Forage Performance and Detection of Marker Trait Associations with Potential for Napier Grass (Cenchrus purpureus) Improvement
Abstract
1. Introduction
2. Materials and Methods
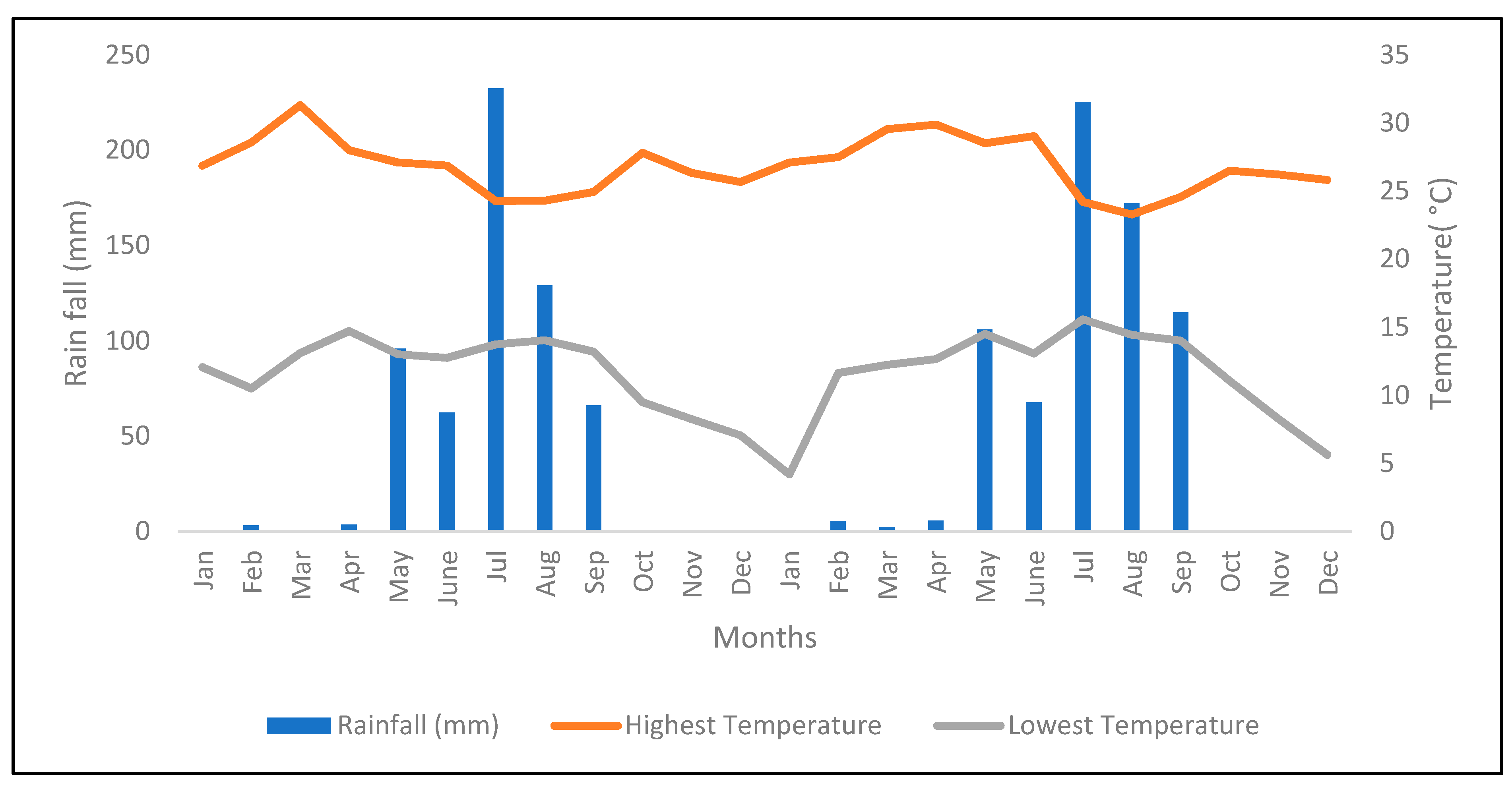
2.1. Description of the Experimental Area
2.2. Planting Materials and Trial Establishment
2.3. Data Collection
2.4. Feed Quality Analysis
2.5. Data Analysis
2.6. Marker-Trait Association Analysis
3. Results
3.1. Forage Biomass Productivity of Napier Grass Genotypes
3.2. Feed Qualities of Napier Grass Genotypes
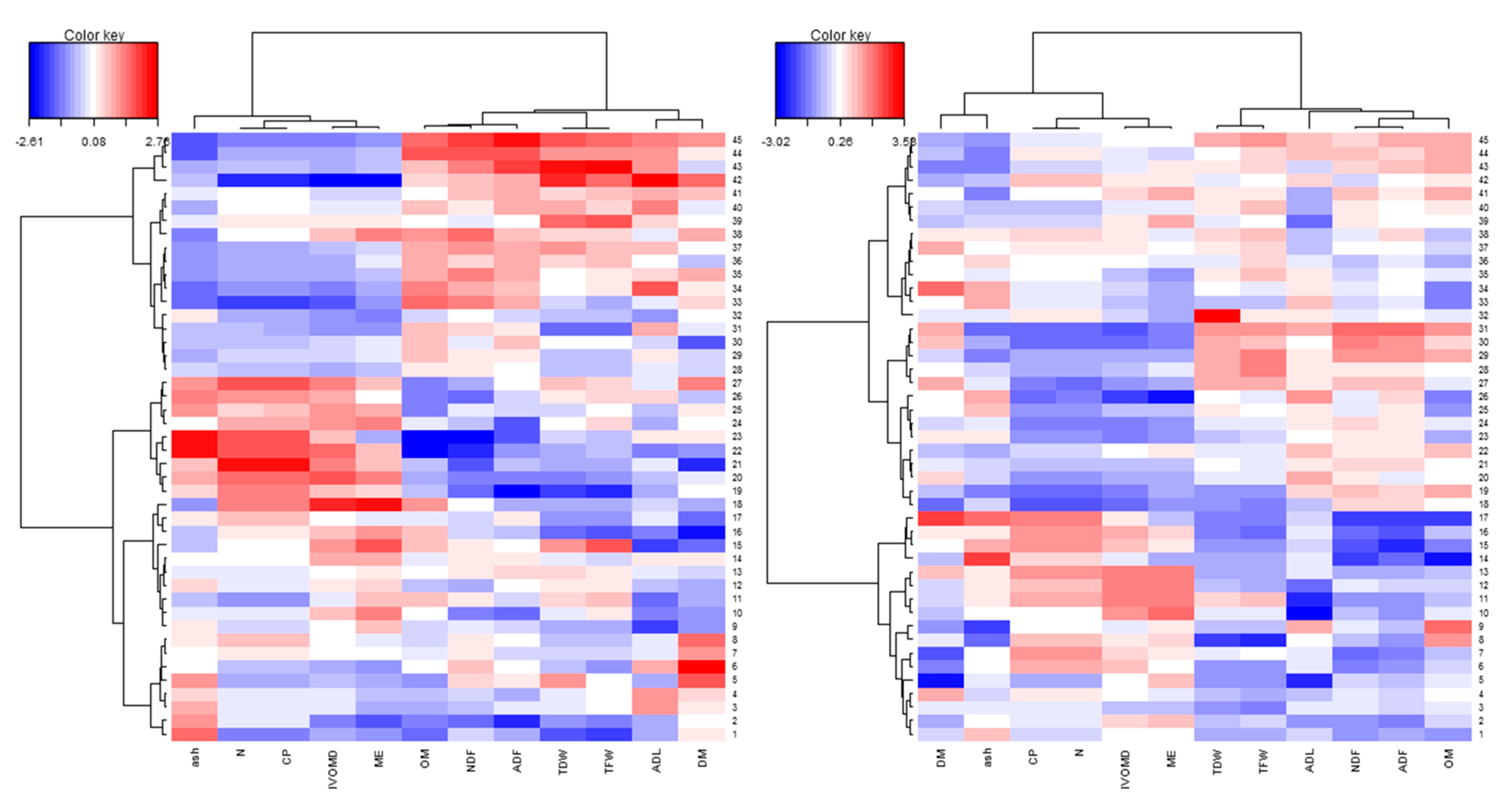
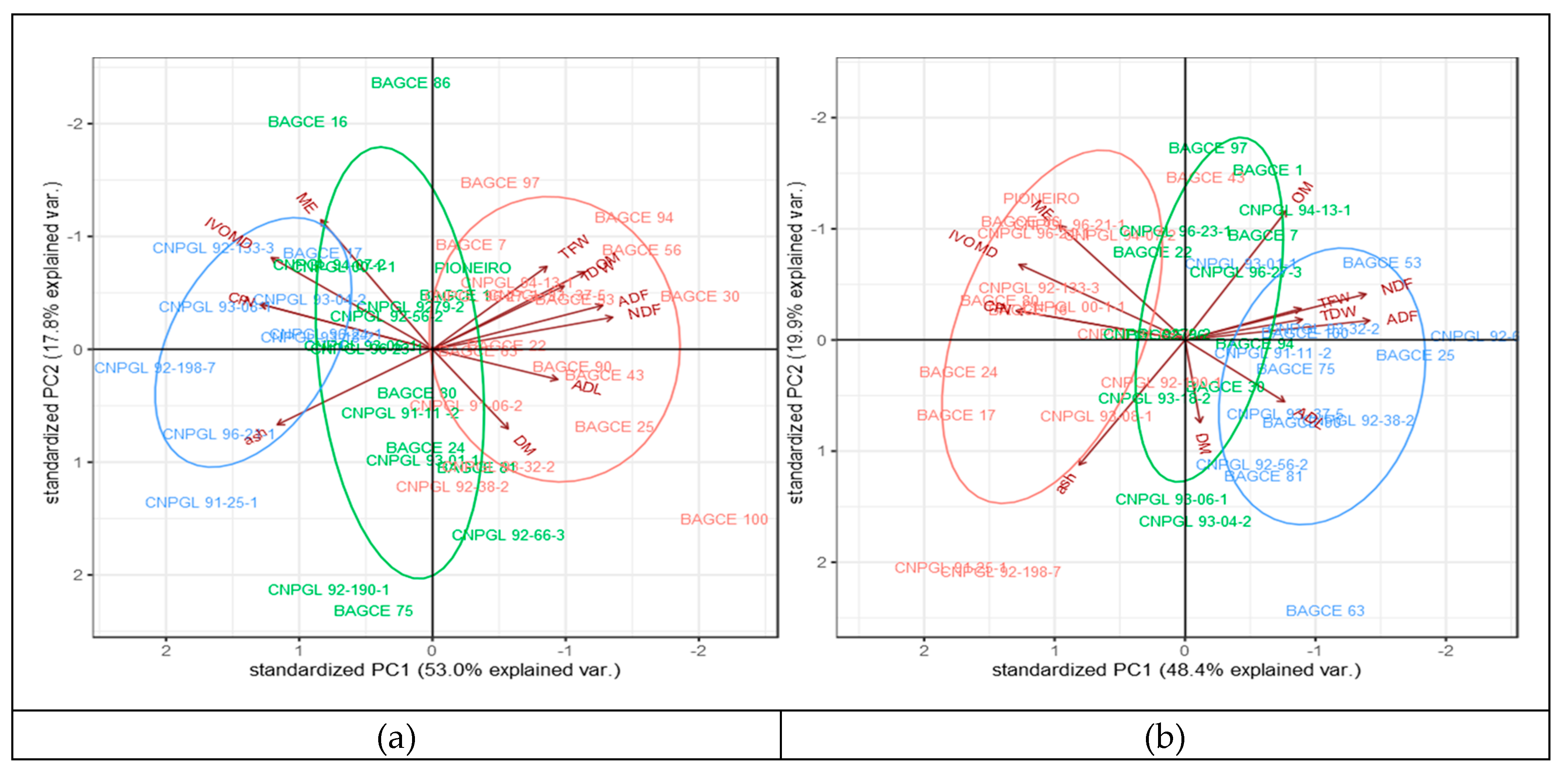
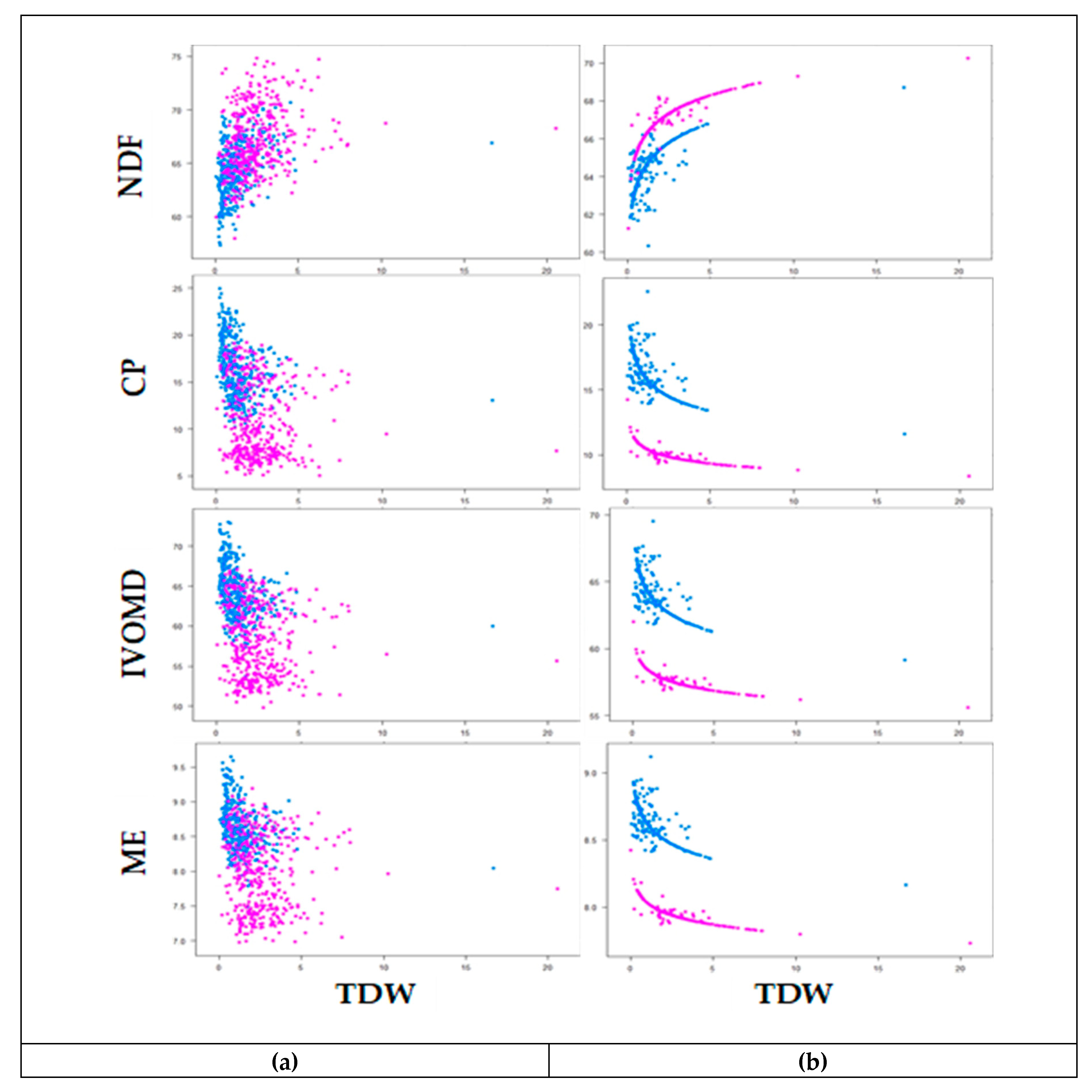
3.3. Phenotypic Variability and Association Between Biomass Yield and Feed Quality Traits
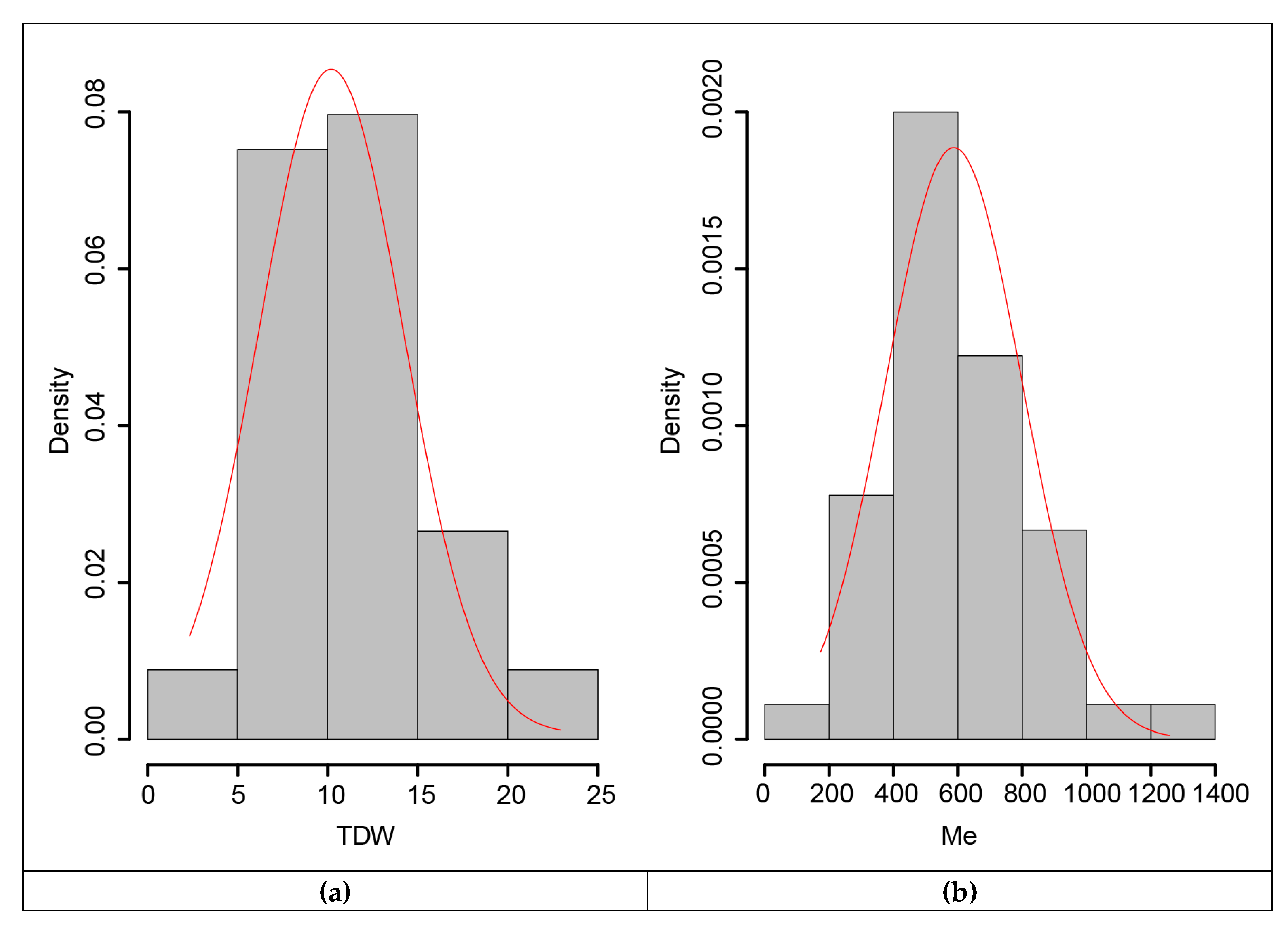
3.4. Cumulative Dry Weight and Metabolizable Energy Production of Genotypes
3.5. Marker-Trait Associations
3.6. Analysis of Nucleotide Sequences Corresponding to the Associated Markers
4. Discussion
4.1. Seasonal Variation Determines Forage Yield of Napier Grass Genotypes
4.2. Forage Yield of Napier Grass Genotypes
4.3. Genotypic Performance for Feed Nutritional Quality
4.4. Markers Associated with Dry Weight Yields and Metabolizable Energy
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Traits | Range | Mean | ||
|---|---|---|---|---|
| Wet | Dry | Wet | Dry | |
| TFW (t/ha) | 5.92–45.56 | 1.4–19.78 | 17.93 | 5.69 |
| TDW (t/ha) | 1.01–8.33 | 0.36–4.01 | 3.38 | 1.2 |
| LSR | 0.55–11.13 | 0.6–18.63 | 2.94 | 4.96 |
| NDF% | 62.64–73.29 | 61.23–73.27 | 68.62 | 66.33 |
| CP% | 7.48–16.95 | 13.25–22.48 | 12.23 | 17.04 |
| ME (MJ/ Kg DM) | 7.3–9.17 | 8.13–9.51 | 8.12 | 8.74 |
| Genotypes | TFW (t/ha) | TDW (t/ha) | LSR | NDF% | CP% | ME (MJ/Kg DM) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wet | Dry | Wet | Dry | Wet | Dry | Wet | Dry | Wet | Dry | Wet | Dry | |
| 14984 | 23.9 | 7.41 | 4.26 | 1.29 | 1.39 | 5.8 | 68.82 | 65.82 | 12.62 | 18.02 | 8.16 | 8.91 |
| 16786 | 13.2 | 14.97 | 2.8 | 3.05 | 3.46 | 2.8 | 69.36 | 67.73 | 10.93 | 15.79 | 7.96 | 8.58 |
| 16837 | 18.9 | 7.27 | 3.7 | 1.55 | 3.81 | 5.1 | 67.69 | 65.43 | 13.14 | 17.3 | 8.23 | 8.73 |
Appendix B
| Number Codes | Genotypes | TFW (t/ha) | TDW (t/ha) | LSR | |||
|---|---|---|---|---|---|---|---|
| Wet | Dry | Wet | Dry | Wet | Dry | ||
| 1 | BAGCE 1 | 20.53 | 6.2 | 3.56 | 1.4 | 1.8 | 5.62 |
| 2 | BAGCE 100 | 28.1 | 7.84 | 5.53 | 1.64 | 2.99 | 4.48 |
| 3 | BAGCE 16 | 13.58 | 3.83 | 2.23 | 0.82 | 2.51 | 3.83 |
| 4 | BAGCE 17 | 20.49 | 3.72 | 3.24 | 0.8 | 2.1 | 7.86 |
| 5 | BAGCE 22 | 21.7 | 5.39 | 3.52 | 1.11 | 1.88 | 5.66 |
| 6 | BAGCE 24 | 18.63 | 2.57 | 2.91 | 0.59 | 3.62 | 6.33 |
| 7 | BAGCE 25 | 13.94 | 6.59 | 2.62 | 1.64 | 2.02 | 8.39 |
| 8 | BAGCE 30 | 26.73 | 7.05 | 4.9 | 1.46 | 1.73 | 7.45 |
| 9 | BAGCE 43 | 19.07 | 3.97 | 3.25 | 0.96 | 1.67 | 8.4 |
| 10 | BAGCE 53 | 22.62 | 8.2 | 4.29 | 1.69 | 1.74 | 4.8 |
| 11 | BAGCE 56 | 25.34 | 7.18 | 4.35 | 1.46 | 1.51 | 2.9 |
| 12 | BAGCE 63 | 15.15 | 5.11 | 2.41 | 1.24 | 4.87 | 5.85 |
| 13 | BAGCE 7 | 29.25 | 6.76 | 4.76 | 1.3 | 2.56 | 11.42 |
| 14 | BAGCE 75 | 7.35 | 2.41 | 1.22 | 0.59 | 3.67 | 7.47 |
| 15 | BAGCE 80 | 16.43 | 3.92 | 2.6 | 0.88 | 4.13 | 2.67 |
| 16 | BAGCE 81 | 18.43 | 4.39 | 4.2 | 0.98 | 3.21 | 4.87 |
| 17 | BAGCE 86 | 28.26 | 6.26 | 4.34 | 1.39 | 1.35 | 5.83 |
| 18 | BAGCE 93 | 19.62 | 4.93 | 3.23 | 1.11 | 1.7 | 5.25 |
| 19 | BAGCE 94 | 32.61 | 5.86 | 5.74 | 2.5 | 2.46 | 7.86 |
| 20 | BAGCE 97 | 20.95 | 5.41 | 3.65 | 1.25 | 1.57 | 5.77 |
| 21 | CNPGL 00-1-1 | 18.69 | 4.49 | 2.97 | 0.95 | 1.6 | 5.35 |
| 22 | CNPGL 91-06-2 | 15.45 | 3.82 | 2.47 | 0.86 | 2.26 | 4.76 |
| 23 | CNPGL 91-11 -2 | 14.86 | 4.57 | 2.51 | 1.21 | 2.31 | 8.03 |
| 24 | CNPGL 91-25-1 | 15.63 | 3.5 | 2.64 | 0.7 | 1.73 | 4.25 |
| 25 | CNPGL 92-133-3 | 14.01 | 5.64 | 2.22 | 1.1 | 1.77 | 5.51 |
| 26 | CNPGL 92-190-1 | 11.17 | 2.59 | 2.01 | 0.65 | 1.75 | 4.92 |
| 27 | CNPGL 92-198-7 | 14.31 | 3.14 | 2.14 | 0.79 | 2.43 | 4.26 |
| 28 | CNPGL 92-38-2 | 14.98 | 7.17 | 2.5 | 1.7 | 2.95 | 4.82 |
| 29 | CNPGL 92-56-2 | 19.16 | 5.25 | 3.38 | 1.37 | 1.81 | 5.98 |
| 30 | CNPGL 92-66-3 | 12.7 | 7.35 | 2.42 | 1.73 | 2.23 | 7.3 |
| 31 | CNPGL 9279-2 | 16.58 | 6.54 | 2.94 | 1.34 | 1.91 | 5.78 |
| 32 | CNPGL 93 -37-5 | 23.01 | 5.39 | 3.64 | 1.14 | 2.01 | 6.47 |
| 33 | CNPGL 93-01-1 | 17.6 | 4.35 | 2.91 | 1.04 | 1.81 | 6.8 |
| 34 | CNPGL 93-04-2 | 21.24 | 4.5 | 3.78 | 1.12 | 2.95 | 3.87 |
| 35 | CNPGL 93-06-1 | 12.45 | 4.27 | 2.07 | 0.88 | 2.19 | 3.6 |
| 36 | CNPGL 93-08-1 | 11.85 | 3.29 | 1.79 | 0.81 | 4.94 | 7.33 |
| 37 | CNPGL 93-18-2 | 21.43 | 6.5 | 3.43 | 1.14 | 2.08 | 4.34 |
| 38 | CNPGL 93-32-2 | 8.96 | 3.16 | 1.65 | 0.7 | 3.04 | 6.11 |
| 39 | CNPGL 94-07-2 | 8.63 | 2.83 | 1.5 | 0.73 | 3.29 | 4.96 |
| 40 | CNPGL 94-13-1 | 21.45 | 7.48 | 4.04 | 1.64 | 3.88 | 4.3 |
| 41 | CNPGL 96-21-1 | 5.79 | 1.42 | 1.05 | 0.32 | 2.11 | 6.55 |
| 42 | CNPGL 96-23-1 | 13.86 | 5.64 | 2.21 | 1.17 | 1.91 | 7.4 |
| 43 | CNPGL 96-24-1 | 18.42 | 3.8 | 2.76 | 0.88 | 2.58 | 6.86 |
| 44 | CNPGL 96-27-3 | 18.81 | 6.84 | 3.19 | 1.37 | 2.27 | 4.56 |
| 45 | PIONEIRO | 21.56 | 4.97 | 3.71 | 1.09 | 1.46 | 7.14 |
| SE | 0.89 | 0.2 | 0.79 | 1.39 | 0.2 | 1.6 | |
| Number Codes | Genotypes | NDF | CP | IVOMD | ME | ||||
|---|---|---|---|---|---|---|---|---|---|
| Wet | Dry | Wet | Dry | Wet | Dry | Wet | Dry | ||
| 1 | BAGCE 1 | 67.51 | 65.74 | 10.65 | 16.16 | 57.94 | 64.21 | 8.01 | 8.69 |
| 2 | BAGCE 100 | 68.13 | 65.26 | 8.99 | 15.44 | 55.54 | 63.6 | 7.72 | 8.62 |
| 3 | BAGCE 16 | 67.32 | 62.5 | 11.87 | 17.88 | 59.82 | 65.12 | 8.24 | 8.67 |
| 4 | BAGCE 17 | 66.47 | 61.72 | 11.41 | 18.11 | 58.9 | 65.35 | 8.11 | 8.68 |
| 5 | BAGCE 22 | 68.06 | 64.21 | 10.72 | 17.55 | 57.55 | 64.77 | 7.95 | 8.69 |
| 6 | BAGCE 24 | 66.63 | 62.24 | 10.59 | 18.24 | 57.69 | 65.59 | 7.92 | 8.72 |
| 7 | BAGCE 25 | 69.09 | 67.15 | 9.14 | 14.76 | 56.33 | 62.57 | 7.88 | 8.49 |
| 8 | BAGCE 30 | 69.75 | 64.23 | 9.75 | 16.85 | 56.72 | 63.49 | 7.88 | 8.5 |
| 9 | BAGCE 43 | 68.49 | 64.57 | 9.85 | 16.64 | 56.77 | 64.26 | 7.9 | 8.69 |
| 10 | BAGCE 53 | 68.59 | 66.74 | 10.09 | 15.65 | 57.26 | 63.23 | 7.95 | 8.54 |
| 11 | BAGCE 56 | 69.49 | 64.47 | 10.13 | 17.32 | 57.08 | 64.62 | 7.93 | 8.64 |
| 12 | BAGCE 63 | 67.44 | 64.63 | 10.43 | 14.82 | 57.46 | 61.92 | 7.96 | 8.34 |
| 13 | BAGCE 7 | 67 | 65.84 | 10.95 | 17.07 | 58.07 | 64.05 | 8 | 8.6 |
| 14 | BAGCE 75 | 66.54 | 65.73 | 9.74 | 14.48 | 57.06 | 62.57 | 7.89 | 8.52 |
| 15 | BAGCE 80 | 67.44 | 63.27 | 10.87 | 18.09 | 57.83 | 66.16 | 7.99 | 8.85 |
| 16 | BAGCE 81 | 67.91 | 65.11 | 10.12 | 15.16 | 57.32 | 62.92 | 7.91 | 8.51 |
| 17 | BAGCE 86 | 67.43 | 63.09 | 10.84 | 17.8 | 58.85 | 66.01 | 8.17 | 8.83 |
| 18 | BAGCE 93 | 68.99 | 65.46 | 10.01 | 15.04 | 57.2 | 62.8 | 7.93 | 8.5 |
| 19 | BAGCE 94 | 68.89 | 64.52 | 10.22 | 17.03 | 57.2 | 63.81 | 7.92 | 8.53 |
| 20 | BAGCE 97 | 69.22 | 66.02 | 10.75 | 16.75 | 58.57 | 65.11 | 8.12 | 8.78 |
| 21 | CNPGL 00-1-1 | 65.59 | 62.94 | 10.51 | 16.56 | 58.57 | 64.98 | 8.12 | 8.75 |
| 22 | CNPGL 91-06-2 | 67.44 | 64.61 | 10.3 | 17 | 57.21 | 64.56 | 7.91 | 8.64 |
| 23 | CNPGL 91-11 -2 | 67.55 | 65.54 | 11.23 | 15.89 | 57.94 | 63.58 | 7.96 | 8.56 |
| 24 | CNPGL 91-25-1 | 63.91 | 61.5 | 12.24 | 18.37 | 58.56 | 64.77 | 7.9 | 8.57 |
| 25 | CNPGL 92-133-3 | 64.85 | 62.31 | 12.67 | 18.06 | 59.13 | 65.01 | 8.06 | 8.7 |
| 26 | CNPGL 92-190-1 | 65.27 | 64.34 | 10.65 | 16.26 | 56.81 | 63.45 | 7.82 | 8.55 |
| 27 | CNPGL 92-198-7 | 64.48 | 61.38 | 12.26 | 17.26 | 59.29 | 64.23 | 8.05 | 8.55 |
| 28 | CNPGL 92-38-2 | 67.25 | 66.07 | 10.06 | 14.84 | 56.89 | 63.09 | 7.85 | 8.53 |
| 29 | CNPGL 92-56-2 | 66.18 | 64.42 | 10.64 | 15.17 | 58.21 | 63.53 | 8.03 | 8.58 |
| 30 | CNPGL 92-66-3 | 68.04 | 67.65 | 10.19 | 14.55 | 57.04 | 62.24 | 7.87 | 8.47 |
| 31 | CNPGL 9279-2 | 67.62 | 64.78 | 10.8 | 17.09 | 58.65 | 64.84 | 8.07 | 8.67 |
| 32 | CNPGL 93 -37-5 | 67.76 | 65.08 | 9.97 | 15.3 | 57.15 | 63.22 | 7.96 | 8.54 |
| 33 | CNPGL 93-01-1 | 66.32 | 65.41 | 10.62 | 16.01 | 57.63 | 64.51 | 7.89 | 8.68 |
| 34 | CNPGL 93-04-2 | 66.07 | 64.54 | 12.21 | 16.41 | 59.12 | 63.77 | 8.06 | 8.54 |
| 35 | CNPGL 93-06-1 | 66.64 | 64 | 11.27 | 16.32 | 57.98 | 63.75 | 7.98 | 8.54 |
| 36 | CNPGL 93-08-1 | 65.46 | 63.09 | 12.09 | 16.26 | 59.52 | 64.46 | 8.12 | 8.67 |
| 37 | CNPGL 93-18-2 | 65.13 | 63.69 | 11.66 | 16.68 | 58.61 | 64.32 | 7.99 | 8.63 |
| 38 | CNPGL 93-32-2 | 67.94 | 66.13 | 10.13 | 14.81 | 56.9 | 62.82 | 7.87 | 8.54 |
| 39 | CNPGL 94-07-2 | 66.74 | 63.92 | 10.89 | 15.54 | 58.48 | 64.51 | 8.09 | 8.73 |
| 40 | CNPGL 94-13-1 | 67.53 | 65.44 | 10.71 | 16.36 | 57.75 | 64.34 | 7.96 | 8.66 |
| 41 | CNPGL 96-21-1 | 65.22 | 63.68 | 11.86 | 17.62 | 58.41 | 64.77 | 8.02 | 8.72 |
| 42 | CNPGL 96-23-1 | 66.98 | 65.2 | 10.35 | 16.03 | 57.95 | 64.79 | 8.04 | 8.77 |
| 43 | CNPGL 96-24-1 | 66.96 | 64.49 | 11.22 | 17.52 | 58.87 | 66.16 | 8.07 | 8.85 |
| 44 | CNPGL 96-27-3 | 67.05 | 65.34 | 10.28 | 15.92 | 57.45 | 64.1 | 7.96 | 8.64 |
| 45 | PIONEIRO | 67.52 | 63.61 | 9.86 | 16.83 | 57.71 | 65.68 | 8.05 | 8.86 |
| SE | 0.71 | 0.75 | 0.64 | 0.91 | 0.75 | 0.97 | 0.09 | 0.11 | |
| Marker ID | Type | Pos. | Marker Allele | MAF | TDW (t/ha/yr) | ME (MJ/t DM/yr) | ||
|---|---|---|---|---|---|---|---|---|
| MM (−log10p) | ANOVA (−log10p) | MM (−log10p) | ANOVA (−log10p) | |||||
| 30280861 | SNP | 0 | A/G | G (0.46) | 4.30 | 5.18 | 4.62 | 5.22 |
| 30283847 | SNP | 0 | A/G | G (0.18) | 3.58 | 3.71 | 3.46 | 3.48 |
| 30285085 | silicoDArT | 0 | 0/1 | 0 (0.27) | 3.39 | 4.54 | 3.50 | 4.67 |
| 23634398 | silicoDArT | 0 | 0/1 | 0 (0.37) | 3.32 | 5.06 | 3.40 | 5.25 |
| 23602557 | silicoDArT | 5 | 0/1 | 0 (0.29) | 3.27 | 5.31 | 3.16 | 5.20 |
| 30288908 | SNP | 4 | G/A | A (0.12) | 3.15 | 4.85 | 3.24 | 5.00 |
| 23603266 | SNP | 0 | A/G | A (0.36) | 3.13 | 4.80 | 3.44 | 4.84 |
| 23590251 | SNP | 4 | C/G | G (0.36) | 3.33 | 5.14 | 3.32 | 5.15 |
| 23642045 | SNP | 0 | A/T | A (0.32) | 3.75 | 4.50 | 3.82 | 4.59 |
| 23619626 | SNP | 0 | T/G | G (0.32) | 3.06 | 4.03 | 3.08 | 4.20 |
| 23603845 | SNP | 0 | C/G | G (0.37) | 3.63 | 4.61 | 3.55 | 4.56 |
| 18160886 | SNP | 3 | G/A | G (0.47) | 3.04 | 4.03 | 3.05 | 4.07 |
| 30288066 | silicoDArT | 3 | 0/1 | 1 (0.30) | 3.03 | 5.38 | 3.01 | 5.43 |
| 23617359 | SNP | 0 | G/C | C (0.49) | 2.98 | 4.41 | 2.95 | 4.43 |
| 30283369 | SNP | 0 | G/T | G (0.49) | 3.15 | 3.00 | 3.16 | 3.08 |
| 23562266 | silicoDArT | 0 | 0/1 | 0 (0.45) | 2.91 | 4.25 | 2.88 | 4.24 |
| 23621857 | silicoDArT | 0 | 0/1 | 0 (0.19) | 2.90 | 4.74 | 2.82 | 4.50 |
| 30272381 | silicoDArT | 0 | 0/1 | 1 (0.16) | 2.85 | 4.91 | 2.76 | 4.74 |
| 23635042 | SNP | 4 | A/G | G (0.16) | 2.83 | 5.34 | 2.85 | 5.44 |
| 23592313 | silicoDArT | 6 | 0/1 | 1 (0.41) | 2.83 | 4.35 | 2.79 | 4.30 |
| 23601388 | silicoDArT | 0 | 0/1 | 1 (0.42) | 2.78 | 4.21 | 2.79 | 4.20 |
| 23613458 | SNP | 0 | C/T | T (0.40) | 2.75 | 3.05 | 2.71 | 3.07 |
| 23558691 | silicoDArT | 3 | 0/1 | 0 (0.36) | 2.72 | 3.51 | 2.66 | 3.49 |
| 23611461 | SNP | 3 | T/C | C (0.19) | 2.69 | 4.43 | 2.79 | 4.60 |
| 30273188 | silicoDArT | 2 | 0/1 | 1 (0.16) | 2.69 | 4.10 | 2.56 | 3.94 |
| 30277167 | SNP | 0 | A/C | C (0.27) | 2.98 | 5.10 | 2.99 | 5.20 |
| 23606642 | silicoDArT | 0 | 0/1 | 1 (0.23) | 2.65 | 3.32 | 2.56 | 3.17 |
| 30285860 | silicoDArT | 7 | 0/1 | 1 (0.29) | 2.65 | 5.24 | 2.65 | 5.30 |
| 23551315 | silicoDArT | 0 | 0/1 | 0 (0.39) | 2.64 | 3.15 | 2.59 | 3.14 |
| 23622292 | SNP | 3 | T/G | T (0.39) | 3.30 | 4.75 | 3.30 | 4.73 |
| 23623489 | SNP | 0 | T/C | C (0.28) | 2.62 | 4.31 | 2.68 | 4.41 |
| 18149799 | silicoDArT | 0 | 0/1 | 1 (0.40) | 2.62 | 4.04 | 2.67 | 4.09 |
| 23553829 | silicoDArT | 0 | 0/1 | 0 (0.20) | 2.62 | 2.83 | 2.60 | 2.88 |
| 23547174 | silicoDArT | 0 | 0/1 | 0 (0.20) | 2.62 | 2.92 | 2.60 | 2.97 |
| 23602556 | SNP | 0 | G/C | C (0.36) | 2.60 | 3.53 | 2.61 | 3.63 |
| 23571406 | silicoDArT | 0 | 0/1 | 0 (0.23) | 2.60 | 4.35 | 2.66 | 4.45 |
| 23601207 | SNP | 0 | A/G | G (0.43) | 2.59 | 4.34 | 2.62 | 4.41 |
| 23637406 | SNP | 1 | T/C | C (0.34) | 2.58 | 3.82 | 2.56 | 3.92 |
| 23554509 | silicoDArT | 0 | 0/1 | 1 (0.27) | 2.57 | 5.34 | 2.54 | 5.33 |
| 23612094 | SNP | 0 | T/C | T (0.46) | 2.56 | 2.55 | NS | NS |
| 23559758 | silicoDArT | 0 | 0/1 | 1 (0.27) | 2.56 | 3.59 | NS | NS |
| 17981556 | SNP | 5 | A/G | G (0.13) | 2.56 | 4.53 | 2.56 | 4.58 |
| 23561721 | silicoDArT | 0 | 0/1 | 1 (0.44) | 2.55 | 4.04 | 2.62 | 4.16 |
| 30278234 | SNP | 0 | G/A | A (0.17) | 3.06 | 4.68 | 3.00 | 4.62 |
| 23593708 | silicoDArT | 0 | 0/1 | 1 (0.35) | 2.53 | 3.75 | 2.53 | 3.69 |
| 23627155 | SNP | 0 | G/A | A (0.32) | 2.52 | 4.36 | 2.54 | 4.41 |
| 23643473 | SNP | 4 | G/C | G (0.34) | 2.52 | 2.65 | NS | NS |
| 23606518 | silicoDArT | 0 | 0/1 | 1 (0.45) | 2.51 | 3.82 | 2.58 | 3.95 |
| 23570280 | silicoDArT | 0 | 0/1 | 1 (0.45) | 2.51 | 3.82 | 2.58 | 3.95 |
| 23644354 | SNP | 0 | A/G | A (0.29) | 2.80 | 4.49 | 2.83 | 4.64 |
| 23617372 | SNP | 0 | C/G | C (0.41) | 2.99 | 4.22 | 2.96 | 4.20 |
| 23630423 | SNP | 5 | A/G | A (0.17) | 3.03 | 4.52 | 2.99 | 4.40 |
| 23614288 | SNP | 3 | C/T | T (0.34) | 3.00 | 5.33 | 3.06 | 5.41 |
| 23601541 | SNP | 0 | C/G | G (0.45) | 2.94 | 4.24 | 2.99 | 4.40 |
| 23593756 | SNP | 0 | A/G | G (0.38) | 2.80 | 3.68 | 2.82 | 3.78 |
| 23640208 | SNP | 3 | C/T | T (0.14) | 2.91 | 4.12 | 2.90 | 4.14 |
| 23634735 | SNP | 0 | A/G | G (0.24) | NS | NS | 2.53 | 5.16 |
| 30277688 | SNP | 0 | C/T | T (0.38) | 2.57 | 4.01 | 2.56 | 4.05 |
| 23603310 | SNP | 0 | T/C | C (0.26) | 2.60 | 5.16 | 2.62 | 5.23 |
| 23644438 | SNP | 0 | C/G | G (0.34) | 2.57 | 4.01 | 2.60 | 4.04 |
| 23637089 | SNP | 3 | C/G | C (0.30) | 2.56 | 4.72 | NS | NS |
| 23634691 | SNP | 0 | T/C | C (0.40) | 2.80 | 3.45 | 2.74 | 3.37 |
| 23610213 | SNP | 0 | G/C | G (0.41) | 2.83 | 3.55 | 2.82 | 3.60 |
| 9976877 | SNP | 0 | G/A | A (0.45) | 2.88 | 4.39 | 2.92 | 4.56 |
| 23641842 | SNP | 5 | T/A | T (0.40) | 2.87 | 4.05 | 2.86 | 4.24 |
| 23641967 | SNP | 3 | A/G | G (0.29) | 2.84 | 4.09 | 2.86 | 4.06 |
| 23624048 | SNP | 0 | G/C | C (0.47) | 2.58 | 4.87 | 2.62 | 5.00 |
| 23633468 | SNP | 5 | G/A | G (0.39) | 2.77 | 3.93 | 2.73 | 4.02 |
| 30284244 | SNP | 0 | A/G | A (0.42) | 2.75 | 3.86 | 2.79 | 4.05 |
| 9972908 | SNP | 0 | T/A | A (0.30) | 2.62 | 5.07 | 2.64 | 5.21 |
| 23591173 | SNP | 4 | C/T | T (0.30) | 2.81 | 4.60 | 2.83 | 4.74 |
| 23633785 | SNP | 3 | A/C | A (0.36) | 2.56 | 3.04 | 2.57 | 3.08 |
| 23603083 | SNP | 0 | G/C | G (0.36) | 2.65 | 3.39 | 2.57 | 3.42 |
| 23637508 | SNP | 4 | C/G | C (0.26) | 2.76 | 4.86 | 2.78 | 4.80 |
| 30275343 | SNP | 6 | A/G | G (0.39) | 2.74 | 4.37 | 2.74 | 4.51 |
| 23634506 | SNP | 6 | A/C | A (0.46) | 2.76 | 4.15 | 2.79 | 4.26 |
| 23600245 | SNP | 0 | G/A | G (0.41) | 2.62 | 4.13 | 2.64 | 4.24 |
| 23639712 | SNP | 5 | A/G | G (0.33) | 2.51 | 5.09 | 2.53 | 5.14 |
| 23643649 | SNP | 0 | A/G | A (0.50) | 2.58 | 2.77 | NS | NS |
| 23612910 | SNP | 7 | G/A | A (0.39) | NS | NS | NS | NS |
| 23612265 | SNP | 0 | C/T | C (0.35) | NS | NS | 2NS | NS |
| 23637849 | SNP | 2 | T/C | T (0.36) | 2.71 | 3.14 | 2.71 | 3.20 |
| 30290093 | SNP | 0 | C/T | C (0.34) | 2.62 | 2.84 | 2.62 | 2.98 |
| 23624218 | SNP | 5 | A/G | A (0.37) | 2.60 | 3.81 | 2.60 | 3.78 |
| 23609995 | SNP | 2 | A/G | A (0.28) | 2.60 | 2.99 | 2.60 | 3.07 |
| 23598317 | SNP | 0 | T/C | C (0.39) | 2.59 | 3.67 | 2.59 | 3.78 |
| 23622543 | SNP | 6 | T/C | T (0.40) | 2.58 | 3.80 | 2.58 | 3.73 |
| 23624444 | SNP | 3 | G/C | G (0.42) | 2.58 | 3.02 | 2.58 | 3.07 |
| 30286830 | SNP | 0 | A/G | G (0.18) | 2.58 | 3.65 | 2.58 | 3.72 |
| 23612073 | SNP | 0 | T/C | T (0.40) | 2.56 | 4.05 | 2.56 | 4.11 |
| 23602072 | SNP | 0 | C/T | T (0.24) | 2.52 | 4.47 | NS | NS |
References
- FAO. Feed Resources. Gateway to Dairy Production and Products. 2019. Available online: http://www.fao.org/dairy-production-products/production/feed-resources/en/ (accessed on 12 November 2019).
- Pandey, P.; Irulappan, V.; Bagavathiannan, M.V.; Senthil-Kumar, M. Impact of Combined Abiotic and Biotic Stresses on Plant Growth and Avenues for Crop Improvement by Exploiting Physio-morphological Traits. Front. Plant Sci. 2017, 8, 537. [Google Scholar] [CrossRef] [PubMed]
- Raza, A.; Razzaq, A.; Mehmood, S.S.; Zou, X.; Zhang, X.; Lv, Y.; Xu, J. Impact of climate change on crops adaptation and strategies to tackle its outcome: A review. Plants 2019, 8, 34. [Google Scholar] [CrossRef] [PubMed]
- Jorge, A.; Lukuyu, B.; Marita, C.; Mwangi, D.M.; Kinuthia, E.; Baltenweck, I.; Poole, J. Assessing the Uptake and Disease Impact of Napier Grass in Kenya; International Livestock Research Institute (ILRI): Nairobi, Kenya, 2014. [Google Scholar]
- Fekede, F.; Getnet, A.; Lulseged, G.; Muluneh, M.; Tadesse, T. Evaluation of Napier grass-vetch mixture to improve total herbage yield in the central highlands. In Proceedings of the 13th Annual Conference of the Ethiopian Society of Animal Production (ESAP), Addis Ababa, Ethiopia, 25–26 August 2005. [Google Scholar]
- Rusdy, M. Elephant grass as forage for ruminant animals. Livest. Res. Rural. Dev. 2016, 28, 49. [Google Scholar]
- Nyambati, E.M.; Muyekho, F.N.; Onginjo, E.; Lusweti, C.M. Production, characterization and nutritional quality of napier grass [Pennisetum purpureum (Schum.)] cultivars in western Kenya. Afr. J. Plant Sci. 2010, 4, 496–502. [Google Scholar]
- Bhandari, A.P.; Sukanya, D.H.; Ramesh, C.R. Application of isozyme data in finger printing Napiergrass (Pennisetum purpureum Schum.) for germplasm management. Genet. Resour. Crop Evol. 2006, 53, 253–264. [Google Scholar] [CrossRef]
- Singh, B.P.; Singh, H.P.; Obeng, E. Elephant grass. In Biofuel Crops: Production, Physiology and Genetics; Singh, B.P., Ed.; CAB International: Fort Valley, GA, USA, 2013; pp. 271–291. [Google Scholar] [CrossRef]
- Negawo, A.; Teshome, A.; Kumar, A.; Hanson, J.; Jones, C. Opportunities for Napier Grass (Pennisetum purpureum) Improvement Using Molecular Genetics. Agronomy 2017, 7, 28. [Google Scholar] [CrossRef]
- Samson, R.; Mani, S.; Boddey, R.; Sokhansanj, S.; Quesada, D.; Urquiaga, S.; Reis, V.; Lem, C. The potential of C4 perennial grasses for developing a global bioheat industry. Crit. Rev. Plant Sci. 2005, 24, 461–495. [Google Scholar] [CrossRef]
- Muktar, M.S.; Teshome, A.; Hanson, J.; Negawo, A.T.; Habte, E.; Domelevo entfellner, J.; Lee, K.; Jones, C.S. Genotyping by sequencing provides new insights into the diversity of Napier grass (Cenchrus purpureus) and reveals variation in genome-wide LD patterns between collections. Sci. Rep. 2019, 9, 1–15. [Google Scholar] [CrossRef]
- Paudel, D.; Kannan, B.; Yang, X.; Harris-Shultz, K.; Thudi, M.; Varshney, R.K.; Altpeter, F.; Wang, J. Surveying the genome and constructing a high-density genetic map of napiergrass (Cenchrus purpureus Schumach). Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef]
- Wang, J.; Chen, Z.; Jin, S.; Hu, Z.; Huang, Y.; Diao, Y. Development and characterization of simple sequence repeat (SSR) markers based on a full-length cDNA library of Napier Grass (Pennisetum purpureum Schum). Genes Genom. 2017, 39, 1297–1305. [Google Scholar] [CrossRef]
- Varshney, R.K.; Shi, C.; Thudi, M.; Mariac, C.; Wallace, J.; Qi, P.; Zhang, H.; Zhao, Y.; Wang, X.; Srivastava, R.K.; et al. Pearl millet genome sequence provides a resource to improve agronomic traits in arid environments. Nat. Biotechnol. 2017, 35, 969–976. [Google Scholar] [CrossRef] [PubMed]
- Kariuki, I.W.; Mwendia, S.W.; Muyekho, F.N.; Ajanga, S.I.; Omayo, D.O. Biomass production and forage quality of head-smut disease Resistant Napier grass accessions. Afr. Crop Sci. J. 2016, 24, 157–165. [Google Scholar] [CrossRef]
- Kabirizi, J.; Muyekho, F.; Mulaa, M.; Kawube, G.; Msangi, R.; Pallangyo, B.; Zziwa, E.; Mugerwa, S.; Ajanga, S.; Lukwago, G.; et al. Napier Grass Feed Resource: Production, Constraints and Implications for Smallholder Farmers in Eastern and Central Africa; EAAPP: Naivasha, Kenya, 2015; ISBN 978-9970-9269-1-6. [Google Scholar]
- Wamalwa, N.I.; Midega, C.A.; Ajanga, S.; Omukunda, N.E.; Muyekho, F.N.; Asudi, G.O.; Mulaa, M.; Khan, Z.R. Screening Napier grass accessions for resistance to Napier grass stunt disease using the loop-mediated isothermal amplification of DNA (LAMP). Crop Prot. 2017, 98, 61–69. [Google Scholar] [CrossRef]
- Kebede, G.; Feyissa, F.; Assefa, G.; Mengistu, A.; Kehaliew, A.; Melese, K.; Mengistu, S.; Estifanos, T.; Wolde, S.; Abera, M. Agronomic performance, dry matter yield stability and herbage quality of Napier grass (Pennisetum purpureum (L.) Schumach) accessions in different agro-ecological zones of Ethiopia. J. Agric. Crop Res. 2017, 5, 49–65. [Google Scholar]
- Gashaw, T.; Asresie, A.; Haylom, M. Climate change and livestock production in Ethiopia. Academia. J. Environ. Sci. 2015, 2, 59–62. [Google Scholar] [CrossRef]
- Negawo, A.T.; Jorge, A.; Hanson, J.; Teshome, A.; Muktar, M.S.; Azevedo, A.L.; Ledo, F.J.; Machado, J.C.; Jones, C.S. Molecular markers as a tool for germplasm acquisition to enhance the genetic diversity of a Napier grass (Cenchrus purpureus syn. Pennisetum purpureum) collection. Trop. Grassl. Forrajes Trop. 2018, 6, 58–69. [Google Scholar] [CrossRef]
- Sunita Choudhary, S.; Hash, C.T.; Sagar, P.; Prasad, K.V.S.V.; Blümmel, M. Near infrared spectroscopy estimation of pearl millet grain composition and feed quality. In Proceedings of the 14th International Conference on NIR Spectroscopy, Bangkok, Thailand, 7 November 2009; Saranwong, S., Ed.; IM Publications: Chichester, UK, 2010. [Google Scholar]
- Velazco, J.G.; Rodriguez, M.X.; Boer, M.P.; Jordans, D.R.; Eilers, P.H.; Malosetti, M.; van Eeuwijk, E. Modelling spatial trends in sorghum breeding field trials using a two-dimensional P-spline mixed model. Theor. Appl. Genet. 2017, 130, 1375–1392. [Google Scholar] [CrossRef]
- Cullis, B.R.; Gleeson, A.C. Spatial analysis of field experiments—An extension to two dimensions. Biometrics 1991, 47, 1449–1460. [Google Scholar] [CrossRef]
- David, B.; Murray, D.; Payne, R.; Soutar, D. Genstat for Windows: Introduction, 19th ed.; VSN International: Hemel Hempstead, UK, 2017. [Google Scholar]
- Singh, R.K.; Chaudhary, B.D. Biometrical Method in Quantitative Genetics Analysis; Kalyani Publishers: New Delhi, India, 1985. [Google Scholar]
- Jahufer, M.Z.; Luo, D. DeltaGen: A Comprehensive Decision Support Tool for Plant Breeders. Crop Sci. 2018, 58, 1–14. [Google Scholar] [CrossRef]
- Lemaire, G.; Belanger, G. Allometries in Plants as Drivers of Forage Nutritive Value: A. Review. Agriculture 2020, 10, 5. [Google Scholar] [CrossRef]
- Rosyara, U.R.; de Jong, W.S.; Douches, D.S.; Endelman, J.B. Software for genome-wide association studies in autopolyploids and its application to potato. Plant Genome 2016, 9, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Yan, H.; Li, J.; Zhou, S.; Liu, T.; Zhang, X.; Huang, L. Genome survey sequencing of purple elephant grass (Pennisetum purpureum Schum Zise) and identification of its SSR markers. Mol. Breed. 2018, 38. [Google Scholar] [CrossRef]
- Zhou, S.; Wang, C.; Frazier, T.P.; Yan, H.; Chen, P.; Chen, Z.; Huang, L.; Zhang, X.; Peng, Y.; Yan, Y.; et al. The first Illumina-based de novo transcriptome analysis and molecular marker development in Napier grass (Pennisetum purpureum). Mol. Breed. 2018, 38, 95. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2014, 14, 1754–1760. [Google Scholar] [CrossRef]
- Shapiro, S.S.; Wilk, M.B. An analysis of variance test for normality (complete samples). Biometrika 1965, 52, 591–611. [Google Scholar] [CrossRef]
- Capstaff, N.M.; Miller, A.J. Improving the Yield and Nutritional Quality of Forage Crops. Front. Plant Sci. 2018, 9, 535. [Google Scholar] [CrossRef]
- Gemiyo, D.; Jimma, A.; Wolde, S. Biomass Yield and Nutritive Value of Ten Napier Grass (Pennisetum purpureum) Accessions at Areka, Southern Ethiopia. World J. Agric. Sci. 2017, 13, 185–190. [Google Scholar] [CrossRef]
- Ball, D.M.; Collins, M.; Lacefield, G.D.; Martin, N.P.; Mertens, D.A.; Olson, K.E.; Putnam, D.H.; Undersander, D.J.; Wolf, M.W. Understanding forage quality. Am. FarmBureau Fed. Publ. 2001, 1, 1–3. [Google Scholar]
- Machado, J.C. (Embrapa, Brazil). Personal Communication. 2019. [Google Scholar]
- Minson, D.J. The Chemical Composition and Nutritive Value of Tropical Grasses. In Tropical Grasses; Skerman, P.J., Cameroon, D.G., Riveros, F., Eds.; Food and Agriculture Organization of the United Nations: Rome, Italy, 1990; pp. 172–180. [Google Scholar]
- Pattanashetti, S.K.; Upadhyaya, H.D.; Blummel, M.; Reddy, K.; Ramanareddy, Y.; Kumar, V.; Singh, S. Genetic variability in Napier grass (Pennisetum purpureum) germplasm conserved at ICRISAT genebank. In Proceedings of the 23rd International Grassland Congress, Delhi, India, 20–24 November 2015. [Google Scholar]
- Van Saun, R.J. Determining forage quality: Understanding feed analysis. Lamalink 2006, 3, 18–19. [Google Scholar]
- Van Soest, P.J. Nutritional Ecology of the Ruminant; Cornell University Press: Ithaca, NY, USA, 1982. [Google Scholar]
- Smith, O.B. Feed Resources for Intensive Smallholder Systems in the Tropics: The Role of Crop Residues. In Proceedings of the XVII International Grassland Congress, Rockhampton, Australia, 18–21 February 1993. [Google Scholar]
- Lazzarini, I.E.; Detmann, C.B.; Sampaio, M.F.; Paulino, S.C.V.; Filho, M.A.; Souza Oliveira, F.A. Intake and digestibility in cattle fed low quality tropical forage and supplemented with nitrogenous compounds. Rev. Bras. Zootec. 2009, 38, 2021–2030. [Google Scholar] [CrossRef]
- Mlay, P.S.; Pereka, E.C.; Phiri, S.; Balthazary, J.; Igusti, T.; Hvelpund, M.R.; Weisbjerg, J.M. Feed value of selected tropical grasses, legumes and concentrates. Vet. Arhiv 2009, 76, 53–63. [Google Scholar]
- Liu, W.; Liu, G.H.; Liao, R.B.; Chang, Y.L.; Huang, X.Y.; Wu, Y.B.; Yang, H.M.; Yan, H.J.; Cai, H.Y. Apparent metabolizable and net energy values of corn and soybean meal for broiler breeding cocks. Poult. Sci. 2017, 96, 135–143. [Google Scholar] [CrossRef]
- Silva, V.B.; Daheri, R.F.; Menzes, B.R.; Oliveira, M.L.; Araujo, M.S.; Novo, A.A. Different stability methods for cultivar recommendation in elephant-grass for energy purposes in Brazil. Cerne 2017, 23, 507–515. [Google Scholar] [CrossRef]
- Khaled, R.A.; Duru, M.; Cruz, P. Are leaf traits suitable for assessing the feeding value of native grass species? In Proceedings of the 20th International Grassland Congress, Dublin, Ireland, 26 June–1 July 2005; O’Mara, F.P., Wilkins, R.J., Mannetje’t, L., Lovett, D.K., Rogers, P.A.M., Boland, T.M., Eds.; Wageningen Academic Publishers: Wageningen, The Netherlands, 2005. [Google Scholar]
- Peiretti, P.G.; Gai, F.; Tassone, S. Nutritional value and fatty acid profile of Niger (Guizotia abyssinica) plant during its growth cycle. Livest. Res. Rural Dev. 2015, 27, 1–10. [Google Scholar]
- Evitayani, L.; Warly, A.; Fariani, T.; Fujihara, T. Seasonal changes in nutritive value of some grass species in west Sumatra, Indonesia. Asian Australas. J. Anim. Sci. 2004, 17, 1663–1668. [Google Scholar] [CrossRef]
- Ford, C.; Morrison, I.; Wilson, J. Temperature effects on lignin, hemicellulose and cellulose in tropical and temperate grasses. Aust. J. Agric. Res. 1979, 30, 621–633. [Google Scholar] [CrossRef]
- Liu, Y.; Wu, Q.; Ge, G.; Han, G.; Jia, Y. Influence of drought stress on afalfa yields and nutritional composition. BMC Plant Biol. 2018, 18, 13. [Google Scholar] [CrossRef]
- Kilian, A.; Wenzl, P.; Huttner, E.; Carling, J.; Xia, L.; Blois, H.; Aschenbrenner-Kilian, M. Diversity arrays technology: A generic genome profiling technology on open platforms. Methods Mol. Biol. 2012, 888, 67–89. [Google Scholar] [CrossRef]
- Victoria, L.S. Genomic Studies of Local Adaptation in Natural Plant Populations. J. Hered. 2018, 109, 3–15. [Google Scholar] [CrossRef]
- Lauria, M.; Molinari, F.; Motto, M. Genetic Strategies to Enhance Plant Biomass Yield and Quality-Related Traits for Bio-Renewable Fuel and Chemical Productions. In Book Plants for the Future; Intech Open: London, UK, 2015; pp. 99–133. [Google Scholar] [CrossRef]
- Schuler, M.A.; Werck-Reichhart, D. Functional Genomics of P450s. Annu. Rev. Plant Biol. 2003, 54, 629–667. [Google Scholar] [CrossRef]
- Kumar, S.; Hash, C.T.; Nepolean, T.; Satayayathi, C.T.; Singh, G.; Mahendrakar, M.D.; Yadav, R.S.; Srivastava, R.K. Mapping QTLs controlling flowering time and important agronomic traits in pearl millet. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
| Genotypes | Scientific Name | Origin | Collection | Genotypes | Scientific Name | Origin | Collection |
|---|---|---|---|---|---|---|---|
| BAGCE 1 | Cenchrus purpureus | Colombia | EMBRAPA_ Collection | CNPGL 91-25-1 | Cenchrus purpureus | NA | EMBRAPA_ elite_line |
| BAGCE 7 | Cenchrus purpureus | Brazil | EMBRAPA_ Collection | CNPGL 92-133-3 * | Cenchrus purpureus | NA | EMBRAPA_ elite_line |
| BAGCE 16 | Cenchrus purpureus | Brazil | EMBRAPA_ Collection | CNPGL 92-190-1 | Cenchrus purpureus | NA | EMBRAPA_ elite_line |
| BAGCE 17 | Cenchrus purpureus | Costa Rica | EMBRAPA_ Collection | CNPGL 92-198-7 | Cenchrus purpureus | NA | EMBRAPA_ elite_line |
| BAGCE 22 | Cenchrus purpureus | NA | EMBRAPA_ Collection | CNPGL 92-38-2 | Cenchrus purpureus | NA | EMBRAPA_ elite_line |
| BAGCE 24 | Cenchrus purpureus | NA | EMBRAPA_ Collection | CNPGL 92-56-2 | Cenchrus purpureus | NA | EMBRAPA_ elite_line |
| BAGCE 25 | Cenchrus purpureus | India | EMBRAPA_ Collection | CNPGL 92-66-3 | Cenchrus purpureus | NA | EMBRAPA_ elite_line |
| BAGCE 30 | Cenchrus purpureus | Brazil | EMBRAPA_ Collection _ | CNPGL 9279-2 | Cenchrus purpureus | NA | EMBRAPA_ elite_line |
| BAGCE 34 | Cenchrus purpureus | Brazil | EMBRAPA_ Collection | CNPGL 93 -37-5 | Cenchrus purpureus | NA | EMBRAPA_ elite_line |
| BAGCE 53 | Cenchrus purpureus | Brazil | EMBRAPA_ Collection | CNPGL 93-01-1 | Cenchrus purpureus | NA | EMBRAPA_ elite_line |
| BAGCE 56 | Cenchrus purpureus | Brazil | EMBRAPA_ Collection | CNPGL 93-04-2 | Cenchrus purpureus | NA | EMBRAPA_ elite_line |
| BAGCE 63 | Cenchrus purpureus | Cuba | EMBRAPA_ Collection | CNPGL 93-06-1 | Cenchrus purpureus | NA | EMBRAPA_ elite_line |
| BAGCE 75 | Cenchrus purpureus | Brazil | EMBRAPA_ Collection | CNPGL 93-08-1 * | Cenchrus purpureus | NA | EMBRAPA_ elite_line |
| BAGCE 80 | Cenchrus purpureus | Brazil | EMBRAPA_ Collection | CNPGL 93-18-2 * | Cenchrus purpureus | NA | EMBRAPA_ elite_line |
| BAGCE 81 | Cenchrus purpureus | Brazil | EMBRAPA_ Collection | CNPGL 93-32-2 | Cenchrus purpureus | NA | EMBRAPA_ elite_line |
| BAGCE 86 | Cenchrus purpureus | NA | EMBRAPA_ Collection | CNPGL 94-07-2 | Cenchrus purpureus | NA | EMBRAPA_ elite_line |
| BAGCE 93 | Cenchrus purpureus | NA | EMBRAPA_ Collection | CNPGL 94-13-1 * | Cenchrus purpureus | NA | EMBRAPA_ elite_line |
| BAGCE 94 | Cenchrus purpureus | NA | EMBRAPA_ Collection | CNPGL 96-21-1 * | Cenchrus purpureus | NA | EMBRAPA_ elite_line |
| BAGCE 97 | Cenchrus purpureus | NA | EMBRAPA_ Collection | CNPGL 96-23-1 | Cenchrus purpureus | NA | EMBRAPA_ elite_line |
| BAGCE 100 | Cenchrus purpureus | Brazil | EMBRAPA_ Collection | CNPGL 96-24-1 | Cenchrus purpureus | NA | EMBRAPA_ elite_line |
| CNPGL 00-1-1 | Cenchrus purpureus | NA | EMBRAPA_ elite_line | CNPGL 96-27-3 * | Cenchrus purpureus | NA | EMBRAPA_ elite_line |
| CNPGL 91-06-2 | Cenchrus purpureus | NA | EMBRAPA_ elite_line | PIONEIRO | Cenchrus purpureus | NA | EMBRAPA_ elite_line |
| CNPGL 91-11 -2 | Cenchrus purpureus | NA | EMBRAPA_ elite_line |
| Trait | Statistics | Genotype | Season | G * S | CV% |
|---|---|---|---|---|---|
| TFW (t/ha) | MS | 295.25 *** | 45,668.33 *** | 150.65 *** | 49 |
| R2% | 10 | 35 | 5 | ||
| TDW (t/ha) | MS | 11.32 *** | 1015.35 *** | 4.26 *** | 41 |
| R2% | 17 | 36 | 7 | ||
| LSR | MS | 25.45 *** | 196.17 *** | 18.44 * | 52 |
| R2% | 22 | 17 | 23 |
| Traits | Range | Mean | GCV% | PCV% | ||||
|---|---|---|---|---|---|---|---|---|
| Wet | Dry | Wet | Dry | Wet | Dry | Wet | Dry | |
| TFW (t/ha) | 5.79–32.61 | 1.42–8.2 | 18.03 | 5.02 | 30.30 | 27.82 | 30.89 | 43.76 |
| TDW (t/ha) | 1.05–5.74 | 0.32–2.5 | 3.08 | 1.14 | 31.52 | 26.36 | 32.51 | 34.29 |
| LSR | 1.35–4.94 | 2.67–11.42 | 2.41 | 5.87 | 39.29 | 37.04 | 90.57 | 93.57 |
| Trait | Leaf and Stem | Leaf | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Statistics | Genotype | Season | G * S | CV% | Genotype | Season | G*S | CV% | |
| DM | MS | 0.3 * | 0.15 * | 0.1 ns | 0.6 | 0.34 * | 0.01 ns | 0.08 ns | 0.6 |
| R2% | 7 | 5 | 2 | 7 | 1 | 2 | |||
| OM | MS | 6.31 *** | 3.43 ns | 2.14 ns | 1.7 | 7.07 *** | 14.35 ** | 1.43 ns | 1.9 |
| R2% | 14 | 1 | 5 | 14 | 1 | 3 | |||
| ash | MS | 6.30 *** | 10.75 ** | 2.17 ns | 7.4 | 7.07 *** | 27.81 *** | 1.46 ns | 7.7 |
| R2% | 13 | 1 | 5 | 14 | 1 | 3 | |||
| NDF | MS | 28.05 *** | 1446.82 *** | 7.04 ns | 3.7 | 28.37 *** | 1582.81 *** | 6.47 ns | 3.7 |
| R2% | 16 | 19 | 4 | 17 | 21 | 4 | |||
| ADF | MS | 23.28 *** | 4517.62 *** | 5.82 ns | 6.1 | 22.61 *** | 4346.15 *** | 5.79 * | 5.8 |
| R2% | 11 | 49 | 3 | 12 | 51 | 3 | |||
| ADL | MS | 0.61 *** | 93.3 *** | 0.15 ns | 10.5 | 0.64 *** | 96.89 *** | 0.14 ns | 10.7 |
| R2% | 7 | 24 | 2 | 7 | 24 | 2 | |||
| N | MS | 0.35 * | 170.81 *** | 0.10 ns | 26.5 | 0.25 * | 163.38 *** | 0.11 ns | 26.2 |
| R2% | 5 | 40 | 2 | 3 | 40 | 1 | |||
| CP | MS | 13.44 * | 6648.85 *** | 3.92 ns | 26.3 | 9.97 * | 6362.67 *** | 4.17 ns | 26 |
| R2% | 4 | 40 | 2 | 3 | 40 | 1 | |||
| IVOMD% | MS | 13.36 * | 8109.84 *** | 3.79 ns | 6.3 | 9.57 * | 8014.08 *** | 4.18 ns | 6.4 |
| R2% | 3 | 41 | 1 | 2 | 41 | 1 | |||
| ME | MS | 0.16* | 85.81 *** | 0.05 ns | 1.5 | 0.13 * | 83.56 *** | 0.05 ns | 5.7 |
| R2% | 3 | 35 | 2 | 2 | 33 | 1 | |||
| Season | Trait | Leaf and Stem | Leaf | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Range | Mean | GCV% | PCV% | Range | Mean | GCV% | PCV% | ||
| Wet Season | DM | 91.00–91.80 | 91.37 | 0.19 | 0.34 | 90.90–91.64 | 91.38 | 0.18 | 0.31 |
| OM | 79.06–82.42 | 81.09 | 1.07 | 1.64 | 78.17–82.58 | 80.94 | 1.06 | 1.64 | |
| ash | 17.70–21.07 | 19.05 | 4.62 | 7.02 | 17.53–21.96 | 19.19 | 4.51 | 6.95 | |
| NDF | 63.91–69.75 | 67.15 | 2.28 | 3.19 | 63.29–69.28 | 67.16 | 2.08 | 3.19 | |
| ADF | 34.70–40.12 | 37.5 | 3.34 | 5 | 34.35–39.45 | 37.52 | 3.11 | 4.81 | |
| ADL | 4.26–5.27 | 4.68 | 4.68 | 8.79 | 4.27–5.31 | 4.74 | 4.99 | 8.85 | |
| N | 1.44–2.03 | 1.71 | 8 | 15.08 | 1.53–1.99 | 1.7 | 6.58 | 13.48 | |
| CP | 9.00–12.67 | 10.71 | 7.98 | 15.04 | 9.57–12.44 | 10.63 | 6.76 | 12.96 | |
| IVOMD | 55.54–59.82 | 57.84 | 1.57 | 3.16 | 55.92–59.57 | 57.47 | 1.41 | 2.85 | |
| ME | 7.72–8.24 | 7.98 | 1.23 | 2.62 | 7.72–8.15 | 7.92 | 1.13 | 2.46 | |
| Dry Season | DM | 91.06–91.75 | 91.4 | 0.15 | 0.28 | 91.02–91.67 | 91.37 | 0.19 | 0.29 |
| OM | 79.42–82.14 | 80.96 | 0.76 | 1.25 | 79.02–81.74 | 80.67 | 0.77 | 1.32 | |
| ash | 18.09–20.73 | 19.28 | 3.12 | 5.18 | 18.46–21.12 | 19.57 | 3.12 | 5.37 | |
| NDF | 61.38–67.65 | 64.48 | 2.55 | 3.55 | 60.90–67.12 | 64.36 | 2.69 | 3.63 | |
| ADF | 29.57–35.62 | 32.78 | 4.86 | 6.86 | 29.66–35.71 | 32.88 | 4.92 | 6.87 | |
| ADL | 3.41–4.29 | 4 | 4.56 | 10.58 | 3.62–4.43 | 4.04 | 4.38 | 10.24 | |
| N | 2.32–2.94 | 2.63 | 6.78 | 13.5 | 2.30–2.90 | 2.6 | 6.74 | 12.21 | |
| CP | 14.48–18.37 | 16.44 | 6.77 | 13.51 | 14.38–18.15 | 16.24 | 6.68 | 12.42 | |
| IVOMD | 61.92–66.16 | 64.17 | 1.49 | 3.53 | 62.04–66.10 | 63.77 | 1.44 | 3.24 | |
| ME | 8.34–8.86 | 8.63 | 1.15 | 3 | 8.32–8.81 | 8.56 | 1.11 | 2.84 | |
| Traits | TDW | TFW | LSR | NDF | ADF | ADL | DM | OM | ash | CP | N | IVOMD | ME |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TDW | 0.86 *** | −0.32 * | 0.52 *** | 0.63 *** | 0.18 | 0.14 | 0.23 | −0.24 | −0.30 * | −0.30 * | −0.31 * | −0.26 | |
| TFW | 0.96 *** | −0.28 | 0.55 *** | 0.66 *** | 0.13 | 0.11 | 0.24 | −0.25 | −0.30 * | −0.30 * | −0.26 | −0.18 | |
| LSR | −0.47 ** | −0.38 * | −0.36 * | −0.14 | −0.07 | −0.17 | 0.14 | 0.13 | 0.31 * | 0.31 * | 0.37 * | 0.31 * | |
| NDF | 0.51 *** | 0.44 ** | −0.48 ** | 0.94 *** | 0.29 * | 0.08 | 0.68 *** | −0.69 *** | −0.75 *** | −0.75 *** | −0.62 *** | −0.36 * | |
| ADF | 0.67 *** | 0.62 *** | −0.12 | 0.83 *** | 0.38 ** | 0.07 | 0.50 *** | −0.52 *** | −0.74 *** | −0.74 *** | −0.68 *** | −0.48 *** | |
| ADL | 0.47 ** | 0.40 ** | −0.36 * | 0.42 ** | 0.57 *** | 0.24 | 0.15 | -0.19 | −0.29 | −0.29 | −0.59 *** | −0.67 *** | |
| DM | 0.24 | 0.11 | −0.15 | 0.38 * | 0.2 | 0.46 ** | −0.27 | 0.27 | 0.03 | 0.03 | −0.13 | −0.30 * | |
| OM | 0.39 ** | 0.38 * | −0.13 | 0.83 *** | 0.67 *** | 0.33 * | −0.02 | −1.00 *** | −0.27 | −0.27 | −0.14 | 0.12 | |
| ash | −0.41 ** | −0.39 ** | 0.18 | −0.83 *** | −0.67 *** | −0.34 * | 0 | −1.00 *** | 0.28 | 0.29 | 0.18 | −0.08 | |
| CP | −0.36 * | −0.27 | 0.22 | −0.73 *** | −0.56 *** | −0.31 * | −0.27 | −0.58 *** | 0.59 *** | 1.00 *** | 0.84 *** | 0.59 *** | |
| N | −0.36 * | −0.27 | 0.22 | −0.73 *** | −0.56 *** | −0.31 * | −0.27 | −0.58 *** | 0.59 *** | 1.00 *** | 0.84 *** | 0.59 *** | |
| IVOMD | −0.29 | −0.17 | 0.12 | −0.57 *** | −0.51 *** | −0.50 *** | −0.38 * | −0.40 ** | 0.42 ** | 0.87 *** | 0.87 *** | 0.91 *** | |
| ME | −0.18 | −0.05 | 0.11 | −0.26 | −0.33 * | −0.58 *** | −0.46 ** | −0.02 | 0.04 | 0.60 *** | 0.60 *** | 0.89 *** |
| Response Variable | Harvest Season | Fixed Variable | Linear Regression | Test1 (p-Value) | R2 (%) |
|---|---|---|---|---|---|
| LSR | Wet | TDW | y = −1.23x + 2.08 | 0.19 | 22 |
| LSR | Dry | TDW | y = −3.1x + 4.46 | <0.001 | 29 |
| NDF | Wet | TDW | y = 1.04x + 1.82 | <0.001 | 23 |
| NDF | Dry | TDW | y = 1.04x + 1.81 | <0.001 | 22 |
| CP | Wet | TDW | y = −1.20x + 1.03 | 0.004 | 38 |
| CP | Dry | TDW | y = −1.32x + 1.21 | <0.001 | 36 |
| IVOMD | Wet | TDW | y = −1.04x + 1.77 | 0.004 | 31 |
| IVOMD | Dry | TDW | y = −1.07x + 1.81 | <0.001 | 32 |
| ME | Wet | TDW | y = −1.02x + 0.91 | 0.013 | 33 |
| ME | Dry | TDW | y = −1.04x + 0.94 | <0.001 | 38 |
| Genotype | TDW | ME | ME Yield | Genotype | TDW | ME | ME Yield | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| (t/ha/year) | (MJ/Kg DM/year) | (MJ/ha/Year) | (t/ha/year) | (MJ/Kg DM/year) | (MJ/ha/Year) | ||||||
| BAGCE 94 | 24.51 | a | 51.35 | 1258.59 | a | BAGCE 93 | 10.42 | lm | 51.28 | 534.39 | klm |
| BAGCE 100 | 20.25 | b | 51.02 | 1033.16 | b | CNPGL 00-1-1 | 10.38 | lm | 52.59 | 545.93 | lmn |
| BAGCE 53 | 18.29 | c | 51.46 | 941.2 | c | CNPGL 93-01-1 | 10.38 | lm | 51.73 | 536.95 | lmn |
| CNPGL 94-13-1 | 16.13 | d | 51.88 | 836.82 | d | BAGCE 17 | 9.89 | mn | 52.39 | 518.14 | mno |
| BAGCE 30 | 16.07 | d | 51.15 | 822.05 | de | CNPGL 96-23-1 | 9.82 | mn | 52.45 | 515.06 | mno |
| BAGCE 7 | 15.75 | d | 51.79 | 815.62 | de | BAGCE 80 | 9.73 | mn | 52.5 | 510.83 | mno |
| BAGCE 56 | 15.65 | d | 51.73 | 809.53 | de | CNPGL 91-11-2 | 9.73 | mn | 51.56 | 501.68 | mnop |
| BAGCE 86 | 15.57 | d | 52.99 | 825.13 | de | BAGCE 63 | 9.71 | mn | 50.91 | 494.34 | mnop |
| BAGCE 81 | 15 | d | 51.27 | 769.05 | e | CNPGL 92-133-3 | 9.06 | no | 52.27 | 473.59 | nopq |
| CNPGL 96-27-3 | 13.53 | e | 51.8 | 700.85 | f | CNPGL 91-25-1 | 9.06 | no | 51.42 | 465.82 | nopq |
| CNPGL 93-04-2 | 13.4 | ef | 51.78 | 693.85 | f | CNPGL 96-24-1 | 9.04 | no | 52.75 | 476.88 | opq |
| CNPGL 93 -37-5 | 13.22 | efg | 51.51 | 680.94 | f | CNPGL 91-06-2 | 8.83 | no | 51.66 | 456.16 | opq |
| BAGCE 97 | 13.06 | efgh | 52.72 | 688.47 | fg | BAGCE 16 | 8.49 | op | 52.72 | 447.59 | pqr |
| BAGCE 1 | 12.99 | efgh | 52.09 | 676.65 | fgh | CNPGL 93-06-1 | 8.14 | opq | 51.54 | 419.54 | qrs |
| BAGCE 22 | 12.83 | efghi | 51.89 | 665.75 | fgh | BAGCE 24 | 7.64 | pq | 51.93 | 396.75 | rs |
| PIONEIRO | 12.47 | efghij | 52.72 | 657.42 | fghi | CNPGL 92-198-7 | 7.22 | qr | 51.8 | 374 | st |
| CNPGL 92-66-3 | 12.23 | fghijk | 51.03 | 624.15 | ghij | CNPGL 92-190-1 | 6.56 | rs | 48.25 | 316.54 | tu |
| CNPGL 92-56-2 | 12.12 | ghijk | 51.83 | 628.23 | ghij | CNPGL 93-08-1 | 6.48 | rs | 52.39 | 339.49 | tu |
| CNPGL 93-18-2 | 11.93 | hijk | 51.85 | 618.57 | hij | CNPGL 93-32-2 | 6.22 | rs | 51.23 | 318.66 | u |
| BAGCE 25 | 11.74 | ijk | 51.13 | 600.27 | ijk | CNPGL 94-07-2 | 5.56 | s | 52.48 | 291.79 | u |
| BAGCE 34 | 11.49 | jkl | 51.77 | 594.84 | jkl | BAGCE 75 | 4.53 | t | 51.23 | 232.07 | v |
| CNPGL 92-38-2 | 11.41 | jkl | 51.13 | 583.39 | jkl | CNPGL 96-21-1 | 3.34 | u | 52.22 | 174.41 | w |
| CNPGL 9279-2 | 11.13 | kl | 52.22 | 581.21 | jkl | ||||||
| Factors | TDW (t/ha/yr) | ME (MJ/t DM/yr) |
|---|---|---|
| Generalized heritability (H2g) | 0.64 | 0.65 |
| ANOVA-p-value (geno) | 2.2 × 10−16 *** | 2.2 × 10−16 *** |
| ANOVA-p-value (rep) | 2.2 × 10−16 *** | 1.89 × 10−10 *** |
| p-value for Shapiro-Wilk normality test | 0.1997 | 0.2677 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Habte, E.; Muktar, M.S.; Abdena, A.; Hanson, J.; Sartie, A.M.; Negawo, A.T.; Machado, J.C.; Ledo, F.J.d.S.; Jones, C.S. Forage Performance and Detection of Marker Trait Associations with Potential for Napier Grass (Cenchrus purpureus) Improvement. Agronomy 2020, 10, 542. https://doi.org/10.3390/agronomy10040542
Habte E, Muktar MS, Abdena A, Hanson J, Sartie AM, Negawo AT, Machado JC, Ledo FJdS, Jones CS. Forage Performance and Detection of Marker Trait Associations with Potential for Napier Grass (Cenchrus purpureus) Improvement. Agronomy. 2020; 10(4):542. https://doi.org/10.3390/agronomy10040542
Chicago/Turabian StyleHabte, Ermias, Meki S. Muktar, Asebe Abdena, Jean Hanson, Alieu M. Sartie, Alemayehu T. Negawo, Juarez Campolina Machado, Francisco José da Silva Ledo, and Chris S. Jones. 2020. "Forage Performance and Detection of Marker Trait Associations with Potential for Napier Grass (Cenchrus purpureus) Improvement" Agronomy 10, no. 4: 542. https://doi.org/10.3390/agronomy10040542
APA StyleHabte, E., Muktar, M. S., Abdena, A., Hanson, J., Sartie, A. M., Negawo, A. T., Machado, J. C., Ledo, F. J. d. S., & Jones, C. S. (2020). Forage Performance and Detection of Marker Trait Associations with Potential for Napier Grass (Cenchrus purpureus) Improvement. Agronomy, 10(4), 542. https://doi.org/10.3390/agronomy10040542





