Introgression of Two Quantitative Trait Loci for Stripe Rust Resistance into Three Chinese Wheat Cultivars
Abstract
1. Introduction
2. Materials and Methods
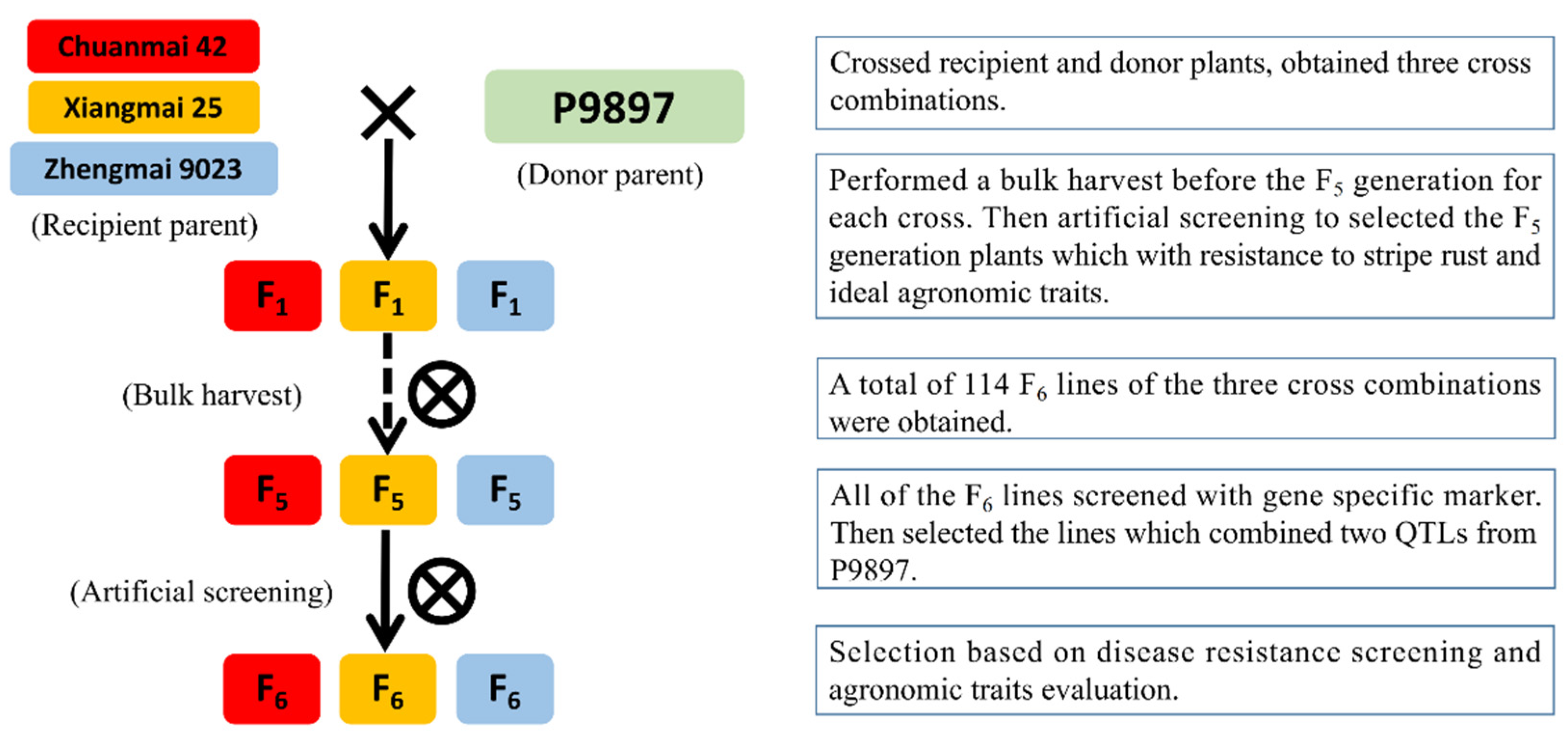
2.1. Plant Material
2.2. Cross Combination and Offspring Screening
2.3. Genotyping Wheat Lines by Molecular Markers
2.4. DNA Extraction and PCR
2.5. Disease Resistance and Agronomic Trait Evaluation in the Field
3. Results
3.1. Quantitative Trait Locus Detection
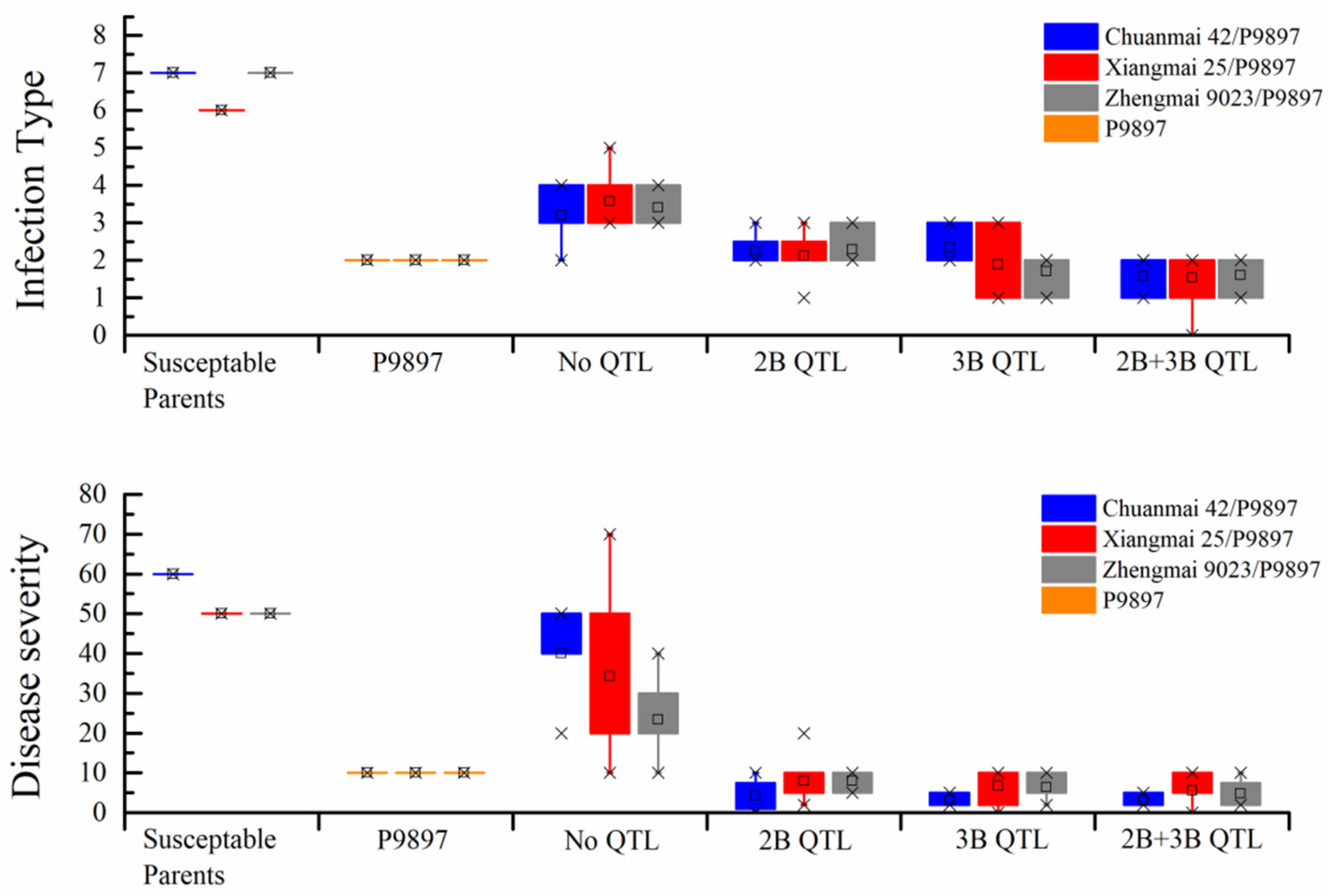
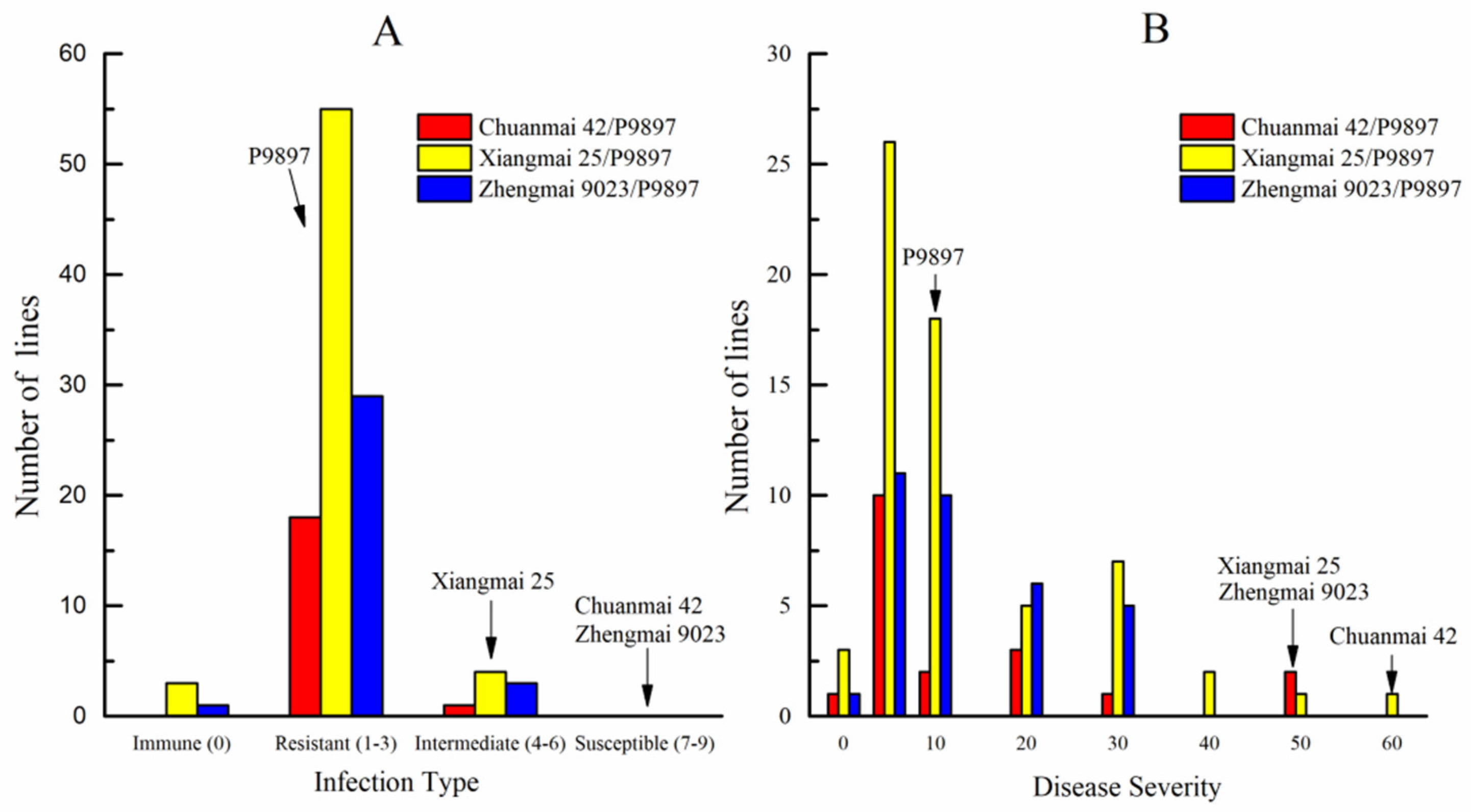
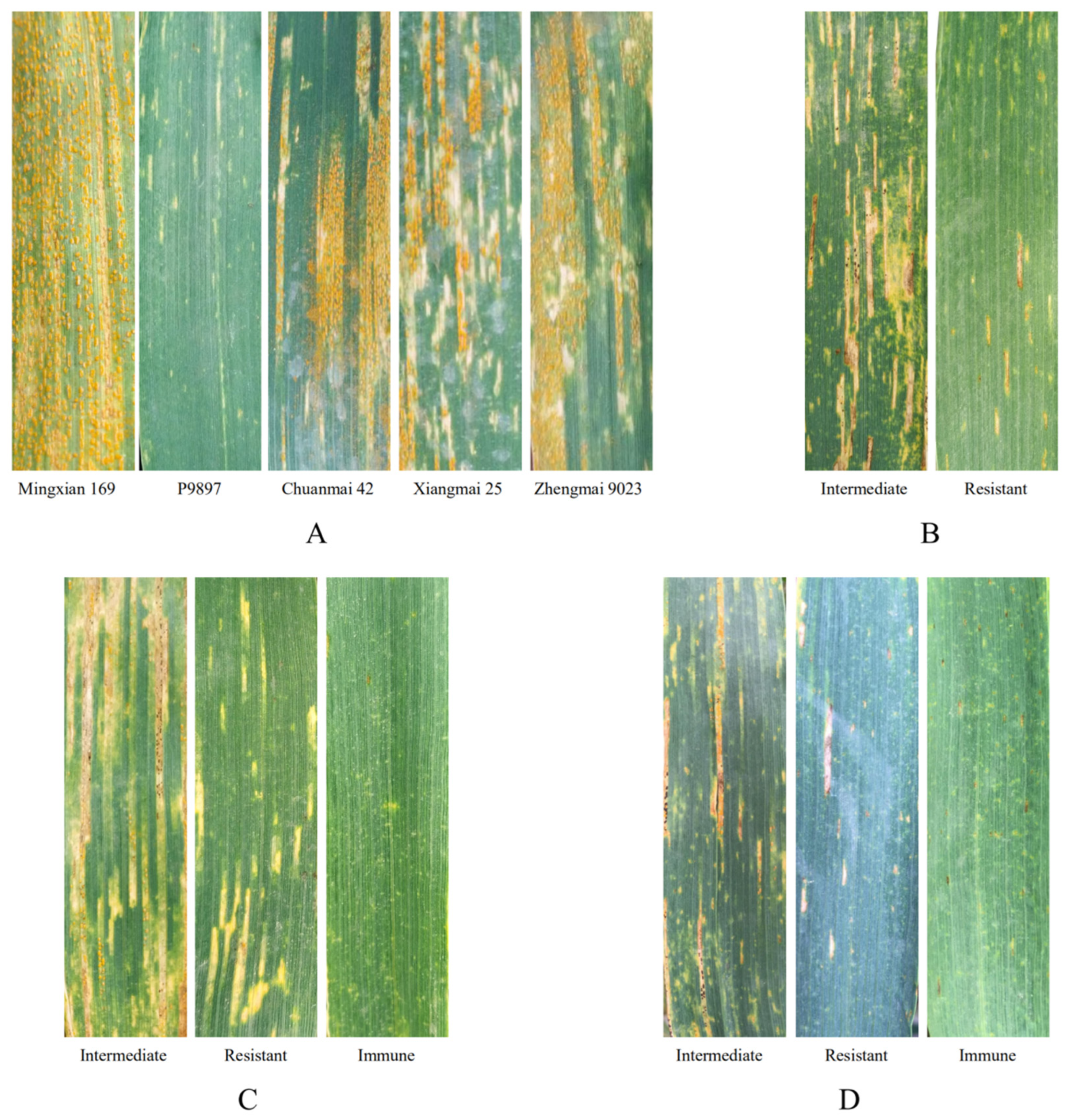
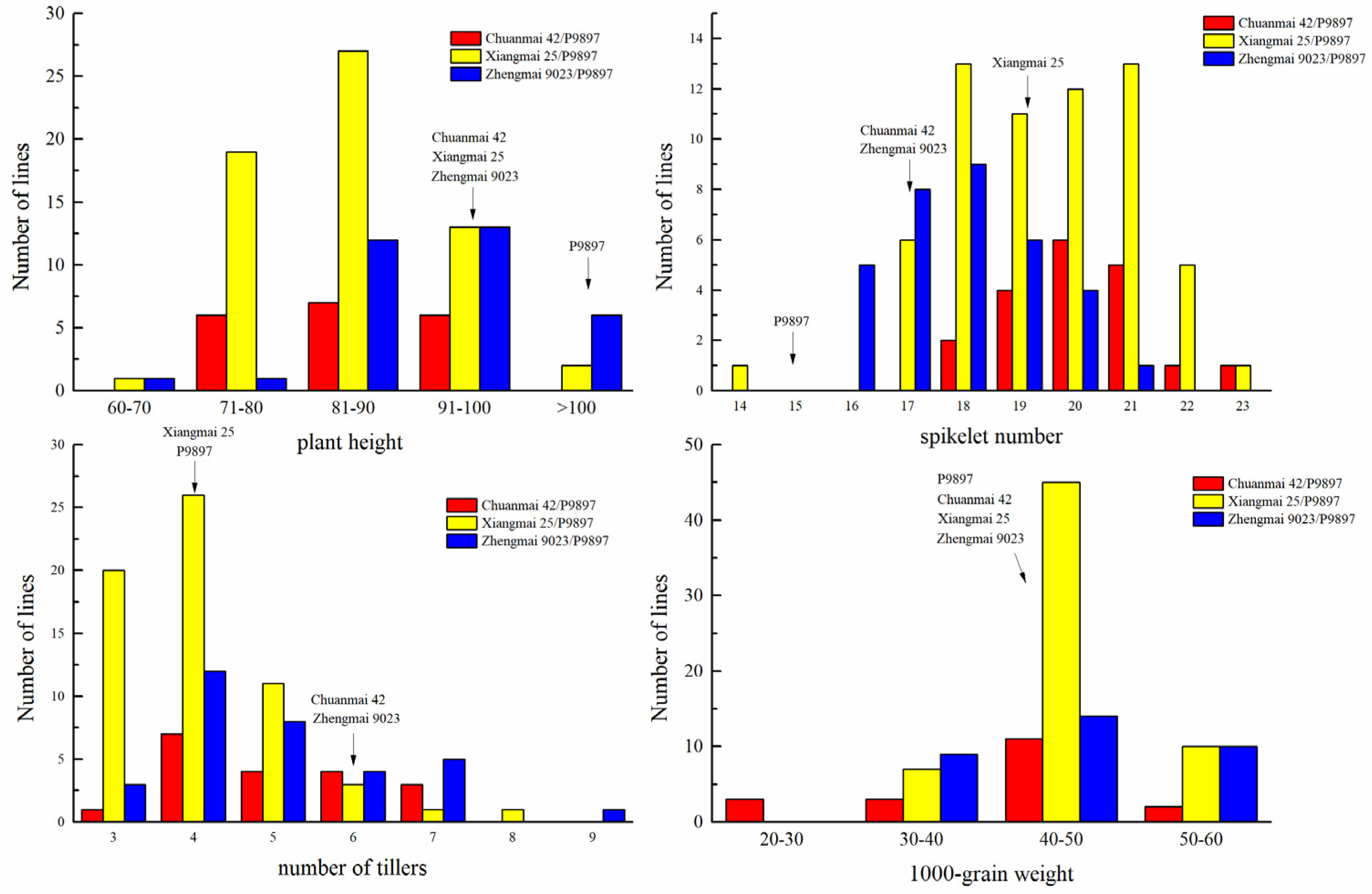
3.2. Evaluation of Disease Resistance and Agronomic Traits
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Liu, J.; Feng, B.; Xu, Z.B.; Fan, X.L.; Jiang, F.; Jin, X.F.; Cao, J.; Wang, F.; Liu, Q.; Yang, L.; et al. A genome-wide association study of wheat yield and quality-related traits in southwest China. Mol. Breed. 2017, 38, 1. [Google Scholar] [CrossRef]
- Savadi, S.; Prasad, P.; Kashyap, P.L.; Bhardwaj, S.C. Molecular breeding technologies and strategies for rust resistance in wheat (Triticum aestivum) for sustained food security. Plant Pathol. 2018, 67, 771–791. [Google Scholar] [CrossRef]
- Singh, R.P.; Hodson, D.P.; Huerta-Espino, J.; Yue, J.; Bhavani, S.; Njau, P.; Herrera-Foessel, S.; Singh, P.K.; Singh, S.; Govindan, V. The emergence of Ug99 races of the stem rust fungus is a threat to world wheat production. Annu. Rev. Phytopathol. 2011, 49, 465–481. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.Q.; Wu, L.R.; Liu, T.G.; Xu, S.C.; Jin, S.L.; Peng, Y.L.; Wang, B.T. Race dynamics, diversity, and virulence evolution in Puccinia striiformis f. sp. tritici, the causal agent of wheat stripe rust in China from 2003 to 2007. Plant Dis. 2009, 93, 1093–1101. [Google Scholar] [CrossRef]
- Wan, A.M.; Zhao, Z.H.; Chen, X.M.; He, Z.H.; Jin, S.L.; Jia, Q.Z.; Yao, G.; Yang, J.X.; Wang, B.T.; Li, G.B.; et al. Wheat stripe rust epidemic and virulence of Puccinia striiformis f. sp. tritici in China in 2002. Plant Dis. 2004, 88, 896–904. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.Q.; Kang, Z.S.; Ma, Z.H.; Xu, S.C.; Jin, S.L.; Jiang, Y.Y. Integrated management of wheat stripe rust caused by Puccinia striiformis f. sp. tritici in China. Sci. Agric. Sin. 2013, 46, 4254–4262. [Google Scholar] [CrossRef]
- Liu, T.G.; Zhang, Z.Y.; Liu, B.; Gao, L.; Peng, Y.L.; Chen, W.Q. Detection of virulence to Yr26 and pathogenicity to Chinese commercial winter wheat cultivars at seedling stage. Acta Phytopathol. Sin. 2015, 45, 41–47. [Google Scholar]
- Zhang, B.; Jia, Q.Z.; Huang, J.; Cao, S.Q.; Sun, Z.Y.; Luo, H.S.; Wang, X.M.; Jin, S.L. Trends and Toxicity Analysis of New Strains G22-9 and G22-14 in Puccinia striiformis f. sp. tritici. Acta Agric. Boreali Occident. Sin. 2015, 24. [Google Scholar] [CrossRef]
- Liu, B.; Liu, T.G.; Zhang, Z.Y.; Jia, Q.Z.; Wang, B.T.; Gao, L.; Peng, Y.L.; Jin, S.L.; Chen, W.Q. Discovery and pathogenicity of CYR34, a new race of Puccinia striiformis f. sp. tritici in China. Acta Phytopathol. Sin. 2017, 47, 681–687. [Google Scholar]
- Oliver, R.P. A reassessment of the risk of rust fungi developing resistance to fungicides. Pest Manag. Sci. 2014, 70, 1641–1645. [Google Scholar] [CrossRef]
- Line, R.F.; Chen, X.M. Successes in breeding for and managing durable resistance to wheat rusts. Plant Dis. 1995, 70, 855–862. [Google Scholar]
- Chen, X.M. Epidemiology and control of stripe rust [Puccinia striiformis f. sp. tritici] on wheat. Can. J. Plant Pathol. 2005, 27, 314–337. [Google Scholar] [CrossRef]
- Bariana, H.S.; Brown, G.N.; Bansal, U.K.; Miah, H.; Standen, G.E.; Lu, M. Breeding triple rust resistant wheat cultivars for Australia using conventional and marker-assisted selection technologies. Aust. J. Agric. Res. 2007, 58, 576–587. [Google Scholar] [CrossRef]
- McDonald, B.A.; Linde, C. The population genetics of plant pathogens and breeding strategies for durable resistance. Euphytica 2002, 124, 163–180. [Google Scholar] [CrossRef]
- Singh, R.P.; Huerta-Espino, J.; Bhavani, S.; Herrera-Foessel, S.A.; Singh, D.; Singh, P.K.; Velu, G.; Mason, R.E.; Jin, Y.; Njau, P.; et al. Race non-specific resistance to rust diseases in CIMMYT spring wheats. Euphytica 2011, 179, 175–186. [Google Scholar] [CrossRef]
- Lu, Y.M.; Lan, C.X.; Liang, S.S.; Zhou, X.C.; Liu, D.; Zhou, G.; Lu, Q.L.; Jing, J.X.; Wang, M.N.; Xia, X.C.; et al. QTL mapping for adult plant resistance to stripe rust in Italian common wheat cultivars Libellula and Strampelli. Theor. Appl. Genet. 2009, 119, 1349–1359. [Google Scholar] [CrossRef]
- Basnet, B.R.; Singh, R.P.; Ibrahim, A.M.H.; Herrera-Foessel, S.A.; Huerta-Espino, J.; Lan, C.X.; Rudd, J.C. Characterization of Yr54 and other genes associated with adult plant resistance to yellow rust and leaf rust in common wheat Quaiu 3. Mol. Breed. 2013, 33, 385–399. [Google Scholar] [CrossRef]
- Lowe, I.; Cantu, D.; Dubcovsky, J. Durable resistance to the wheat rusts: Integrating systems biology and traditional phenotype-based research methods to guide the deployment of resistance genes. Euphytica 2011, 179, 69–79. [Google Scholar] [CrossRef]
- Brown, J.K. Durable resistance of crops to disease: A Darwinian perspective. Annu. Rev. Phytopathol. 2015, 53, 513–539. [Google Scholar] [CrossRef]
- Niks, R.E.; Qi, X.Q.; Marcel, T.C. Quantitative resistance to biotrophic filamentous plant pathogens: Concepts, misconceptions, and mechanisms. Annu. Rev. Phytopathol. 2015, 53, 445–470. [Google Scholar] [CrossRef]
- Bariana, H.; McIntosh, R. Genetics of adult plant stripe rust resistance in four Australian wheats and the French cultivar ‘Hybride-de-Bersée’. Plant Breed. 1995, 114, 485–491. [Google Scholar] [CrossRef]
- Singh, R.P.; Huerta-Espino, J.; Rajaram, S. Achieving near immunity to leaf and stripe rusts in wheat by combining slow rusting resistance genes. Acta Phytopathol. Entomol. Hung. 2000, 35, 133–139. [Google Scholar]
- Elkot, A.F.; Chhuneja, P.; Kaur, S.; Saluja, M.; Keller, B.; Singh, K. Marker assisted transfer of two powdery mildew resistance genes PmTb7A.1 and PmTb7A.2 from Triticum boeoticum (Boiss.) to Triticum aestivum (L.). PLoS ONE 2015, 10, e0128297. [Google Scholar] [CrossRef][Green Version]
- Collard, B.C.Y.; Jahufer, M.Z.Z.; Brouwer, J.B.; Pang, E.C.K. An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: The basic concepts. Euphytica 2005, 142, 169–196. [Google Scholar] [CrossRef]
- Khan, M.A.; Kiran, U.; Ali, A.; Abdin, M.Z.; Zargar, M.Y.; Ahmad, S.; Sofi, P.A.; Gulzar, S. Molecular markers and marker-assisted selection in crop plants. In Plant Biotechnology: Principles and Applications; Springer: Singapore, 2017; pp. 295–328. [Google Scholar]
- Yoon, J.B.; Kwon, S.W.; Ham, T.H.; Kim, S.; Thomson, M.; Hechanova, L.; Jena, K.K.; Park, Y. Marker-assisted breeding. In Current Technologies in Plant Molecular Breeding; Springer: Dordrecht, The Netherlands, 2015; pp. 95–144. [Google Scholar]
- Ren, M.J.; Xu, R.H. Molecular Marker-assisted Selection and Identification of Guinong 19, a New Wheat Variety. Guizhou Agric. Sci. 2009, 37, 10–13. [Google Scholar] [CrossRef]
- Bie, T.D.; Zhao, R.H.; Zhu, S.Y. Development and characterization of marker MBH1 simultaneously tagging genes Pm21 and PmV conferring resistance to powdery mildew in wheat. Mol. Breed. 2015, 35, 189. [Google Scholar] [CrossRef]
- Wu, H.Y.; Zhang, B.Q.; Gao, D.R.; Cheng, S.H. A New Wheat Variety with High Quality and Weak Gluten Resistance-Yangmai 22. J. Triticeae Crops 2014, 34. [Google Scholar] [CrossRef]
- Okada, Y.; Kashiwazaki, S.; Kanatani, R.; Arai, S.; Ito, K. Effects of barley yellow mosaic disease resistant gene rym1 on the infection by strains of barley yellow mosaic virus and barley mild mosaic virus. Theor. Appl. Genet. 2003, 106, 181–189. [Google Scholar] [CrossRef]
- Pradhan, S.K.; Nayak, D.K.; Mohanty, S.; Behera, L.; Barik, S.R.; Pandit, E.; Lenka, S.; Anandan, A. Pyramiding of three bacterial blight resistance genes for broad-spectrum resistance in deepwater rice variety, Jalmagna. Rice 2015, 8, 19–33. [Google Scholar] [CrossRef]
- Xu, H.X.; Cao, Y.W.; Xu, Y.F.; Ma, P.T.; Ma, F.F.; Song, L.P.; Li, L.H.; An, D.G. Marker-assisted development and evaluation of near-isogenic lines for broad-spectrum powdery mildew resistance gene Pm2b introgressed into different genetic backgrounds of wheat. Front Plant Sci. 2017, 8, 1322–1331. [Google Scholar] [CrossRef]
- Wessels, E.; Botes, W.C. Accelerating resistance breeding in wheat by integrating marker-assisted selection and doubled haploid technology. S. Afr. J. Plant Soil 2014, 31, 35–43. [Google Scholar] [CrossRef]
- Zhou, X.L.; Han, D.J.; Chen, X.M.; Mu, J.M.; Xue, W.B.; Zeng, Q.D.; Wang, Q.L.; Huang, L.L.; Kang, Z.S. QTL mapping of adult-plant resistance to stripe rust in wheat line P9897. Euphytica 2015, 205, 243–253. [Google Scholar] [CrossRef]
- Chen, D.Q.; Zhang, D.R.; Tang, Q.Y.; Wang, Z.S.; Jiang, Q.B.; Zhou, F.J. Development and application of high yield and good quality wheat cultivar Xiangmai 25. Hubei Agric. Sci. 2009, 12, 19. [Google Scholar]
- Xu, W.G.; Hu, L.; Wang, G.S.; Zhang, L.; Dong, H.B.; Li, Y.; Zhang, J.Z.; Zan, X.C.; Qi, X.L.; Li, C.X.; et al. Breeding strategies of high quality and wide regional adaptability wheat variety Zhengmai 9023 and relevant thoughts on wheat high yield breeding. Henan Acad. Agric. Sci. 2009, 9, 14–18. [Google Scholar] [CrossRef]
- Tang, Y.L.; Li, J.; Wu, Y.Q.; Wei, H.T.; Li, C.S.; Yang, W.Y.; Chen, F. Identification of QTL for yield-related traits in the ecombinant inbred line population derived from the cross between a synthetic hexaploid wheat-derived variety Chuanmai 42 and a Chinese elite variety Chuannong 16. Agric. Sci. China 2011, 10, 1665–1680. [Google Scholar] [CrossRef]
- Liu, T.G.; Peng, Y.L.; Chen, W.Q.; Zhang, Z.Y. First detection of virulence in Puccinia striiformis f. sp. tritici in China to resistance genes Yr24 (=Yr26) present in wheat cultivar Chuanmai 42. Plant Dis. 2010, 94, 1163. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, Y.; Han, D.; Kang, Z.; Li, G.; Cao, A.; Chen, P. SSR and STS markers for wheat stripe rust resistance gene Yr26. Euphytica 2007, 159, 359–366. [Google Scholar] [CrossRef]
- Guyomarc’h, H.; Sourdille, P.; Charmet, G.; Edwards, K.J.; Bernard, M. Characterisation of polymorphic microsatellite markers from Aegilops tauschii and transferability to the D-genome of bread wheat. Theor. Appl. Genet. 2002, 109, 1105–1114. [Google Scholar] [CrossRef]
- Roder, M.S.; Korzun, V.; Wendehake, K.; Plaschke, J.; Tixier, M.H.; Leroy, P.; Ganal, M.W. A microsatellite map of wheat. Genetics 1998, 149, 2007–2023. [Google Scholar] [CrossRef]
- Somers, D.J.; Isaac, P.; Edwards, K. A high-density microsatellite consensus map for bread wheat. Theor. Appl. Genet. 2004, 109, 1105–1114. [Google Scholar] [CrossRef]
- Liu, S.X.; Anderson, J.A. Marker assisted evaluation of fusarium head blight resistant wheat germplasm. Crop Sci. 2003, 43, 760–766. [Google Scholar] [CrossRef]
- Cuc, L.M.; Mace, E.S.; Crouch, J.H.; Quang, V.D.; Long, T.D.; Varshney, R.K. Isolation and characterization of novel microsatellite markers and their application for diversity assessment in cultivated groundnut (Arachis hypogaea). BMC Plant Biol. 2008, 8, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.M.; Line, R.F.; Leung, H. Genome scanning for resistance-gene analogs in rice, barley, and wheat by high-resolution electrophoresis. Theor. Appl. Genet. 1998, 97, 345–355. [Google Scholar] [CrossRef]
- Zhou, X.L.; Han, D.J.; Chen, X.M.; Gou, H.L.; Guo, S.J.; Rong, L.; Wang, Q.L.; Huang, L.L.; Kang, Z.S. Characterization and molecular mapping of stripe rust resistance gene Yr61 in winter wheat cultivar Pindong 34. Theor. Appl. Genet. 2014, 127, 2349–2358. [Google Scholar] [CrossRef] [PubMed]
- Bassam, B.J.; Caetano-Anolles, G.; Gresshoff, P.M. Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal. Biochem. 1991, 196, 80–83. [Google Scholar] [CrossRef]
- Line, R.F.; Qayoum, A. Virulence, Aggressiveness, Evolution and Distribution of Races of Puccinia striiformis (the Cause of Stripe of Wheat) in North America, 1968–1987; Technical Bulletin-United States Department of Agriculture: Beltsville, MD, USA, 1992. [Google Scholar]
- Peterson, R.F.; Campbell, A.B.; Hannah, A.E. A diagrammatic scale for estimating rust intensity of leaves and stem of cereals. Can. J. Res. 1948, 26, 496–500. [Google Scholar] [CrossRef]
- Yuan, F.P.; Zeng, Q.D.; Wu, J.H.; Wang, Q.L.; Yang, Z.J.; Liang, B.P.; Kang, Z.S.; Chen, X.H.; Han, D.J. QTL mapping and validation of adult plant resistance to stripe rust in Chinese wheat landrace Humai 15. Front. Plant Sci. 2018, 9, 968–981. [Google Scholar] [CrossRef]
- Li, M.Z.; Li, Q.; Chao, K.X.; Shen, X.X.; Fan, Y.; Wang, Y.; Wang, B.T. Molecular detection of stripe rust resistance genes in 115 wheat varieties(lines) from Shaanxi Province. Acta Phytopathol. Sin. 2015, 45, 632–640. [Google Scholar] [CrossRef]
- Ren, Y.; Li, S.R.; Zhou, S.Q.; Du, X.Y.; He, Y.J.; Wei, Y.M.; Zheng, Y.L. Evaluation of resistance to stripe rust of 134 wheat Cultivars and lines from Sichuan Province. J. Triticeae Crops. 2014, 34, 847–853. [Google Scholar] [CrossRef]
- Li, G.Q.; Li, Z.F.; Yang, W.Y.; Zhang, Y.; He, Z.H.; Xu, S.C.; Singh, R.P.; Qu, Y.Y.; Xia, X.C. Molecular mapping of stripe rust resistance gene YrCH42 in Chinese wheat cultivar Chuanmai 42 and its allelism with Yr24 and Yr26. Theor. Appl. Genet. 2006, 112, 1434–1440. [Google Scholar] [CrossRef]
- Wu, J.H.; Zeng, Q.D.; Wang, Q.L.; Liu, S.J.; Yu, S.Z.; Mu, J.M.; Huang, S.; Sela, H.; Distelfeld, A.; Huang, L.L.; et al. SNP-based pool genotyping and haplotype analysis accelerate fine-mapping of the wheat genomic region containing stripe rust resistance gene Yr26. Theor. Appl. Genet. 2018, 131, 1481–1496. [Google Scholar] [CrossRef] [PubMed]
- Klymiuk, V.; Yaniv, E.; Huang, L.; Raats, D.; Fatiukha, A.; Chen, S.S.; Feng, L.H.; Frenkel, Z.; Krugman, T.; Lidzbarsky, G.; et al. Cloning of the wheat Yr15 resistance gene sheds light on the plant tandem kinase-pseudokinase family. Nat Commun. 2018, 9, 3735–3747. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.K.; Varshney, R.K.; Sharma, P.C.; Ramesh, B. Molecular markers and their applications in wheat breeding. Plant Breed. 1999, 119, 369–390. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, L.; Shi, H.; Chern, M.; Yu, H.; Yi, H.; He, M.; Yin, J.J.; Zhu, X.B.; Li, Y.; et al. A single transcription factor promotes both yield and immunity in rice. Science 2018, 361, 1026–1028. [Google Scholar] [CrossRef]
- Yaniv, E.; Raats, D.; Ronin, Y.; Korol, A.B.; Grama, A.; Bariana, H.; Dubcovsky, J.; Schulman, A.H.; Fahima, T. Evaluation of marker-assisted selection for the stripe rust resistance gene Yr15, introgressed from wild emmer wheat. Mol. Breed. 2015, 35, 43–55. [Google Scholar] [CrossRef]
- Chen, F.S.; Mu, J.M.; Huang, C.; Liu, J.Y.; Yao, J.B.; Meng, J. Comparison between pedigree and bulk method in wheat breeding. J. Nanjing Agric. Technol. Coll. 2003, 19, 1008–1895. [Google Scholar] [CrossRef]
| Marker | Primer Sequence | Tm d (°C) | References |
|---|---|---|---|
| Xcfd73a | F:GATAGATCAATGTGGGCCGT R:AACTGTTCTGCCATCTGAGC | 60 | [40] |
| Xgwm120 | F:GATCCACCTTCCTCTCTCTC R:GATTATACTGGTGCCGAAAC | 60 | [41] |
| Xbarc87b | F:GCTCACCGGGCATTGGGATCA R:GCGATGACGAGATAAAGGTGGAGAAC | 55 | [42] |
| Xbarc133 | F: AGCGCTCGAAAAGTCAG R:GGCAGGTCCAACTCCAG | 50 | [43] |
| WE-173c | F:GGGACAAGGGGAGTTGAAGC R:GAGAGTTCCAAGCAGAACAC | 55 | [39] |
| Xbarc181 | F:CGCTGGAGGGGGTAAGTCATCAC R:CGCAAATCAAGAACACGGGAGAAAGAA | 58 | [42] |
| Cross Combination | No QTL | 2B QTL a | 3B QTL b | 2B+3B QTL | Contains QTL Totals |
|---|---|---|---|---|---|
| Chuanmai 42/P9897 | 5 | 4 | 3 | 7 | 14 |
| Xiangmai 25/P9897 | 13 | 25 | 9 | 15 | 49 |
| Zhengmai 9023/P9897 | 10 | 8 | 10 | 5 | 23 |
| Cross Combination | Immune (IT = 0) | Resistant (IT = 1–3) | Intermediate (IT = 4–6) | Susceptible (IT = 7–9) |
|---|---|---|---|---|
| Chuanmai 42/P9897 | 0 | 18 | 1 | 0 |
| Xiangmai 25/P9897 | 3 | 55 | 4 | 0 |
| Zhengmai 9023/P9897 | 1 | 29 | 3 | 0 |
| Parent/Line | Trait | QTL a | ||||
|---|---|---|---|---|---|---|
| IT | PH | SPN | TLN | TGW | ||
| Mingxian169 | 9 | 120 | 15 | 5 | 38.13 | 0 |
| P9897 | 2 | 112 | 15 | 4 | 40.47 | 2BS+3BL |
| Chuanmai 42 | 7 | 100 | 18 | 6 | 41.03 | Yr26 |
| Xiangmai 25 | 6 | 93 | 21 | 4 | 44.37 | ? b |
| Zhengmai 9023 | 7 | 91 | 15 | 6 | 42.37 | ? |
| Chuanmai 42/P9897-2 | 2 | 74 | 21 | 5 | 44.93 | 2BS+3BL+Yr26 |
| Chuanmai 42/P9897-3c | 2 | 81 | 20 | 7 | 45.37 | 2BS+3BL+Yr26 |
| Chuanmai 42/P9897-4 | 2 | 77 | 21 | 7 | 33.67 | 2BS+3BL |
| Chuanmai 42/P9897-5 | 1 | 80 | 20 | 3 | 34.60 | 2BS+3BL+Yr26 |
| Chuanmai 42/P9897-7 | 2 | 80 | 23 | 6 | 33.37 | 2BS+3BL+Yr26 |
| Chuanmai 42/P9897-9 | 1 | 80 | 21 | 6 | 33.37 | 2BS+3BL+Yr26 |
| Chuanmai 42/P9897-14c | 2 | 88 | 19 | 5 | 40.77 | 2BS+3BL+Yr26 |
| Xiangmai 25/P9897-25c | 2 | 88 | 18 | 4 | 50.47 | 2BS+3BL |
| Xiangmai 25/P9897-26c | 1 | 90 | 17 | 7 | 42.60 | 2BS+3BL |
| Xiangmai 25/P9897-27c | 2 | 90 | 20 | 5 | 44.43 | 2BS+3BL |
| Xiangmai 25/P9897-34 | 0 | 117 | 21 | 5 | 41.03 | 2BS+3BL |
| Xiangmai 25/P9897-39c | 1 | 90 | 21 | 4 | 49.27 | 2BS+3BL |
| Xiangmai 25/P9897-40c | 2 | 94 | 21 | 4 | 43.20 | 2BS+3BL |
| Xiangmai 25/P9897-41 | 2 | 95 | 22 | 3 | 47.80 | 2BS+3BL |
| Xiangmai 25/P9897-42 | 2 | 95 | 22 | 3 | 36.50 | 2BS+3BL |
| Xiangmai 25/P9897-56c | 2 | 80 | 17 | 4 | 43.03 | 2BS+3BL |
| Xiangmai 25/P9897-59c | 2 | 83 | 21 | 4 | 47.80 | 2BS+3BL |
| Xiangmai 25/P9897-60c | 0 | 80 | 20 | 6 | 42.80 | 2BS+3BL |
| Xiangmai 25/P9897-62 | 2 | 80 | 18 | 3 | 41.46 | 2BS+3BL |
| Xiangmai 25/P9897-63 | 2 | 75 | 18 | 3 | 43.48 | 2BS+3BL |
| Xiangmai 25/P9897-65 | 2 | 70 | 14 | 3 | 38.75 | 2BS+3BL |
| Xiangmai 25/P9897-78c | 2 | 85 | 22 | 4 | 46.27 | 2BS+3BL |
| Zhengmai 9023/P9897-92c | 1 | 95 | 17 | 6 | 51.00 | 2BS+3BL |
| Zhengmai 9023/P9897-96 | 2 | 90 | 19 | 6 | 39.93 | 2BS+3BL |
| Zhengmai 9023/P9897-102 | 1 | 86 | 20 | 5 | 39.87 | 2BS+3BL |
| Zhengmai 9023/P9897-105c | 2 | 97 | 17 | 7 | 44.20 | 3BL+3BL |
| Zhengmai 9023/P9897-107 | 2 | 103 | 19 | 4 | 50.50 | 2BS+3BL |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, T.; Zhong, X.; Yang, Q.; Zhou, X.; Li, X.; Yang, S.; Hou, L.; Yao, Q.; Guo, Q.; Kang, Z. Introgression of Two Quantitative Trait Loci for Stripe Rust Resistance into Three Chinese Wheat Cultivars. Agronomy 2020, 10, 483. https://doi.org/10.3390/agronomy10040483
Hu T, Zhong X, Yang Q, Zhou X, Li X, Yang S, Hou L, Yao Q, Guo Q, Kang Z. Introgression of Two Quantitative Trait Loci for Stripe Rust Resistance into Three Chinese Wheat Cultivars. Agronomy. 2020; 10(4):483. https://doi.org/10.3390/agronomy10040483
Chicago/Turabian StyleHu, Tian, Xiao Zhong, Qiang Yang, Xinli Zhou, Xin Li, Suizhuang Yang, Lu Hou, Qiang Yao, Qingyun Guo, and Zhensheng Kang. 2020. "Introgression of Two Quantitative Trait Loci for Stripe Rust Resistance into Three Chinese Wheat Cultivars" Agronomy 10, no. 4: 483. https://doi.org/10.3390/agronomy10040483
APA StyleHu, T., Zhong, X., Yang, Q., Zhou, X., Li, X., Yang, S., Hou, L., Yao, Q., Guo, Q., & Kang, Z. (2020). Introgression of Two Quantitative Trait Loci for Stripe Rust Resistance into Three Chinese Wheat Cultivars. Agronomy, 10(4), 483. https://doi.org/10.3390/agronomy10040483






