Digital Count of Corn Plants Using Images Taken by Unmanned Aerial Vehicles and Cross Correlation of Templates
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Site
2.2. Image Acquisition and Processing
2.3. Image Color Pre-Processing
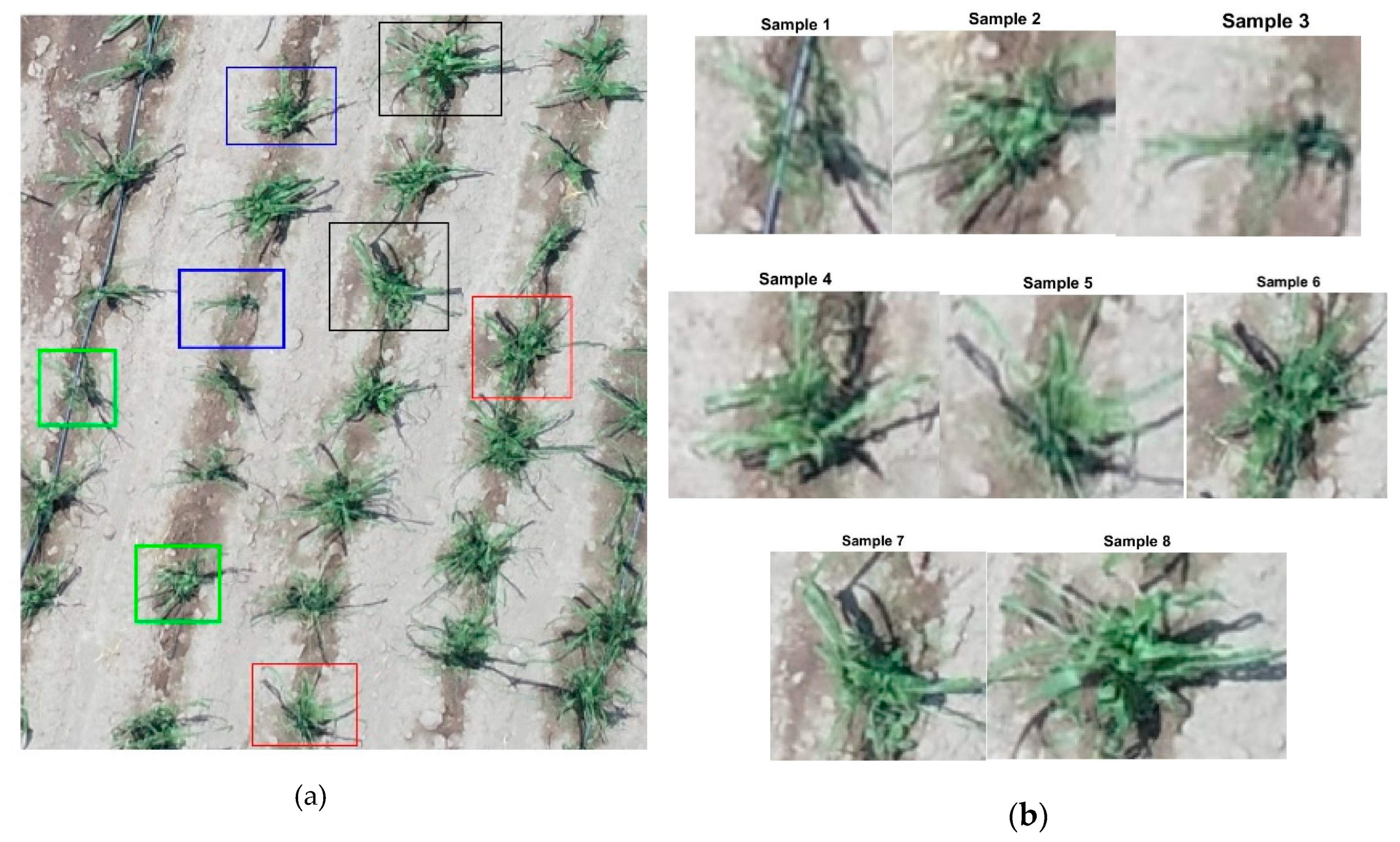
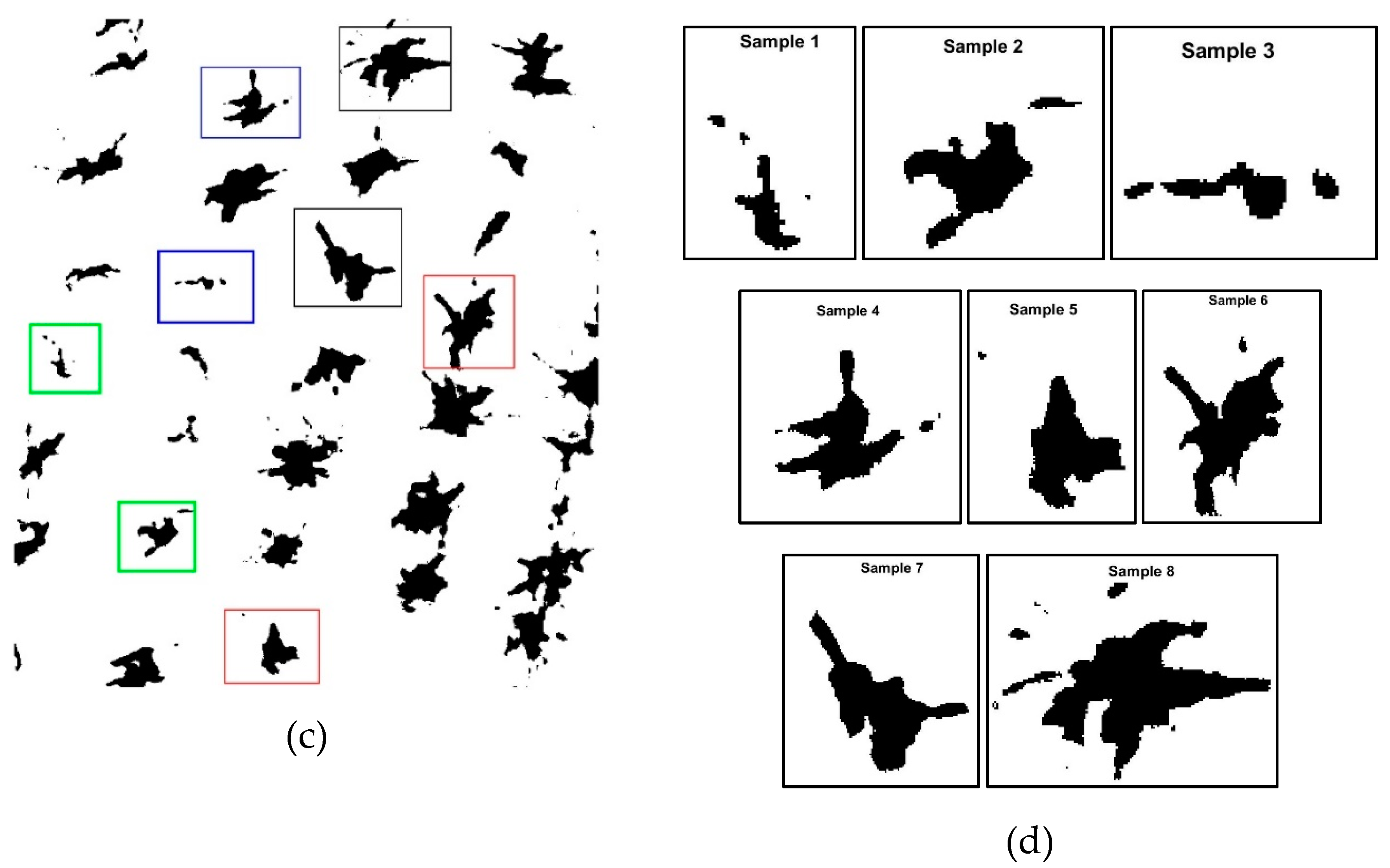
2.4. Selection of Samples or Templates (Plants)
2.5. Normalized Cross Correlation
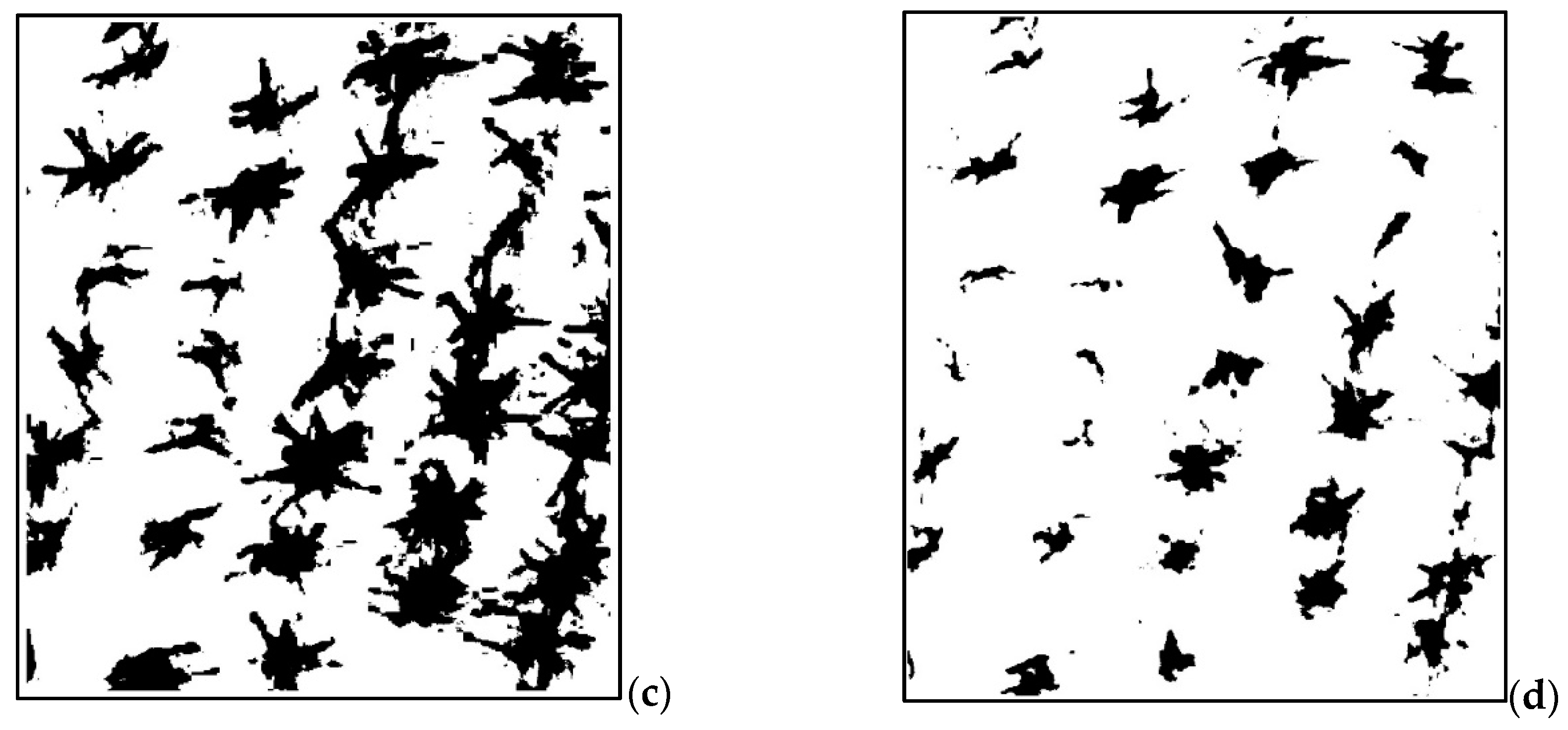
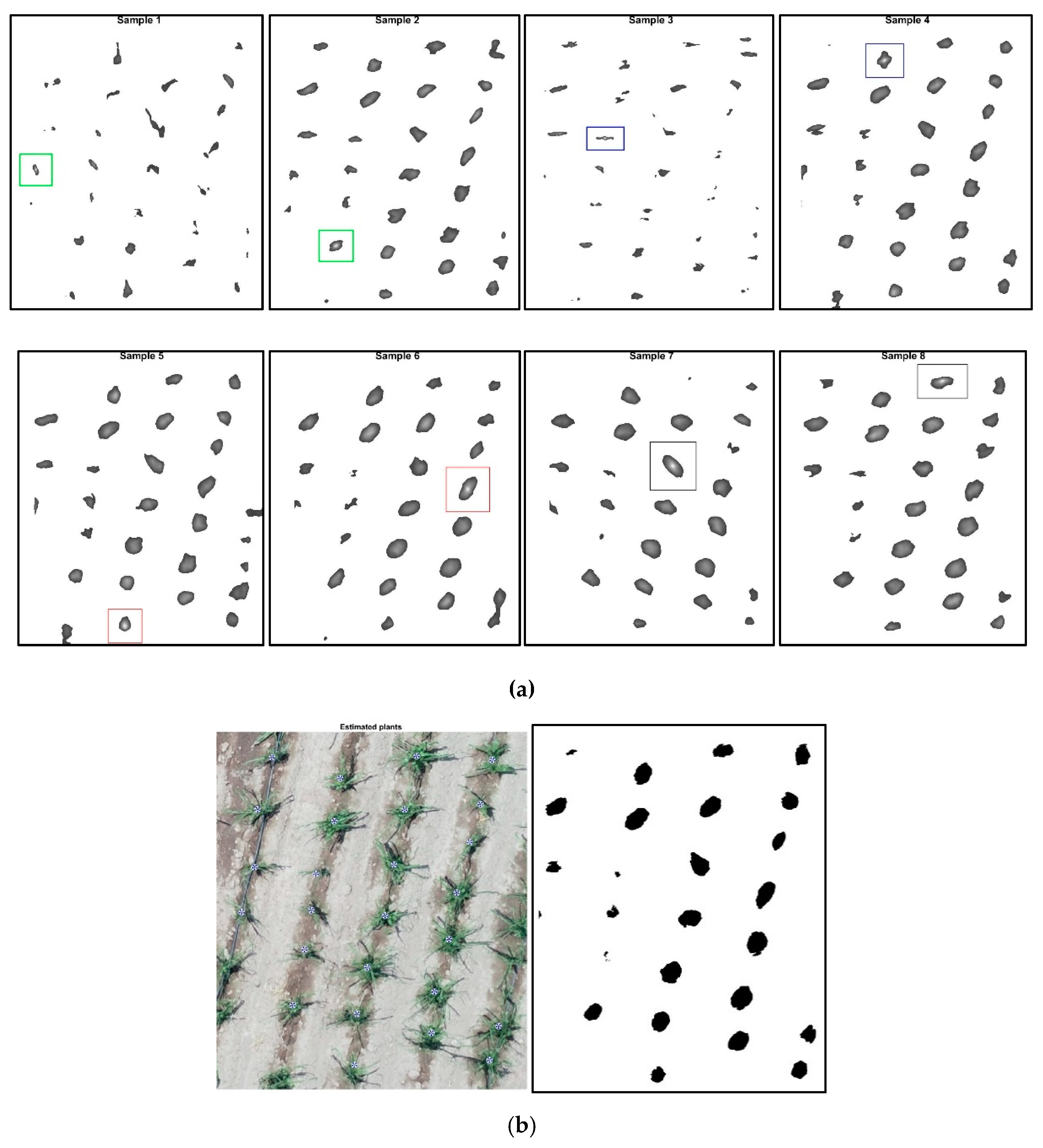
2.6. Description of the Analysis Process and Plant Counting
2.7. Data Analysis
3. Results and Discussion
3.1. Normalized Cross Correlation in Counting Corn Plants
3.2. Precision in Plant Counting
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lunven, P. El Maiz en la Nutrición Humana; Organización de las Naciones Unidas para la Agricultura y la Alimentación: Roma, Italy, 1993. [Google Scholar]
- Assefa, Y.; Prasad, P.; Carter, P.; Hinds, M.; Bhalla, G.; Schon, R.; Jeschke, M.; Paszkiewicz, S.; Ciampitti, I.A. Yield responses to planting density for us modern corn hybrids: A synthesis-analysis. Crop. Sci. 2016, 56, 2802–2817. [Google Scholar] [CrossRef]
- Tollenaar, M. Is low plant density a stress in maize? Low Plant Density Stress Maize 1992, 37, 305–311. [Google Scholar]
- Ciampitti, I.A.; Camberato, J.J.; Murrell, S.T.; Vyn, T.J. Maize nutrient accumulation and partitioning in response to plant density and nitrogen Rate: I. macronutrients. Agron J. 2013, 105, 783–795. [Google Scholar] [CrossRef]
- Yang, G.; Liu, J.; Zhao, C.; Li, Z.; Huang, Y.; Yu, H.; Xu, B.; Yang, X.; Zhu, D.; Zhang, X.; et al. Unmanned Aerial Vehicle Remote Sensing for Field-Based Crop Phenotyping: Current Status and Perspectives. Front. Plant Sci. 2017, 8, 1111. Available online: https://www.frontiersin.org/articles/10.3389/fpls.2017.01111/full (accessed on 4 February 2020). [CrossRef]
- Torres-Sánchez, J.; Peña, J.M.; de Castro, A.I.; López-Granados, F. Multi-temporal mapping of the vegetation fraction in early-season wheat fields using images from UAV. Comput. Electron. Agric. 2014, 103, 104–113. [Google Scholar] [CrossRef]
- Zhang, C.; Kovacs, J.M. The application of small unmanned aerial systems for precision agriculture: A review. Precis Agric. 2012, 13, 693–712. [Google Scholar] [CrossRef]
- Hengl, T. Finding the right pixel size. Comput. Geosci. 2006, 32, 1283–1298. [Google Scholar] [CrossRef]
- Nex, F.; Remondino, F. UAV for 3D mapping applications: A review. Appl. Geomat. 2014, 6, 1–15. [Google Scholar] [CrossRef]
- Torres-Sánchez, J.; López Granados, F.; Castro AI de Peña Barragán, J.M. Configuration and specifications of an unmanned aerial vehicle (UAV) for early site specific weed management. PLoS ONE 2013. [Google Scholar] [CrossRef]
- Peña, J.M.; Torres-Sánchez, J.; Serrano-Pérez, A.; De Castro, A.I.; López-Granados, F. Quantifying efficacy and limits of Unmanned Aerial Vehicle (UAV) technology for weed seedling detection as affected by sensor resolution. Sensors 2015, 15, 5609–5626. [Google Scholar] [CrossRef]
- Koh, J.C.O.; Hayden, M.; Daetwyler, H.; Kant, S. Estimation of crop plant density at early mixed growth stages using UAV imagery. Plant Methods 2019, 15, 64. [Google Scholar] [CrossRef] [PubMed]
- Colomina, I.; Molina, P. Unmanned aerial systems for photogrammetry and remote sensing: A review. ISPRS J. Photogramm. Remote Sens. 2014, 92, 79–97. [Google Scholar] [CrossRef]
- Yao, H.; Qin, R.; Chen, X. Unmanned aerial vehicle for remote sensing applications—A review. Remote Sens. 2019, 11, 1443. [Google Scholar] [CrossRef]
- Maes, W.H.; Steppe, K. Perspectives for remote sensing with unmanned aerial vehicles in precision agriculture. Trends Plant Sci. 2019, 24, 152–164. [Google Scholar] [CrossRef]
- López-Granados, F.; Torres-Sánchez, J.; De Castro, A.-I.; Serrano-Pérez, A.; Mesas-Carrascosa, F.-J.; Peña, J.-M. Object-based early monitoring of a grass weed in a grass crop using high resolution UAV imagery. Agron. Sustain. Dev. 2016, 36, 67. [Google Scholar] [CrossRef]
- Holman, F.H.; Riche, A.B.; Michalski, A.; Castle, M.; Wooster, M.J.; Hawkesford, M.J. High Throughput field phenotyping of wheat plant height and growth rate in field plot trials using uav based remote sensing. Remote Sens. 2016, 8, 1031. [Google Scholar] [CrossRef]
- Maresma, Á.; Ariza, M.; Martínez, E.; Lloveras, J.; Martínez-Casasnovas, J.A. Analysis of vegetation indices to determine nitrogen application and yield prediction in maize (Zea mays L.) from a standard UAV service. Remote Sens. 2016, 8, 973. [Google Scholar] [CrossRef]
- Madec, S.; Baret, F.; De Solan, B.; Thomas, S.; Dutartre, D.; Jezequel, S.; Hemmerlé, M.; Colombeau, G.; Comar, A. High-throughput phenotyping of plant height: Comparing unmanned aerial vehicles and ground LiDAR estimates. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef]
- Marcial, M.D.J.; Gonzalez-Sanchez, A.; Jimenez-Jimenez, S.I.; Ontiveros-Capurata, R.E.; Ojeda-Bustamante, W. Estimation of vegetation fraction using RGB and multispectral images from UAV. Int. J. Remote Sens. 2019, 40, 420–438. [Google Scholar] [CrossRef]
- Liu, S.; Baret, F.; Andrieu, B.; Burger, P.; Hemmerlé, M. Estimation of Wheat Plant Density at Early Stages Using High Resolution Imagery. Front. Plant Sci. 2017. Available online: https://www.frontiersin.org/articles/10.3389/fpls.2017.00739/full (accessed on 4 February 2020). [CrossRef]
- Gnädinger, F.; Schmidhalter, U. digital counts of maize plants by Unmanned Aerial Vehicles (UAVs). Remote Sens. 2017, 9, 544. [Google Scholar] [CrossRef]
- Zhao, B.; Zhang, J.; Yang, C.; Zhou, G.; Ding, Y.; Shi, Y.; Zhang, N.; Xie, J.; Liao, Q. Rapeseed seedling stand counting and seeding performance evaluation at two early growth stages based on unmanned aerial vehicle imagery. Front. Plant Sci. 2018, 9, 1362. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Chu, T.; Landivar, J.; Yang, C.; Maeda, M. Monitoring cotton (Gossypium hirsutum L.) germination using ultrahigh-resolution UAS images. Precis. Agric. 2017, 19, 1–17. [Google Scholar] [CrossRef]
- Sankaran, S.; Quirós, J.J.; Knowles, N.R.; Knowles, L.O. High-resolution aerial imaging based estimation of crop emergence in potatoes. Am. J. Potato Res. 2017, 94, 658–663. [Google Scholar] [CrossRef]
- Jin, X.; Liu, S.; Baret, F.; Hemerlé, M.; Comar, A. Estimates of plant density of wheat crops at emergence from very low altitude UAV imagery. Remote Sens. Environ. 2017, 198, 105–114. [Google Scholar] [CrossRef]
- Zhang, J.; Basso, B.; Richard, F.P.; Putman, G.; Shuai, G. Estimating Plant Distance in Maize Using Unmanned Aerial Vehicle (UAV). Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5909920/ (accessed on 16 March 2020).
- Shuai, G.; Martinez-Feria, R.A.; Zhang, J.; Li, S.; Price, R.; Basso, B. Capturing maize stand heterogeneity across yield-stability zones using Unmanned Aerial Vehicles (UAV). Sensors 2019, 19, 4446. [Google Scholar] [CrossRef] [PubMed]
- Kitano, B.T.; Mendes, C.C.T.; Geus, A.R.; Oliveira, H.C.; Souza, J.R. Corn plant counting using deep learning and UAV images. IEEE Geosci. Remote Sens. Lett. 2019, 1–5. [Google Scholar] [CrossRef]
- Ribera, J.; Chen, Y.; Boomsma, C.; Delp, E.J. Counting plants using deep learning. In Proceedings of the 2017 IEEE Global Conference on Signal and Information Processing (GlobalSIP), Montreal, QC, Canada, 14–16 November 2017. [Google Scholar] [CrossRef]
- Wang, C.; Guo, X.; Zhao, C. Detection of corn plant population and row spacing using computer vision. In Proceedings of the 2011 Second International Conference on Digital Manufacturing Automation, Zhangjiajie, China, 5–7 August 2011; pp. 405–408. [Google Scholar] [CrossRef]
- Gracia-Romero, A.; Kefauver, S.; Vergara-Díaz, O.; Zaman-Allah, M.; Prasanna, B.M.; Cairns, J.E.; Araus, J.L. Comparative performance of ground vs. aerially assessed RGB and multispectral indices for early-growth evaluation of maize performance under phosphorus fertilization. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef]
- Guerrero, J.M.; Pajares, G.; Montalvo, M.; Romeo, J.; Guijarro, M. Support Vector Machines for crop/weeds identification in maize fields. Expert Syst. Appl. 2012, 39, 11149–11155. [Google Scholar] [CrossRef]
- Guijarro, M.; Pajares, G.; Riomoros, I.; Herrera, P.J.; Burgos-Artizzu, X.P.; Ribeiro, A. Automatic segmentation of relevant textures in agricultural images. Comput. Electron. Agric. 2011, 75, 75–83. [Google Scholar] [CrossRef]
- Zaman-Allah, M.; Vergara, O.; Araus, J.L.; Tarekegne, A.; Magorokosho, C.; Zarco-Tejada, P.J.; Hornero, A.; Albà, A.H.; Das, B.; Craufurd, P.; et al. Unmanned aerial platform-based multi-spectral imaging for field phenotyping of maize. Plant Methods 2015, 11, 35. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Elazab, A.; Bort, J.; Vergara, O.; Serret, M.D.; Araus, J.L. Low-cost assessment of wheat resistance to yellow rust through conventional RGB images. Comput. Electron. Agric. 2015, 116, 20–29. [Google Scholar] [CrossRef]
- Diaz, O.V.; Zaman-Allah, M.; Masuka, B.; Hornero, A.; Zarco-Tejada, P.; Prasanna, B.M.; Cairns, J.E.; Araus, J.L. A novel remote sensing approach for prediction of maize yield under different conditions of nitrogen fertilization. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef]
- Yousfi, S.; Kellas, N.; Saidi, L.; Benlakehal, Z.; Chaou, L.; Siad, D.; Herda, F.; Karrou, M.; Vergara, O.; Gracia, A.; et al. Comparative performance of remote sensing methods in assessing wheat performance under Mediterranean conditions. Agric. Water Manag. 2016, 164, 137–147. [Google Scholar] [CrossRef]
- Robertson, A.R. The CIE 1976 color-difference formulae. Color Res. Appl. 1977, 2, 7–11. [Google Scholar] [CrossRef]
- Mendoza, F.; Dejmek, P.; Aguilera, J.M. Calibrated color measurements of agricultural foods using image analysis. Postharvest Biol. Technol. 2006, 41, 285–295. [Google Scholar] [CrossRef]
- Macedo-Cruz, A.; Pajares, G.; Santos, M.; Villegas-Romero, I. Digital image sensor-based assessment of the status of oat (Avena sativa L.) crops after frost damage. Sensors 2011, 11, 6015–6036. [Google Scholar] [CrossRef]
- Cheng, G.; Han, J. A survey on object detection in optical remote sensing images. ISPRS J. Photogramm. Remote Sens. 2016, 117, 11–28. [Google Scholar] [CrossRef]
- Tiede, D.; Krafft, P.; Füreder, P.; Lang, S. Stratified template matching to support refugee camp analysis in OBIA workflows. Remote Sens. 2017, 9, 326. [Google Scholar] [CrossRef]
- Nuijten, R.J.G.; Kooistra, L.; De Deyn, G.B. Using Unmanned Aerial Systems (UAS) and Object-Based Image Analysis (OBIA) for measuring plant-soil feedback effects on crop productivity. Drones 2019, 3, 54. [Google Scholar] [CrossRef]
- Ahuja, K.K.; Tuli, P. Object recognition by template matching using correlations and phase angle method. Int. J. Adv. Res. Comput. Commun. Eng. 2013, 2, 3. [Google Scholar]
- Kalantar, B.; Mansor, S.B.; Shafri, H.Z.M.; Halin, A.A. Integration of template matching and object-based image analysis for semi-automatic oil palm tree counting in UAV images. In Proceedings of the 37th Asian Conference on Remote Sensing, Colombo, Sri Lanka, 17–21 October 2016. [Google Scholar]
- Schanda, J. Colorimetry: Understanding the CIE System; John Wiley & Sons: Hoboken, NJ, USA, 2007. [Google Scholar]
- Recky, M.; Leberl, F. Windows Detection Using K-means in CIE-Lab Color Space. In Proceedings of the 20th International Conference on Pattern Recognition, Istanbul, Turkey, 23–26 August 2010; pp. 356–359. [Google Scholar] [CrossRef]
- Van Der Meer, F.D.; De Jong, S.M.; Bakker, W. Imaging Spectrometry: Basic Analytical Techniques. In Imaging Spectrometry: Basic Principles and Prospective Applications; Remote Sensing and Digital Image Processing; Springer: Dordrecht, The Netherlands, 2001; pp. 17–61. [Google Scholar] [CrossRef]
- Lewis, J.P. Fast Template Matching. In Proceedings of the Vision Interface 95, Quebec City, QC, Canada, 15–19 May 1995; pp. 120–123. [Google Scholar]
- Lindoso Muñoz, A. Contribución al Reconocimiento de Huellas Dactilares Mediante Técnicas de Correlación y Arquitecturas Hardware Para el Aumento de Prestaciones. February 2009. Available online: https://e-archivo.uc3m.es/handle/10016/5571 (accessed on 4 February 2020).
- Peña, J.M.; Torres-Sánchez, J.; de Castro, A.I.; Kelly, M.; López-Granados, F. Weed mapping in early-season maize fields using object-based analysis of Unmanned Aerial Vehicle (UAV) images. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Gée, C.H.; Bossu, J.; Jones, G.; Truchetet, F. Crop/weed discrimination in perspective agronomic images. Comput. Electron. Agric. 2008, 60, 49–59. [Google Scholar] [CrossRef]
- Swain, K.C.; Nørremark, M.; Jørgensen, R.N.; Midtiby, H.S.; Green, O. Weed identification using an automated active shape matching (AASM) technique. Biosyst. Eng. 2011, 110, 450–457. [Google Scholar] [CrossRef]



| DAS | Date | Development Stages | Sensor | N° of Images | Area (m2) | Pixel Size |
|---|---|---|---|---|---|---|
| 23 | April 27 | V2 | Sequoia_4.9_4608 × 3456 (RGB) | 41 | 3545 | 0.56 |
| 44 | May 18 | V5 | Sequoia_4.9_4608 × 3456 (RGB) | 157 | 6309 | 0.53 |
| 44 | May 18 | V5 | DJI FC6310_8.8_5472 × 3648 (RGB) | 272 | 10,067 | 0.49 |
| 44 | May 18 | V5 | CanonPowerShotS100_5.2_4000 × 3000 (RGB) | 120 | 6977 | 0.88 |
| 67 | June 8 | V9 | CanonPowerShotS100_5.2_4000 × 3000 (RGB) | 80 | 13,543 | 1.05 |
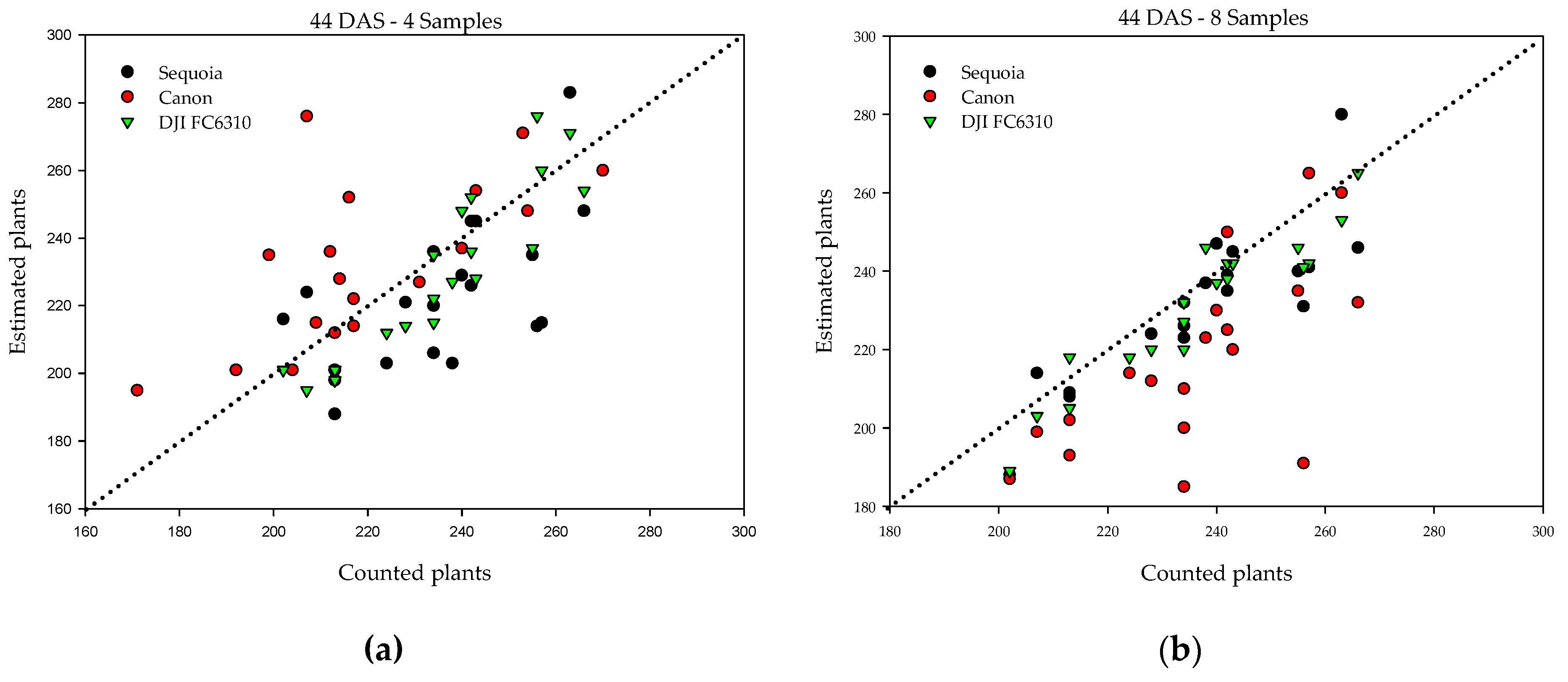
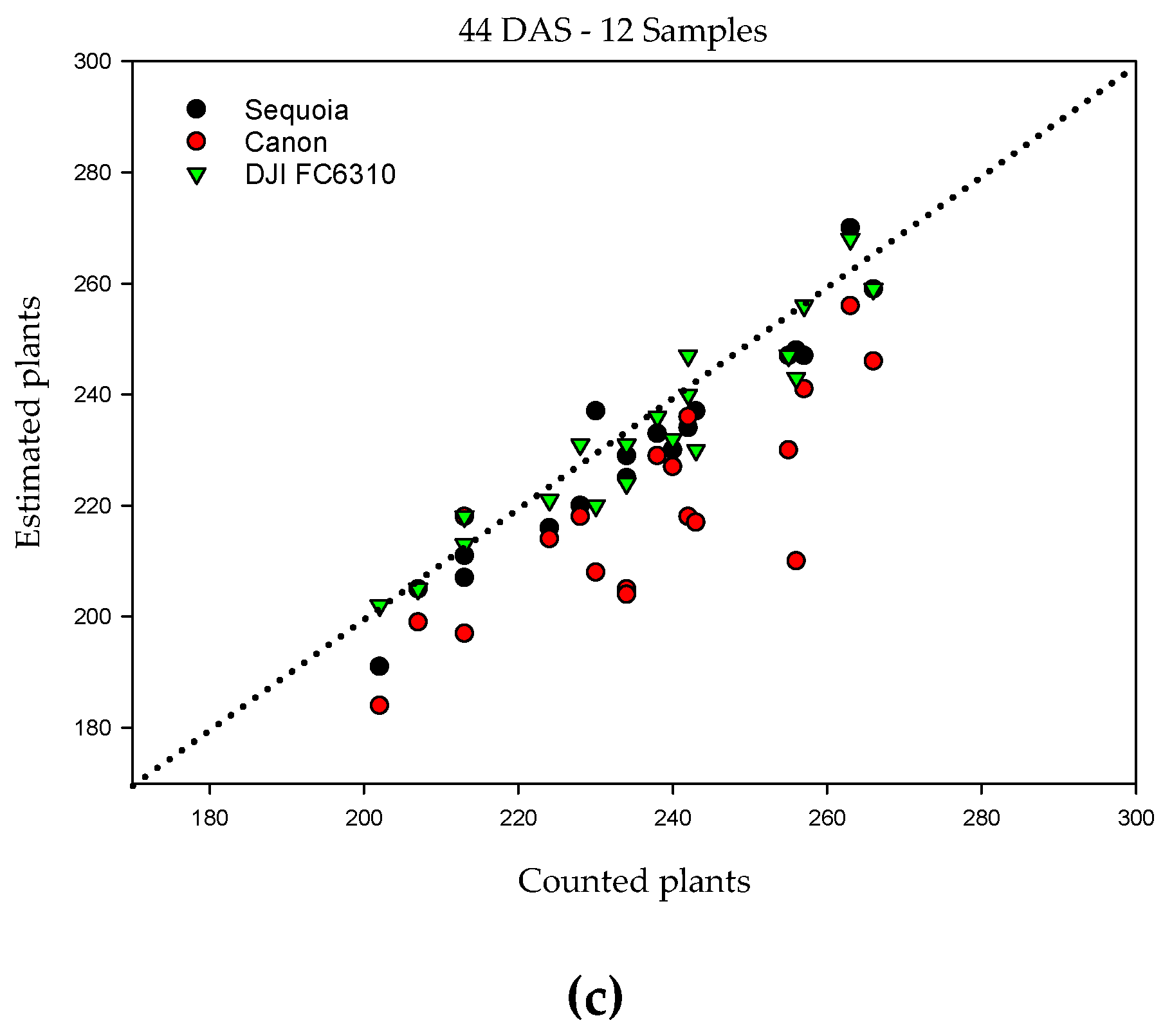
| Samples | Pixel Size (cm) | Ps (%) | RMSE | r | r2 | MAE (%) |
|---|---|---|---|---|---|---|
| 4 | 0.53 | 95 | 21.8 | 0.59 | 0.35 | 7.7 |
| 0.88 | 93 | 22.9 | 0.74 | 0.54 | 8.2 | |
| 0.49 | 97 | 12.2 | 0.91 | 0.82 | 4.7 | |
| 8 | 0.53 | 98 | 11.5 | 0.86 | 0.74 | 3.9 |
| 0.88 | 92 | 25.5 | 0.68 | 0.46 | 8.6 | |
| 0.49 | 98 | 8.4 | 0.94 | 0.89 | 3.0 | |
| 12 | 0.53 | 98 | 7.5 | 0.97 | 0.93 | 3.0 |
| 0.88 | 93 | 20.6 | 0.81 | 0.65 | 7.5 | |
| 0.49 | 99 | 6.6 | 0.95 | 0.90 | 2.2 |
| DAS | Sensor | Samples | Ps(%) | RMSE | r | r2 | MAE (%) |
|---|---|---|---|---|---|---|---|
| 23 | Sequoia_4.9_4608 × 3456 (RGB) | 4 | 91 | 25.87 | 0.99 | 0.98 | 11.0 |
| 8 | 93 | 19.17 | 0.82 | 0.67 | 7.2 | ||
| 12 | 97 | 13.17 | 0.88 | 0.77 | 4.7 | ||
| 44 | DJI FC6310_8.8_5472 × 3648 (RGB) | 4 | 95 1 | 12.24 | 0.91 | 0.82 | 4.7 |
| 8 | 98 | 8.41 | 0.94 | 0.89 | 3.0 | ||
| 12 | 98 | 6.61 | 0.95 | 0.90 | 2.2 | ||
| 67 | CanonPowerShotS100_5.2_4000 × 3000 (RGB) | 4 | 87 | 43.05 | 0.08 | 0.01 | 14.2 |
| 8 | 74 | 93.15 | 0.23 | 0.05 | 25.7 | ||
| 12 | 83 | 46.62 | 0.40 | 0.16 | 17.6 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
García-Martínez, H.; Flores-Magdaleno, H.; Khalil-Gardezi, A.; Ascencio-Hernández, R.; Tijerina-Chávez, L.; Vázquez-Peña, M.A.; Mancilla-Villa, O.R. Digital Count of Corn Plants Using Images Taken by Unmanned Aerial Vehicles and Cross Correlation of Templates. Agronomy 2020, 10, 469. https://doi.org/10.3390/agronomy10040469
García-Martínez H, Flores-Magdaleno H, Khalil-Gardezi A, Ascencio-Hernández R, Tijerina-Chávez L, Vázquez-Peña MA, Mancilla-Villa OR. Digital Count of Corn Plants Using Images Taken by Unmanned Aerial Vehicles and Cross Correlation of Templates. Agronomy. 2020; 10(4):469. https://doi.org/10.3390/agronomy10040469
Chicago/Turabian StyleGarcía-Martínez, Héctor, Héctor Flores-Magdaleno, Abdul Khalil-Gardezi, Roberto Ascencio-Hernández, Leonardo Tijerina-Chávez, Mario A. Vázquez-Peña, and Oscar R. Mancilla-Villa. 2020. "Digital Count of Corn Plants Using Images Taken by Unmanned Aerial Vehicles and Cross Correlation of Templates" Agronomy 10, no. 4: 469. https://doi.org/10.3390/agronomy10040469
APA StyleGarcía-Martínez, H., Flores-Magdaleno, H., Khalil-Gardezi, A., Ascencio-Hernández, R., Tijerina-Chávez, L., Vázquez-Peña, M. A., & Mancilla-Villa, O. R. (2020). Digital Count of Corn Plants Using Images Taken by Unmanned Aerial Vehicles and Cross Correlation of Templates. Agronomy, 10(4), 469. https://doi.org/10.3390/agronomy10040469





