technology/21731614-nutrition-and-geneticsafrica-improving-plants-africans-eat (accessed on 5 December 2018).
- UN. United Nations Sustainable Development Goals. United Nations. Available online: https://Sustainabledevelopment.Un.Org/ (accessed on 15 January 2019).
- Hendre, P.S.; Muthemba, S.; Kariba, R.; Muchugi, A.; Fu, Y.; Chang, Y.; Song, B.; Liu, H.; Liu, M.; Liao, X.; et al. African Orphan Crops Consortium (AOCC): Status of Developing Genomic Resources for African Orphan Crops. Planta 2019, 250, 989–1003. [Google Scholar] [CrossRef] [PubMed]
- LATINCROP. An Integrated Strategy for the Conservation and Use of Underutilized Latin American Agrobiodiversity. 2019. Available online: http://www.latincrop.org/ (accessed on 16 April 2019).
- Dawson, I.K.; Powell, W.; Hendre, P.; Ban, J.; Hickey, J.M.; Kindt, R.; Hoad, S.; Hale, I. Tansley review the role of genetics in mainstreaming the production of new and orphan crops to diversify food systems and support human nutrition. New Phytol. 2019, 224, 37–54. [Google Scholar] [CrossRef] [PubMed]
- Tadele, Z.; Bartels, D. Promoting orphan crops research and development. Planta 2019, 250, 675–676. [Google Scholar] [CrossRef] [PubMed]
- Kahane, R.; Hodgkin, T.; Jaenicke, H.; Hoogendoorn, C.; Hermann, M.; Dyno Keatinge, J.D.H.; D’Arros Hughes, J.; Padulosi, S.; Looney, N. Agrobiodiversity for food security, health and income. Agron. Sustain. Dev. 2013, 33, 671–693. [Google Scholar] [CrossRef]
- Barbieri, R.L.; Gomes, J.C.C.; Alercia, A.; Padulosi, S. Agricultural biodiversity in Southern Brazil: Integrating efforts for conservation and use of neglected and underutilized species. Sustainability 2014, 6, 741–757. [Google Scholar] [CrossRef]
- Ndidi, U.S.; Ndidi, C.U.; Aimola, I.A.; Bassa, O.Y.; Mankilik, M.; Adamu, Z. Effects of processing (boiling and roasting) on the nutritional and antinutritional properties of bambara groundnuts (Vigna subterranea [L.] Verdc.) from Southern Kaduna. Nigeria 2014, 2014, 472129. [Google Scholar] [CrossRef]
- Murevanhema, Y.Y.; Jideani, V.A. Potential of bambara groundnut (Vigna subterranea (L.) Verdc.) milk as a probiotic beverage—A review. Crit. Rev. Food Sci. Nutr. 2013, 53, 954–967. [Google Scholar] [CrossRef]
- Oyeyinka, S.A.; Tijani, T.S.; Oyeyinka, A.T.; Arise, A.K.; Balogun, M.A.; Kolawole, F.L.; Obalowu, M.A.; Joseph, J.K. Value added snacks produced from bambara groundnut (Vigna subterranea) paste or flour. LWT Food Sci. Technol. 2018, 88. [Google Scholar] [CrossRef]
- Mazahib, A.M.; Nuha, M.O.; Salawa, I.S.; Babiker, E.E. Some nutritional attributes of bambara groundnut as influenced by domestic processing. Int. Food Res. J. 2013, 20, 1165–1171. [Google Scholar]
- Bamshaiye, O.M.; Adegbola, J.A.; Bamishaiye, E.I. Bambara groundnut: An under-utilized nut in Africa. Adv. Agric. Biotechnol. 2011, 1, 60–72. [Google Scholar]
- Mabhaudhi, T.; Chibarabada, T.P.; Chimonyo, V.G.P.; Modi, A.T. Modelling climate change impact: A case of bambara groundnut (Vigna subterranea). Phys. Chem. Earth 2018, 105, 25–31. [Google Scholar] [CrossRef]
- Obidiebube, E.A.; Eruotor, P.G.; Akparobi, S.O.; Okolie, H.; Obasi, C.C. Assessment of bambara groundnut (Vigna subterranea (L.) Verdc.) varieties for adaptation to rainforest agro-ecological zone of anambra state of nigeria. Can. J. Agric. Crop. 2020, 5, 1–6. [Google Scholar] [CrossRef]
- Dalziel, J. Voandzeia Thou. In The Useful Plants of West Tropical Africa; Crown Agents: London, UK, 1937; pp. 269–271. [Google Scholar]
- Hepper, F.N. The bambara groundnut (Voandzeia subterranea) and kersting’s groundnut (Kerstingiella Geocarpa) wild in West Africa. Kew Bull. 1963, 16, 395–407. [Google Scholar] [CrossRef]
- Goli, A. Characterization and Evaluation of IITA’s Bambara Groundnut Collection. In Proceedings of the Workshop on Conser-Vation and Improvement of Bambara Groundnut (Vigna subtarranea (L.) Verdc.); Begemann, J.H., Mush-Onga, J., Eds.; Internatinal Plant Genetic Resources Institute (IPGRI): Harare, Zimbabwe, 1995. [Google Scholar]
- Basu, S.; Roberts, J.A.; Azam-Ali, S.N.; Mayes, S. Bambara Groundnut. Genome mapping and molecular breeding in plants. In Pulses, Sugar and Tuber; Kole, C.M., Ed.; Springer: New York, NY, USA, 2007; pp. 159–173. [Google Scholar]
- Cheng, A.; Raai, M.N.; Amalina, N.; Zain, M.; Massawe, F.; Singh, A. In search of alternative proteins: Unlocking the potential of underutilized tropical legumes. Food Secur. 2019, 11, 1205–1215. [Google Scholar] [CrossRef]
- Goli, A. Bambara groundnut (Vigna subterranea (L.) Verdc.). In Promoting the Con-Servation and Use of Underutilized and Neglected Crops; Heller, J., Begemann, F., Mushonda, J., Eds.; Int. Plant Genet. Resour. Institute: Rome, Italy, 1997; Volume 9, p. 167. [Google Scholar]
- Olayide, O.E.; Donkoh, S.A.; Gershon, I.; Ansah, K.; Adzawla, W.; Reilly, P.J.O.; Mayes, S.; Feldman, A.; Halimi, R.A.; Nyarko, G.; et al. Handbook of Climate Change Resilience; Leal Filho, W., Ed.; Springer Nature Switzerland AG: Basel, Switzerland, 2018. [Google Scholar]
- Atoyebi, J.O.; Oyatomi, O.; Osilesi, O.; Adebawo, O.; Abberton, M. Morphological characterisation of selected African accessions of bambara morphological characterisation of selected African accessions of bambara groundnut (Vigna subterranea (L.) Verdc.). Int. J. Plant Res. 2017, 7, 29–35. [Google Scholar] [CrossRef]
- Gbaguidi, A.A.; Dansi, A.; Dossou-Aminon, I.; Gbemavo, D.S.J.C.; Orobiyi, A.; Sanoussi, F.; Yedomonhan, H. Agromorphological diversity of local bambara groundnut (Vigna subterranea (L.) Verdc.) collected in benin. Genet. Resour. Crop Evol. 2018, 65, 1159–1171. [Google Scholar] [CrossRef]
- Feldman, A.; Ho, W.K.; Massawe, F.; Mayes, S. Bambara Groundnut is a Climate-Resilient Crop: How Could a Drought-Tolerant and Nutritious Legume Improve Community Resilience in the Face of Climate Change? Springer Nature: Basel, Switzerland, 2019; pp. 151–167. [Google Scholar] [CrossRef]
- Abu, H.B.; Buah, S.S.J. Characterization of bambara groundnut landraces and their evaluation by farmers in the upper west region of ghana. J. Dev. Sustain. Agric. 2011, 6, 64–74. [Google Scholar]
- Tadele, Z.; Assefa, K. Increasing food production in Africa by boosting the productivity of understudied crops. Agronomy 2012, 2, 240–283. [Google Scholar] [CrossRef]
- Foyer, C.H.; Lam, H.; Nguyen, H.T.; Siddique, K.H.M.; Varshney, R.K.; Colmer, T.D.; Cowling, W.; Bramley, H.; Mori, T.A.; Hodgson, J.M.; et al. Health and sustainable food production. Nat. Plants 2016, 2, 1–10. [Google Scholar] [CrossRef]
- Cullis, C.; Kunert, K.J. Unlocking the potential of orphan legumes. J. Exp. Bot. 2017, 68, 1895–1903. [Google Scholar] [CrossRef]
- Mubaiwa, J. Managing the Hard-to-Cook (HTC) Phenomenon in Bambara Groundnut (Vigna subterranea (L.) Verdc.) In Processing for Resource Limited Communities in Zimbabwe. Ph.D. Thesis, Wageningen University, Wageningen, The Netherlands, 2018. [Google Scholar]
- Khan, F.; Azman, R.; Chai, H.H.; Mayes, S.; Lu, C. Genomic and transcriptomic approaches towards the genetic improvement of an underutilised crops: The case of Bambara groundnut. Afr. Crop Sci. J. 2016, 24, 429–458. [Google Scholar] [CrossRef]
- Enwere, N.J.; Hung, Y. Some chemical and physical properties of bambara groundnut (Voundzeia Subterrunea Thouars) Seed and Products. Int. J. Food Sci. Nutr. 1996, 47, 469–475. [Google Scholar] [CrossRef] [PubMed]
- Hillocks, R.J.; Bennett, C.; Mponda, O.M. Bambara Nut: A review of utilisation, market potential and crop improvement. Afr. Crop Sci. J. 2012, 20, 1–16. [Google Scholar]
- Massawe, F.J.; Mwale, S.S.; Roberts, J.A. Breeding in bambara groundnut (Vigna subterranea (L.) Verdc.): Strategic considerations. Afr. J. Biol. 2005, 4, 463–471. [Google Scholar]
- Olukolu, B.A.; Mayes, S.; Stadler, F.; Ng, N.Q.; Fawole, I.; Dominique, D.; Azam-Ali, S.N.; Abbott, A.G.; Kole, C. Genetic diversity in bambara groundnut (Vigna subterranea (L.) Verdc.) as revealed by phenotypic descriptors and dart marker analysis. Genet. Resour. Crop Evol. 2012, 59, 347–358. [Google Scholar] [CrossRef]
- Zeven, A.C. Landraces: A review of definitions and classifications. Euphytica 1998, 104, 127–139. [Google Scholar] [CrossRef]
- Massawe, F.J.; Dickinson, M.; Roberts, J.A.; Azam-Ali, S.N. Genetic diversity in bambara groundnut (Vigna subterranea (L.) Verdc.) landraces revealed by AFLP markers. Genome 2002, 45, 1175–1180. [Google Scholar] [CrossRef]
- Mwale, S.S.; Azam-Ali, S.N.; Massawe, F.J. Growth and development of bambara groundnut (Vigna subterranea) in response to soil moisture 1. dry matter and yield. Eur. J. Agron. 2007, 26, 345–353. [Google Scholar] [CrossRef]
- Mubaiwa, J.; Fogliano, V.; Chidewe, C.; Bakker, E.J.; Linnemann, A.R. Utilization of bambara groundnut (Vigna subterranea (L.) Verdc.) for sustainable food and nutrition security in semi-arid regions of zimbabwe. PLoS ONE 2018, 13, e0204817. [Google Scholar] [CrossRef]
- Mayes, S.; Ho, W.K.; Chai, H.H.; Gao, X.; Kundy, A.C.; Mateva, K.I.; Zahrulakmal, M.; Hahiree, M.K.I.M.; Kendabie, P.; Licea, L.C.S.; et al. Bambara groundnut: An exemplar underutilised legume for resilience under climate change. Planta 2019, 250, 803–820. [Google Scholar] [CrossRef]
- Abdullah, N.; Rafii Yusop, M.; Ithnin, M.; Saleh, G.; Latif, M.A. Genetic Variability of Oil Palm Parental Genotypes and Performance of Its’ Progenies as Revealed by Molecular Markers and Quantitative Traits. Comptes Rendus. Biol. 2011, 334, 290–299. [Google Scholar] [CrossRef] [PubMed]
- Ogundele, O.M.; Minnaar, A.; Emmambux, M.N. Effects of micronisation and dehulling of pre-soaked bambara groundnut seeds on microstructure and functionality of the resulting flours. Food Chem. 2017, 214, 655–663. [Google Scholar] [CrossRef]
- Oladosu, Y.; Rafii, M.Y.; Abdullah, N.; Malek, M.A.; Rahim, H.A.; Hussin, G.; Ismail, M.R.; Latif, M.A.; Kareem, I. Genetic Variability and Diversity of Mutant Rice Revealed by Quantitative Traits and Molecular Markers. Agrociencia 2015, 49, 249–266. [Google Scholar]
- F.A.O. of the United Nations. FAOSTAT Statistics Database; F.A.O. of the United Nations: Roma, Italy, 2017. [Google Scholar]
- Waziri, P.M.; Massawe, F.J.; Wayah, S.B.; Sani, J.M. Ribosomal DNA variation in landraces of bambara groundnut. Afr. J. Biotechnol. 2013, 12, 5395–5403. [Google Scholar] [CrossRef][Green Version]
- Bonny, B.S.; Seka, D.; Adjoumani, K.; Koffi, K.G.; Kouonon, L.C.; Sie, R.S. Evaluation of the diversity in qualitative traits of bambara groundnut germplasm (Vigna subterranea (L.) Verdc.) of Côte d ’Ivoire. Afr. J. Biotechnol. 2019, 18, 23–36. [Google Scholar] [CrossRef]
- Hamrick, J.L.; Godt, M.J.W. Allozyme diversity in cultivated crops. Crop Sci. 1997, 37, 26–30. [Google Scholar] [CrossRef]
- Karikari, S.K.; Tabona, T.T. constitutive traits and selective indices of bambara groundnut (Vigna subterranea (L.) verdc) landraces for drought tolerance under Botswana conditions. Phys. Chem. Earth 2004, 29, 1029–1034. [Google Scholar] [CrossRef]
- Ellstrand, N.C.; Elam, D.R. Population genetic consequences of small population size: Implications for plant conservation. Annu. Rev. Ecol. Syst. 1993, 24, 217–242. [Google Scholar] [CrossRef]
- Massawe, F.; Mayes, S. Genetic diversity and population structure of bambara groundnut (Vigna subterranea (L.) Verdc.): Synopsis of the past two decades of analysis and implications for crop improvement programmes. Genet. Resour. Crop Evol. 2016, 63, 925–943. [Google Scholar] [CrossRef]
- Oladosu, Y.; Rafii, M.Y.; Abdullah, N.; Abdul Malek, M.; Rahim, H.A.; Hussin, G.; Abdul Latif, M.; Kareem, I. Genetic Variability and Selection Criteria in Rice Mutant Lines as Revealed by Quantitative Traits. Sci. World J. 2014. [Google Scholar] [CrossRef]
- Bonny, B.S.; Adjoumani, K.; Seka, D.; Koffi, K.G.; Kouonon, L.C.; Koffi, K.K.; Zoro Bi, I.A. Agromorphological divergence among four agro-ecological populations of bambara groundnut (Vigna subterranea (L.) Verdc.) in Côte d’Ivoire. Ann. Agric. Sci. 2019, 64, 103–111. [Google Scholar] [CrossRef]
- Ntundu, W.H.; Shillah, S.A.; Marandu, W.Y.F.; Christiansen, J.L. Morphological diversity of bambara groundnut [Vigna subterranea (L.) Verdc.] landraces in Tanzania. Genet. Resour. Crop Evol. 2006, 53, 367–378. [Google Scholar] [CrossRef]
- Mabhaudhi, T.; Reilly, P.O.; Walker, S.; Mwale, S. Opportunities for underutilised crops in Southern Africa’s post—2015 sustainability opportunities for underutilised crops in Southern Africa’s post—2015 Development Agenda. Sustainability 2016, 8, 302. [Google Scholar] [CrossRef]
- Molosiwa, O.; Basu, S.M.; Stadler, F.; Azam-Ali, S.; Mayes, S. Assessment of genetic variability of bambara groundnut (Vigna subterranea (L.) Verde.) accessions using morphological traits and molecular markers. Acta Hortic. 2013, 979, 779–790. [Google Scholar] [CrossRef]
- Molosiwa, O.O. Genetic Diversity and Population Structure Analysis of Bambara Groundnuts (Vigna subterranea (L.) Verdc.) Landraces Using Morpho-Agronomic Characters and SSR Markers. Ph.D. Thesis, University of Nottingham, Nottingham, UK, 2012; pp. 1–285. [Google Scholar]
- Hoque, A.; Begum, S.; Robin, A.; Hassan, L. Partitioning of rice (Oryza sativa, L.) genotypes based on Morphometric Diversity. Am. J. Exp. Agric. 2015, 7, 242–250. [Google Scholar] [CrossRef]
- Eckert, A.J.; Van Heerwaarden, J.; Wegrzyn, J.L.; Nelson, C.D.; Ross-Ibarra, J.; González-Martínez, S.C.; Neale, D.B. Patterns of population structure and environmental associations to aridity across the range of loblolly pine (Pinus Taeda, L., Pinaceae). Genetics 2010, 185, 969–982. [Google Scholar] [CrossRef]
- Linnemann, A.R.; Craufurd, P.Q. Effects of temperature and photoperiod on phenological development in three genotypes of bambara groundnut (Vigna subterranea). Ann. Bot. 1994, 74, 675–681. [Google Scholar] [CrossRef]
- Collinson, S.T.; Azam-Ali, S.N.; Chavula, K.M.; Hodson, D.A. Growth, develop- ment and yield of bambara groundnut (Vigna subterranea) in Response to Soil Moisture. J. Agric. Sci. 1996, 126, 307–318. [Google Scholar] [CrossRef]
- Brink, M.; Sibuga, K.P.; Tarimo, A.J.P.; Ramolemana, G. Quantifying photothermal influences on reproductive development in bambara groundnut (Vigna subterranea): Models and Their Vali-Dation. Field. Crop. Res. 2000, 66, 1–14. [Google Scholar] [CrossRef]
- Wu, S.; Ning, F.; Zhang, Q.; Wu, X.; Wang, W. Enhancing omics research of crop responses to drought under field conditions many but limited useful data from omics analysis of. Front. Plant Sci. 2017, 8, 1–5. [Google Scholar] [CrossRef]
- Shego, A.; van Rensburg, W.S.J.; Adebola, P.O. Aassessment of genetic variability in bambara groundnut (Vigna subterrenea L. verdc.) using morphological quantitative traits. Acad. J. Agric. Res. 2013, 1, 45–51. [Google Scholar]
- Unigwe, A.E.; Gerrano, A.S.; Adebola, P.; Pillay, M. Morphological Variation in Selected Accessions of Bambara Groundnut (Vigna subterranea, L. Verdc.) in South Africa. J. Agric. Sci. 2016, 8, 69. [Google Scholar] [CrossRef]
- IPGRI/IITA/BAMNET. Descriptors for bambara groundnut (Vigna subterranea); International Plant Genetic Resources Institute: Rome, Italy; International Institute of Tropical Agriculture: Ibadan, Nigeria; The International Bambara Groundnut Network: Bonn, Germany, 2000; ISBN 92-9043-461-9. [Google Scholar]
- Ntundu, W.H.; Bach, I.C.; Christiansen, J.L.; Andersen, S.B. Analysis of genetic diversity in bambara groundnut [Vigna subterranea (L.) Verdc.] landraces using amplified fragment length polymorphism (AFLP) Markers. Afr. J. Biotechnol. 2004, 3, 220–225. [Google Scholar]
- Aliyu, S.; Massawe, F.; Mayes, S. SSR marker development, genetic diversity and population structure analysis of bambara groundnut [Vigna subterranea (L.) Verdc.] landraces. Genet. Resour. Crop Evol. 2015, 62, 1225–1243. [Google Scholar] [CrossRef]
- Mayes, S.; Ho, W.K.; Kendabie, P.; Chai, H.H.; Aliyu, S.; Feldman, A.; Halimi, R.A.; Massawe, F.; Azam-Ali, S. Applying molecular genetics to underutilised species—Problems and opportunities. Malays. Appl. Biol. 2015, 44, 1–9. [Google Scholar]
- Aliyu, S.; Massawe, F.; Mayes, S. Beyond Landraces: Developing improved germplasm resources for underutilized species—A case for bambara groundnut. Biotechnol. Genet. Eng. Rev. 2015, 30, 127–141. [Google Scholar] [CrossRef]
- Aliyu, S.; Massawe, F. Microsatellites based marker molecular analysis of ghanaian bambara groundnut (Vigna subterranea (L.) Verdc.) landraces alongside morphological haracterization. Genet. Resour. Crop Evol. 2013, 60, 777–787. [Google Scholar] [CrossRef]
- Basu, S.; Mayes, S.; Davey, M.; Robert, J.A.; Azam-Ali, S.N.; Mithen, R.; Pasquet, R.S. Inheritance of domestication traits in bambara groundnut (Vigna subterranea (L.) Verdc.). Euphytica 2007, 157, 59–68. [Google Scholar] [CrossRef]
- Karikari, S.K. Variability between local and exotic bambara groundnut landraces in Botswana. Afr. Crop Sci. J. 2000, 8, 145–152. [Google Scholar] [CrossRef]
- Golestan, F.S.; Rafii, M.Y.; Ismail, M.R.; Mahmud, T.M.M.; Rahim, H.A.; Asfaliza, R.; Malek, M.A.; Latif, M.A. Biochemical, Genetic and Molecular Advances of Fragrance Characteristics in Rice. Crit. Rev. Plant Sci. 2013, 32, 445–457. [Google Scholar]
- Ghafoor, A.; Sharif, A.; Ahmad, Z.; Zahid, M.A.; Rabbani, M.A. Genetic diversity in blackgram (Vigna mungo, L. Hepper). Field Crop. Res. 2001, 69, 183–190. [Google Scholar] [CrossRef]
- Yuliawati, Y.; Wahyu, Y.; Surahman, M.; Rahayu, A. Genetic variation and agronomic characters of bambara groundnut (Vigna subterranea, L. Verdc.) lines results of pure line selection from Sukabumi Lanras. J. Agronida 2019, 4, 152–161. [Google Scholar] [CrossRef]
- Malik, M.F.A.; Ashraf, M.; Qureshi, A.S.; Ghafoor, A. Assessment of genetic variability, correlation and path analyses for yield and its components in soybean. Pakistan J. Bot. 2007, 39, 405–413. [Google Scholar]
- Linnemann, A.R.; Westphal, E.; Wessel, M. Photoperiod regulation of development and growth in bambara groundnut (Vigna subterranea). Field Crop. Res. 1995, 40, 39–47. [Google Scholar] [CrossRef]
- Brink, M. Rates of progress towards flowering and podding in bambara groundnut (Vigna subterranea) as a function of temperature and photoperiod. Ann. Bot. 1997, 80, 505–513. [Google Scholar] [CrossRef][Green Version]
- Brink, M. Development, growth and dry matter partitioning in bambara groundnut (Vigna subterranea) as influenced by photo-period and shading. J. Agric. Sci. Camb. 1999, 133, 159–166. [Google Scholar] [CrossRef][Green Version]
- Jorgensen, S.T.; Aubanton, M.; Harmonic, C.; Dieryck, C.; Jacobsen, S.; Simonsen, H.; Ntundu, W.; Stadler, F.; Basu, S.; Christiansen, J. Identification of photoperiod neutral lines of bambara groundnut (Vigna subterranea) from Tanzania. IOP Conf. Ser. Earth Env. Sci. 2009, 6, 20–23. [Google Scholar] [CrossRef]
- Presidor, K.; Massawe, F.; Mayes, S. Developing genetic mapping resources from landrace-derived genotypes that differ for photoperiod sensitivity in bambara groundnut (Vigna subterranea, L.). Asp. Appl. Biol. 2015, 124, 49–55. [Google Scholar]
- Choudhary, G.; Ranjitkumar, N.; Surapaneni, M.; Deborah, A.D.; Anuradha, G.; Siddiq, E.A.; Vemireddy, L.R. Molecular genetic diversity of major indian rice cultivars over decadal periods. PLoS ONE 2013, 8, e66197. [Google Scholar] [CrossRef]
- Huynh, B.; Close, T.J.; Roberts, P.A.; Hu, Z.; Wanamaker, S.; Lucas, M.R.; Chiulele, R.; Cissé, N.; David, A.; Hearne, S.; et al. Gene pools and the genetic architecture of domesticated cowpea. Plant Genome 2013, 6, 1–8. [Google Scholar] [CrossRef]
- Ahmad, N.; Basu, S.; Redjeki, E.; Murchie, E.; Massawe, F.; Azam-Ali, S.; Kilian, A.; Mayes, S. Developing genetic mapping and marker-assisted techniques in bambara groundnut (Vigna subterranea, L.) breeding. Acta Hortic. 2013, 979, 437–449. [Google Scholar] [CrossRef]
- Somta, P.; Chankaew, S.; Rungnoi, O.; Srinives, P. Genetic diversity of the bambara groundnut (Vigna subterranea (L.) Verdc.) as assessed by SSR Markers. Genome 2011, 54, 898–910. [Google Scholar] [CrossRef]
- Ho, W.K.; Muchugi, A.; Muthemba, S.; Kariba, R.; Mavenkeni, B.O.; Hendre, P.; Song, B.; Deynze, A.V.; Massawe, F.; Mayes, S. Use of microsatellite markers for the assessment of bambara groundnut breeding system and varietal purity before genome sequencing. Genome 2016, 59, 427–431. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, N.S.; Redjeki, E.S.; Ho, W.K.; Aliyu, S.; Mayes, K. Construction of a genetic linkage map and QTL analysis in bambara groundnut (Vigna subterranea (L.) Verdc.). Genome 2016, 59, 459–472. [Google Scholar] [CrossRef] [PubMed]
- Chai, H.H.; Ho, W.K.; Graham, N.; May, S.; Massawe, F.; Mayes, S. A cross-species gene expression marker-based genetic map and QTL analysis in bambara groundnut. Genes (Basel) 2017, 8, 84. [Google Scholar] [CrossRef]
- Pasquet, R.S.; Schwedes, S.; Gepts, P. Isozyme diversity in bambara groundnut. Crop Sci. 1999, 39, 1228–1236. [Google Scholar] [CrossRef]
- Rungnoi, O.; Suwanprasert, J.; Somta, P.; Srinives, P. Molecular genetic diversity of bambara groundnut (Vigna subterranea, L. Verdc.) revealed by RAPD and ISSR marker analysis. SABRAO J. Breed. Genet. 2012, 44, 87–101. [Google Scholar]
- Massawe, F.J.; Roberts, J.A.; Azam-Ali, S.N.; Davey, M.R. Genetic diversity in bambara groundnut (Vigna subterranea (L.) Verdc.) landraces assessed by Random Amplified Polymorphic DNA (RAPD) markers. Genet. Resour. Crop Evol. 2003, 50, 737–741. [Google Scholar] [CrossRef]
- Abberton, M.; Batley, J.; Bentley, A.; Bryant, J.; Cai, H.; Cockram, J.; de Oliveira, A.C.; Cseke, L.J.; Dempewolf, H.; De Pace, C.; et al. Global agricultural intensification during climate change: A role for genomics. Plant Biotechnol. J. 2016, 14, 1095–1098. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Amatriaín, M.; Mirebrahim, H.; Xu, P.; Wanamaker, S.I.; Luo, M.; Alhakami, H.; Alpert, M.; Atokple, I.; Batieno, B.J.; Boukar, O.; et al. Genome resources for climate-resilient cowpea, an essential crop for food security. Plant J. 2017, 89, 1042–1054. [Google Scholar] [CrossRef]
- Hiremath, P.J.; Kumar, A.; Penmetsa, R.V.; Farmer, A.; Schlu, J.A.; Chamarthi, S.K.; Whaley, A.M.; Carrasquilla-garcia, N.; Gaur, P.M.; Up, H.D.; et al. Large-Scale development of cost-effective SNP marker assays for diversity assessment and genetic mapping in chickpea and comparative mapping in legumes. Plant Biotechnol. J. 2012, 10, 716–732. [Google Scholar] [CrossRef] [PubMed]
- Igwe, D.O.; Afiukwa, C.A. Competency assessment of directed amplified minisatellite dna and start codon targeted markers for genetic diversity study in accessions of Vigna subterranea (L.) Verdcourt. J. Crop Sci. Biotechnol. 2017, 20, 263–278. [Google Scholar] [CrossRef]
- Fatimah, S.; Ariffin; Ardiarini, N.R.; Kuswanto, K. Genetic diversity of madurese bambara groundnut (Vigna subterranea, L. Verdc.) lines based on morphological and RAPD markers. Sabrao J. Breed. Genet. 2018, 50, 101–114. [Google Scholar]
- Massawe, F.J.; Schenkel, W.; Basu, S.; Temba, E.M.T. Artificial hybridisation in bambara groundnut (Vigna subterranea (L.) Verdc.). In Proceedings of the International Bambara Groundnut Symposium, Francistown, Botswana, 8–12 September 2003; pp. 193–209. [Google Scholar]
- Shiyam, J.O.; Nkor, N.N.; Binang, W.B.; Effa, E.D. Yield response of bambara groundnut (Voandzeia subterrenea (L.) thours.) varieties to organomineral fertilizer in the coastal forest of southeastern nigeria. Scirea J. Agric. 2016, 1, 91–106. [Google Scholar]
- Chang, Y.; Liu, H.; Liu, M.; Liao, X.; Sahu, S.K.; Fu, Y.; Song, B.; Cheng, S.; Kariba, R.; Muthemba, S.; et al. The draft genomes of five agriculturally important african orphan crops. Gigascience 2018, 8, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Musa, M.; Massawe, F.; Mayes, S.; Alshareef, I.; Singh, A. Nitrogen fixation and n-balance studies on bambara groundnut (Vigna subterranea, L. Verdc.) landraces grown on tropical acidic soils of malaysia. Commun. Soil Sci. Plant Anal. 2016, 47, 533–542. [Google Scholar] [CrossRef]
- Taffouo, V.D.; Wamba, O.F.; Yombi, E.; Nono, G.V.; Akoe, A. Growth, yield, water status and ionic distribution response of three bambara groundnut (Vigna subterranean (L.) Verdc.) landraces grown under saline conditions. Int. J. Bot 2010, 6, 53–58. [Google Scholar]
- Temegne, N.C.; Gouertoumbo, W.F.; Wakem, G.A.; Nkou, F.T.D.; Youmbi, E.; Ntsomboh-Ntsefong, G. Origin and ecology of bambara groundnut (Vigna subterranea (L.) Verdc.: A review. J. Ecol. Nat. Resour. 2018, 1–10. [Google Scholar] [CrossRef]
- Vurayai, R.; Emongor, V.; Moseki, B. Physiological responses of bambara groundnut (Vigna subterranea, L. Verdc) to short periods of water stress during different developmental stages. Asian J. Agric. Sci. 2011, 3, 37–43. [Google Scholar]
- Mabhaudhi, T.; Modi, A.T. Growth, phenological and yield responses of a bambara groundnut (Vigna subterranea (L.) Verdc.) landrace to imposed water stress under field conditions. South Afr. J. Plant Soil 2013, 30, 69–79. [Google Scholar] [CrossRef]
- Milla, R.; García-Palacios, P.; Matesanz, S. Looking at past domestication to secure ecosystem services of future croplands. J. Ecol. 2017, 105, 885–889. [Google Scholar] [CrossRef]
- Ouedraogo, M.; Ouedraogo, J.T.; Tignere, J.B.; Balma, D.; Dabire, C.B.; Konate, G. Characterization and evaluation of accessions of bambara groundnut (Vigna subterranea (L.) verdcourt) from Burkina Faso. Sci. Nat. 2008, 5, 191–197. [Google Scholar] [CrossRef]
- Goli, A.E.; Begemann, F.; Ng, N.Q. Characterization and evaluation of IITA’s bambara groundnut collection. In conservation and improvement of bambara groudnut (Vigna subterranea (L.) Verdc.); Heller, J., Begemann, F., Mushonga, J., Eds.; Int. Plant Genet. Resour. Institute: Harare, Zimbabwe, 1997; pp. 101–118. [Google Scholar]
- Berchie, J.N.; Sarkodie-Addo, J.; Adu-Dapaah, H.; Agyemang, A.; Addy, S.; Asare, S.; Donkor, J. Yield evaluation of three early maturing bambara groundnut (Vigna subterranea, L. Verdc.) landraces at the CSIR-Crops Research Institute, Fumesua-Kumasi, Ghana. J. Agron. 2010, 9, 175–179. [Google Scholar] [CrossRef]
- Berchie, J.N.; Opoku, M.; Adu-Dapaah, H.; Agyemang, A.; Sarkodie-Addo, J.; Asare, E.; Akuffo, H. Evaluation of five bambara groundnut (Vigna subterranea (L.) Verdc.) landraces to heat and drought stress at Tono-Navrongo, Upper East Region of Ghana. Afr. J. Agric. Res. 2012, 7, 250–256. [Google Scholar] [CrossRef]
- Abejide, D.R.; Falusi, O.A.; Gana, A.S.; Adebola, M.O.; Daudu, O.A.Y.; Salihu, B.Z. evaluation of seed yield of Nigerian bambara groundnut [Vigna subterranea (L.) Verdc.] landraces under varying water conditions. Notulae Scientia Biologicae 2018, 10, 233–239. [Google Scholar] [CrossRef]
- Ibrahin, H.D.; Ogunwusi, A.A. Industrial Potentials of Bambara Nut. J. Poverty Invest. Dev. 2016, 22, 12–18. [Google Scholar]
- Laary, J.K.; Ofori, K.; Kumaga, F. The influence of soil moisture status on reproductive growth and development of bambara groundnut (Vigna subterranea (L.) Verdc.) landraces in Ghana. J. Agric. Biol. Sci. 2012, 7, 845–851. [Google Scholar]
- Al-Shareef, I.; Sparkes, D.; Azam-A, S. Temperature and drought stress effects on growth and development of bambara groundnut (Vigna subterranea, L.). Exp. Agric. 2014, 50, 72–89. [Google Scholar] [CrossRef]
- Chibarabada, T.P.; Modi, A.T.; Mabhaudhi, T. Water use character-istics of a bambara groundnut (Vigna subterranea, L. Verdc.) landrace during seedling establishment. Water SA 2015, 41, 472–482. [Google Scholar] [CrossRef]
- Nautiyal, P.C.; Kulkarni, G.; Singh, A.L.; Basu, M. Evaluation of water-deficit stress tolerance in bambara groundnut landraces for cultivation in sub-tropical environments in India. Indian J. Plant Physiol. 2017, 22, 190–196. [Google Scholar] [CrossRef]
- Collinson, S.T.; Clawson, E.J.; Azam-A, S.N.; Black, C.R. Effects of soil moisture deficits on the water relations of bambara groundnut (Vigna subterranea L. Verde.). J. Exp. Bota 1997, 48, 877–884. [Google Scholar] [CrossRef]
- Touré, Y.; Koné, M.; Kouakou Tanoh, H.; Koné, D. Agromorphological and phenological variability of 10 bambara groundnut [Vigna subterranea (L.) Verdc. (Fabaceae)] landraces cultivated in the Ivory Coast. Tropicultura 2012, 30, 216–221. [Google Scholar]
- Ofori, G.; Oduro, I.; Ellis, O.W.; Dapaah, H.K. Assessment of vitamin a content and sensory attributes of new sweet potato (Ipomoea batatas) genotypes in Ghana. Afr. J. Food Sci. 2009, 3, 184–192. [Google Scholar]
- Onwubiko, N.C.; Uguru, M.I.; Chimdi, G.O. Selection for Yield improvement in bambara groundnut (Vigna subterranea, L. Verdc.). J. Plant Breed. Genet. 2019, 7, 41–53. [Google Scholar] [CrossRef]
- Maréchal, R. Etude Taxonomique d’un groupe complexe d’espèces des genres phaseolus et Vigna (Papilionaceae) sur la base de données morphologiques et polliniques, traitées par l’analyse informatique. Boissiera 1978, 28, 1–273. [Google Scholar]
- Doku, E.V.; Karikari, S.K. Operational selection in wild bambara groundnut. Ghana J. Sci. 1971, 11, 45–56. [Google Scholar]
- Amadou, H.I.; Bebeli, P.J.; Kaltsikes, P.J. Genetic diversity in bambara groundnut (Vigna subterranea, L.) germplasm revealed by RAPD markers. Genome 2001, 44, 995–999. [Google Scholar] [CrossRef]
- Basu, S.; Roberts, J.A.; Azam-Ali, S.N.; Mayes, S. Development of microsatellite markers for bambara groundnut (Vigna subterranea, L. Verdc.) an underutilized African legume crop species. Mol. Ecol. Notes 2007, 7, 1326–1328. [Google Scholar] [CrossRef]
- Kilian, A.; Wenzl, P.; Huttner, E.; Carling, J.; Xia, L.; Caig, V.; Heller-uszynska, K.; Jaccoud, D.; Hopper, C.; Aschenbrenner-kilian, M.; et al. Diversity arrays technology: A generic genome proiling technology on open platforms. Methods Mol. Biol. 2012, 67–89. [Google Scholar] [CrossRef]
- Egea, L.A.; Mérida-garcía, R.; Kilian, A.; Hernandez, P.; Dorado, G. Assessment of genetic diversity and structure of large garlic (Allium sativum) germplasm bank, by diversity arrays technology platform (DArTseq). Front. Plant Sci. 2017, 8, 1–9. [Google Scholar] [CrossRef]
- Edet, O.U.; Gorafi, Y.S.A.; Shuhei, N.; Tsujimoto, H. DArTseq-Based analysis of genomic relationships among species of tribe Triticeae. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Bohra, A.; Dubey, A.; Saxena, R.K.; Penmetsa, R.V.; Poornima, K.N.; Kumar, N.; Farmer, A.D.; Srivani, G.; Upadhyaya, H.D.; Gothalwal, R.; et al. Analysis of BAC-End Sequences (BESs) and development of BES-SSR markers for genetic mapping and hybrid purity assessment in pigeonpea (Cajanus Spp.). BMC Plant Biol. 2011, 11, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Bonthala, V.S.; Mayes, K.; Moreton, J.; Blythe, M. Identification of gene modules associated with low temperatures response in bambara groundnut by network-based analysis. PLoS ONE 2016, 11, e0148771. [Google Scholar] [CrossRef] [PubMed]
- Mayes, S.; Basu, S.M.; Molosiwa, O.; Redjeki, E.S.; Ahmad, N.S.; Khan, F.; Zehra, S.; Noah, S.; Mayes, K.; Roberts, J.; et al. Molecular analysis of bambara groundnut, an underutilised African legume crop as part of the BAMLINK Project–What lessons can we learn? Acta Hort. 2013, 9798, 451–458. [Google Scholar] [CrossRef]
- Akpinar, B.A.; Lucas, S.J.; Budak, H.; Akpýnar, B.A. Genomics approaches for crop improvement against abiotic stress. Sci. World J. 2013. [Google Scholar] [CrossRef]
- Mabhaudhi, T.; Vimbayi, G.P.; Chimonyo, T.P.C.; Modi, A.T. Developing a roadmap for improving neglected and underutilized crops: A case study of South Africa. Front. Plant Sci. 2017, 2143. [Google Scholar] [CrossRef]
- Dhanapal, A.P. Genomics of crop plant genetic resources. Adv. Biosci. Biotechnol. 2012, 3, 378–385. [Google Scholar] [CrossRef][Green Version]
- Van Damme, V.; Gómez-Paniagua, H.; de Vicente, M.C. The GCP molecular marker toolkit, an instrument for use in breeding food security crops. Mol Breed. 2011, 28, 597–610. [Google Scholar] [CrossRef]
- Fu, Y.B. Understanding crop genetic diversity under modern plant breeding. Theor. Appl. Genet. 2015, 128, 2131–2142. [Google Scholar] [CrossRef]
- Chai, H.H.; Lai, H.; Guo, H.; Basu, S.M.; Massawe, F.; Graham, N.; Mayes, S. Developing XSpecies approaches for genomics and transcriptomics using resources developed in major species for research in bambara. Acta Hortic. 2013, 979, 773–778. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).