Abstract
Domesticated crops suffer from major genetic bottlenecks while wild relatives retain higher genomic diversity. Wild soybean (Glycine soja Sieb. & Zucc.) is the presumed ancestor of cultivated soybean (Glycine max [L.] Merr.), and is an important genetic resource for soybean improvement. Among the East Asian habitats of wild soybean (China, Japan, Korea, and Northeastern Russia), the Korean peninsula is of great importance based on archaeological records, domestication history, and higher diversity of wild soybeans in the region. The collection and conservation of these wild soybean germplasms should be put on high priority. Chung’s Wild Legume Germplasm Collection maintains more than 10,000 legume accessions with an intensive and prioritized wild soybean germplasm collection (>6000 accessions) guided by the international code of conduct for plant germplasm collection and transfer. The center holds a library of unique wild soybean germplasms collected from East Asian wild habitats including the Korean mainland and nearby islands. The collection has revealed interesting and useful morphological, biochemical, and genetic diversity. This resource could be utilized efficiently in ongoing soybean improvement programs across the globe.
1. Introduction
Soybean (Glycine max [L.] Merr.) is an economically important legume worldwide with diverse applications in food, feed, and biofuel industries [1,2]. Increasing per capita food consumption and limited land and water resources, coupled with a growing population and changing dietary habits, are projected to escalate the pressure on global food supplies [3,4]. It is estimated that food consumption is growing faster than production, particularly in Asia, which will lead to grain and soybean deficits. This deficit in food supply must be addressed through a global effort. Additionally, the global climate change, together with the above-mentioned factors, has outpaced the forecasted crop yields, while at the same time having a negative effect on the state of diversity in the farmer’s field [5]. To alleviate negative effects due to climate change on crops and ecosystems and to increase crop yields, the resilience and sustainability of current agricultural systems have become pressing issues [6]. The development of new soybean varieties and cropping systems with the ability to adapt to a changing environment and new socio-economic conditions is crucial for yield enhancement. While the genetic improvement of crops plays an important role, the genetic base of major crops is narrow, due to genetic bottlenecks and human selection [7]. Therefore, it is essential to investigate novel sources of genetic diversity and to expand gene pools by exploiting crop wild relatives (CWRs) [8,9,10]. The search for novel genetic diversity will result in an increased demand for novel breeding material from the world’s gene banks.
The wild (Glycine soja Sieb. & Zucc.) and cultivated (G. max) soybeans are both annual plants belonging to one of the two subgenera of the genus Glycine, and are distinguished from other wild perennial Glycine species. It has been shown that wild soybean can be used in the genetic improvement of cultivated soybean [11].
The taxonomic and phylogenetic knowledge based on the concept of gene pools provides important information for wild x cultivated soybean hybridization. The primary gene pool (GP-1) includes G. max and its wild progenitor, G. soja, resulting in the production of vigorous hybrids, normal meiotic chromosome pairing, and fertile offspring. As far as secondary (GP-2) and tertiary (GP-3) gene pools of G. max are concerned, all the 26 perennial species are included in GP-3, while no Glycine species has been found in GP-2 [11]. Some researchers have previously attempted to hybridize G. max with five GP-3 member species, but their success was limited [12]. Among GP-3 members, only three perennial species (Glycine argyrea, Glycine canescens, and Glycine tomentella) have been successfully hybridized with cultivated soybean. The F1 hybrids were either sterile or could not develop beyond the amphidiploid stage [13]. Further information could be found in the paper by Singh ([14] and references therein).
Wild soybean accessions, the wild relatives of cultivated soybean, are mainly found in East Asia [15]. They harbor abundant and unique gene/allele resources, due to their widespread distributions in complex geographic topographies with diverse microclimates [16]. Cultivated soybeans have lower adaptive potential to climatic changes and reduced genetic diversity when compared to wild soybeans [3,17]. More than half of the genetic variations in soybean were lost due to habitat fragmentation and genetic bottlenecks [18]. Therefore, wild soybeans are important genetic resources for gaining a deeper understanding of the genetic diversity, genomic variations, and environmental adaptations of soybean [10,19].
Preliminary assessments of germplasm conservation in many parts of the world have revealed substantial gaps, which should be filled by expanding the collection of wild soybean germplasm resources from the main distribution areas [20,21]. Such efforts are being adopted by many countries and germplasm centers around the globe, including Korea, which is one of the main distribution areas of wild soybeans.
2. Role of Wild Soybean in Soybean Improvement Programs
While the members of the GP-3 gene pool are of limited use in soybean breeding programs, wild soybean offers a great potential for providing novel genes/alleles. Resequencing and comparative genomics of wild and cultivated soybeans and landraces have identified higher allelic diversity in wild soybean [17,22]. Recently, a high-quality reference genome for wild soybean has been completed to provide an important tool for exploring the gene/allele resources in wild soybean [23]. Many useful QTLs/genes have been identified by characterizing wild soybean or using genetic populations resulting from crosses between wild and cultivated soybeans [10,23,24]. These QTLs/genes are useful in improving biotic and abiotic stress tolerance, as well as enhancing the yield and quality of soybean.
For example, the stacking of QTLs from both wild and cultivated soybeans was applied to increase the resistance against soybean cyst nematode (SCN) [25]. Using a genome-wide association study (GWAS) in combination with RNA-Seq analysis, the genetic architecture and gene regulatory networks of SCN resistance were delineated [26,27]. In response to environmental changes, genes and transcription factors identified from wild soybean have been reported to enhance drought stress tolerance via the regulation of the abscisic acid signaling pathway [28]. In addition, a salt tolerance gene was identified from wild soybean using a combination of whole-genome de novo sequencing, QTL mapping by resequencing-based high-density markers, and functional studies [24].
Wild soybeans are also genetic resources for yield improvement and nutritional enhancement. For instance, the whole-genome resequencing of a recombinant inbred line population has identified a protein phosphatase 2C-1 (PP2C-1) allele from the wild soybean ZYD7. This allele appeared to regulate seed weight and seed size [29]. Furthermore, using the whole-genome shotgun (WGS) approach, 29 SNPs located in 10 different wild soybean chromosomes were found to be associated with seven seed composition traits [30]. Wild soybean is also a valuable genetic resource for the enhancement of isoflavone contents in the soybean seed through conventional breeding and modern biotechnological techniques [31]. Many genes involved in the soysaponin biosynthetic pathway have been identified in wild soybean [32], including the newly discovered Sg-5 gene, which is responsible for soysaponin A biosynthesis [33,34]. A summary of the traits identified and/or characterized in wild soybean by screening or hybridization with cultivated soybean is given in Table 1 [modified from 1,10,14]. While the introgression from G. soja has resulted in many useful traits, it may also result in linkage drag and introduce unwanted traits such as vining, susceptibility to lodging, lack of complete leaf abscission, hard seed coat, pod bursting, and seed shattering. Rapid progress in biotechnology involving genetic transformation and genome editing might be useful for circumventing these obstacles [1].

Table 1.
List of genes and QTLs found to be associated with specific traits using either wild soybean or a cross between wild and cultivated soybeans.
3. Recent Studies on Genome-Wide Patterns of Genetic Diversity in Wild Soybean
Recent developments in high-throughput next-generation sequencing techniques such as whole-genome sequencing and DNA microarrays have enabled large-scale molecular variation studies [18]. These advancements enable soybean researchers to explore the domestication-related traits and to understand how genome-wide genetic variations were shaped by domestication. The first whole-genome sequence of Korean wild soybean (IT182932) was completed by Kim et al. [35], showing that the G. soja/G. max complex might be 0.27 million years old, dating back much earlier than the more recent event of soybean domestication (6000–9000 years ago). Another study used nine cultivated and five wild Korean soybean accessions in a resequencing approach to assemble unmapped reads to contigs [36]. In-depth genomic mining of wild soybean will be expedited by the recent release of a reference-grade wild soybean genome by our group [23]. Following the study by Chung et al. [36], a resequencing of semi-wild soybeans reported a hybridization origin, revealing a complex G. soja population structure and introgression [37]. Li et al. [38] reported the construction and analysis of a pan-genome of wild soybean based on sequencing and de novo genome assembly. Two other notable studies have also employed a whole-genome resequencing approach to identify the patterns of genetic diversity and selection in wild and cultivated soybeans [17,22]. Lam et al. [22] resequenced a total of 14 cultivated and 17 wild soybean genomes with >90% genome coverage, and identified a higher allelic diversity among wild soybean genomes relative to the cultivated ones. Human selection, coupled with genetic bottlenecks, has reduced the genetic diversity of cultivated soybean to nearly half of that of the wild soybean. In another study [17], resequencing was performed in 302 wild and cultivated soybean accessions including landraces. A total of 230 selective sweeps and 162 copy number variants were reported. GWAS revealed associations between 10 selected genomic regions and nine domestication-related traits.
4. Distribution and Conservation of Wild Soybean in Korea
4.1. Geographical Distribution of Wild Soybean in Korea
Wild soybean grows in moist habitats from 0 to 2650 MASL (meters above sea level), spanning subtropical and subarctic zones (between 24° N and 53° N latitude) [16], and is restricted to East Asia. Occasionally, it can also be found in dry and salt-affected areas. In the Korean peninsula, wild soybeans are distributed throughout the mainland as well as on nearby islands. They can be found growing almost everywhere in Korea, including farmlands, roadsides, and river banks, and from mountain tops to deep valley bottoms. There are over 3000 islands off the coast of the Korean peninsula and nearly 400 are inhabited. A few of these inhabited islands have already been surveyed for wild soybean distribution [77].
4.2. Archaeological Records and Background
Despite the importance of soybean to the world economy, the history of soybean has been mostly restricted to phylogenetics and historical documents mentioning the domestication of soybean in East Asia. G. soja is the only wild member of the subgenus Soja native to the East Asian countries mentioned above [15,78]. Many reports suggest that soybean was first domesticated in China around 5000 years ago, but this statement is still controversial [79]. However, it is generally agreed that the ancient geographical region comprising China, Japan, and Korea (the CJK region) harbored one of the three pioneering societies to domesticate and cultivate this important legume [80]. In an attempt to rectify the domestication history of soybean, Lee et al. [81] pointed out that the differences between domesticated plants and their wild relatives/progenitors are not always clear, and therefore it is sometimes impossible to discern between wild and domesticated soybean in archaeological specimens which are not always well preserved. Despite their different appearances and growing conditions, cultivated soybean plants can cross breed with wild soybean plants and produce fertile F1 hybrids, making it quite challenging to understand and interpret the archaeological seed records [11]. There is still an ongoing debate over the archaeological records in southeastern Korea and China ([81] and references therein).
Figure 1 shows the archaeological sites in Korea. The Nam River valley contains multiple sites, e.g., Oun 1, Okbang 1, 2, 4, 6, 9, Sangchon B, and Pyeonggeodong, dating back to the Chulmun (8000~3500 CAL. B.P.), Mumun (~3500-2000 CAL. B.P.), and Three Kingdom Periods (2000~1200 CAL. B.P.). Other sites include Daundong in Ulsan and Dongsamdong shell midden in Busan. A detailed examination and comparison of archaeological seed records showed that the morphology of the seeds in the Korean soybean archaeological records resembles that of smaller domesticated varieties in more modern times [81,82] because there is a range of seed morphologies among soybean cultivars with ovate seeds. However, the report is based on archaeobotanical seed size measurements. A genomic approach could be more rewarding in terms of species identification. Another report [82] did not mention wild soybean in Korean flotation samples at all. Due to a higher number of landraces, as well as the presence of wild soybean in the area, the authors were not able to link any archaeological findings to a particular species (most probably G. max) or a soybean landrace. However, these studies confirm that the Korean peninsula is a region of great importance for early soybean domestication.
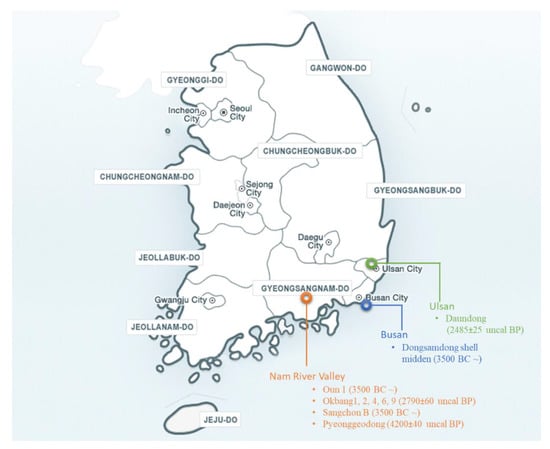
Figure 1.
Map showing the archaeological sites of soybeans in Korea. Bullets under each city show the names of the archaeological sites while the dates in brackets show the carbon dating information on charred soybean seeds. If there is no carbon dating of the soybean samples, then the reported chronological year is given [81,83].
5. Korea: A Region of High Wild Soybean Diversity
Wild soybean propagates by self-pollination with a limited outcrossing potential (2.4–19%) [84]. Short-distance dispersal is attributed to pod dehiscence while long-distance dispersal may occur via migratory birds, water, and mammals [85]. During the interglacial cycle, wild soybean survived in multiple cryptic refugia (at least two refugia have been inferred: one in Northeast Asia including the Korean peninsula, and another in the Yangtze River basin) in its native habitats (the CJK region) [86]. The geographical distribution is characterized by three climatic zones, i.e., subtropical, warm temperate, and cool temperate zones. Such a wide distribution area, with a temperature range of at least 20 °C, requires that wild soybean be able to adapt to a wide range of growing conditions [87]. The Korean peninsula was one of the wild soybean refugia. Owing to its geographical location in the West Pacific, as well as to periodic warm/wet and cold/dry spells during the Quaternary Period, a rich wild soybean diversity is expected to be present in this region [86]. It has been reported that wild soybean is distributed across the entire Korean mainland territory and nearby islands [77].
Archaeological records suggest that wild soybean could have been present in Korea for more than 5000 years (as discussed above in Section 4.2). Many studies based on molecular markers, agronomic characteristics, and isozymes reported that Korean wild soybean is rich in genetic diversity [88,89,90,91,92,93]. Based on the protein Kunitz trypsin inhibitor variations in 172 wild soybeans, it is proposed that Korea should be regarded as a subcenter of soybean diversity [89]. Based on microsatellite markers in the Korean wild soybean, a high genetic diversity index was obtained, supporting previous reports [94,95,96]. A chloroplast microsatellite-based study of 604 wild soybean accessions from 43 locations across China, Japan, and Korea suggested that Korea may be a subcenter of diversity. However, nuclear microsatellite markers revealed that the Yangtze River basin in China had the highest genetic diversity [97].
The wild soybean populations growing on the islands off the Korean peninsula have a much higher risk of extinction, compared to the mainland populations. The risk factors include the accumulation of different mutations, adaptation to the microenvironment of a particular island, loss of genetic diversity, and inbreeding [98,99,100]. In this regard, there is a greater urgency to collect and conserve as many wild soybean germplasms as possible from the islands. Lee et al. [77] studied the genetic diversity of wild soybean accessions collected from 24 inhabited islands and reported that these accessions showed a similar degree of genetic diversity to that among the mainland accessions. It was thus concluded that, regardless of the collection site, i.e., mainland versus island habitats, the genetic diversity in Korean wild soybean is high, meaning that the Korean wild soybean accessions from connected as well as small isolated regions are equally important wild genetic resources, and their conservation programs should be of equally high priority.
6. Collection of Wild Soybean Germplasm in Korea
The second report on the state of the world’s plant genetic resources for food and agriculture stated that public awareness of the importance of crop diversity and the use of CWRs is growing in both developing and developed countries. A 20% increase in the number of global accessions conserved ex situ has been observed since 1996. Major ex situ soybean collections are reported to be present in China, Russia, Ukraine, and the United States of America, with a total of 229,944 accessions. However, 23% of the total collection is held by only two organizations, i.e., the Institute of Crop Germplasm Resources at The Chinese Academy of Agricultural Sciences (ICGR-CAAS) and SOY (USA) [5]. Among these soybean germplasm collections, the majority of the wild accessions are held by organizations in China, USA, Korea, Japan, and Russia (Table 2). According to the second report on plant genetic resources, there has been a significant increase in nationally designated protected areas since 1928. However, the report did not include priority genetic reserve locations for wild soybean relatives. The most likely reason for the omission is that this report based its information on another report by Maxted and Kell which focused only on 12 major food crops, excluding soybean [101]. A recent report on global conservation priorities classified soybean wild relatives as medium priority [21]. This classification is based on the consideration given to the situations of all food crops. Therefore, a classification system based solely on the rankings within legumes or oilseeds could help advocate for a higher priority for conserving wild soybean.

Table 2.
Global soybean germplasm collections by institute.
Wild soybean, being able to hybridize with cultivated soybean, becomes an extended source of diversity to broaden the gene pool [11]. The utilization of wild soybean is expected to increase as a result of ongoing improvements in the information on the species’ genome and genetic diversity, and advances in breeding tools [21]. This expectation is based on the assumption that the wild accessions will be readily available for research and soybean breeding, which requires their conservation as germplasms in gene banks. Urbanization, road construction, deforestation, intensive agriculture, climate change, and soil erosion are all threatening wild soybean in its natural habitats. Since wild soybean is mainly distributed in the CJK region, greater conservation efforts should be made by the local authorities in charge of biological conservation in these countries. In East Asia, major wild soybean germplasm centers in these three countries include the Chinese Crop Germplasm Information System, in China, the National Institute of Agrobiological Sciences Genebank and the Legume Base, in Japan, and the National Agrobiodiversity Center and the Chung’s Wild Legume Germplasm Collection (CWLGC) (described in this work), in Korea. In Korea, there are >6000 accessions in CWLGC and 3229 accessions at the National Agrobiodiversity Center.
Soybean breeding in Korea started in the early 1900s and, since then, more than 178 varieties/cultivars have been registered. Major soybean breeding work has been completed in the past three decades, particularly after the establishment of the National Agrobiodiversity Center in 1987 [102]. Cultivated soybean has received greater attention regarding germplasm collection when compared to wild soybean. This is quite logical because most of the germplasm centers were busy collecting, breeding, and documenting major crops during their early establishment. However, the recent trend of utilizing CWRs in breeding and preliminary assessments of the comprehensiveness of the conservation of CWRs in gene banks have reported substantial gaps [21]. A report by FAO on the state of diversity indicated that the number of accessions of CWRs has substantially increased since 1996. However, they are still under-represented [5]. Considering this scenario, as well as the limited number of wild soybean accessions collected from Korea, large-scale individual as well as collective efforts are needed to enhance wild soybean collections. Although information is available about the conservation status of this species in Korea, lack of information and access to wild soybean in North Korea is a big limitation. Table 3 details the conservation efforts by the CWLGC in Korea, placing its major focus on wild soybean collection.

Table 3.
List of wild legume germplasm accessions at Chung’s Wild Legume Germplasm Collection.
6.1. Chung’s Wild Legume Germplasm Collection
Founded by Professor Gyuhwa Chung in 1983 and located in the Yeosu campus of the Chonnam National University, the CWLGC now possesses the most comprehensive collection of wild soybean in Korea. Guided by the international code of conduct for plant germplasm collecting and transfer (http://www.fao.org/3/x5586e/x5586e0k.htm), the main focus of this center is on the direct collection, acquisition, conservation, evaluation, characterization, documentation, and distribution of wild legume germplasms. CWLGC followed the guidelines of the National Agrobiodiversity Center, Genebank of Rural Development Administration, Korea, and became a local sub-bank in 2007. The list of species and the respective number of accessions available at CWLGC is shown in Table 3. The wild legume accessions were directly collected by the CWLGC team from Korea, Japan, China, and Russia, or acquired from Australia, India, Taiwan, and Zambia. The wild soybean collections include 5050 accessions originating from Korea.
6.2. In situ Conservation of G. soja at CWLGC
In the last 30 years, a major effort at CWLGC has been dedicated to wild soybean germplasm collection, acquisition, development, and characterization. In situ wild soybean germplasm propagation and characterization at CWLGC is done throughout the year and is ongoing. About 500-1000 accessions/year are propagated and characterized at two outdoor planting and propagation sites: Yeosu (Chonnam Province) and Jinju (Gyeongsangnam Province) (Figure 2).

Figure 2.
Wild soybean germplasm. (a) Wild soybean germplasm distribution and collection. (b) Different stages of in situ propagation and characterization at CWLGC.
One important feature of CWLGC is that it holds wild soybean accessions collected from ~130 islands along the coast of the Korean peninsula. These accessions could be used to address questions related to the adaptations to microclimatic conditions in geographically isolated areas. With increasing land exploitations by humans and decreasing wild soybean habitats in Korea, CWLGC is focusing on extensive surveys and germplasm collections. The germplasm characterization performed at CWLGC has revealed interesting and useful morphological and biochemical variations. Examples of the morphological diversity in leaf shape and root architecture are shown in Figure 3.
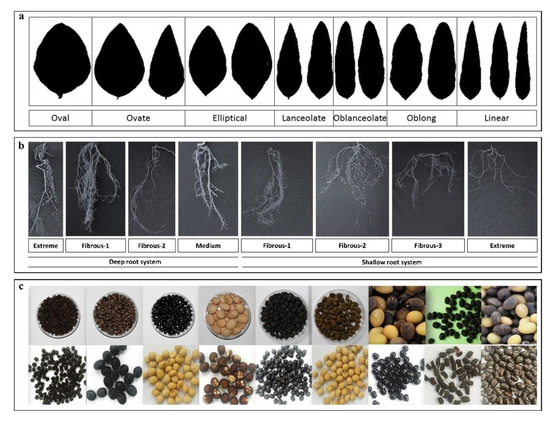
Figure 3.
Morphological diversity of soybeans in the CWLGC collection. (a) Leaf shape variations in G. soja accessions. (b) Root structure variations in G. soja accessions. (c) Samples of the legume seed collection at CWLGC.
The germplasm collection at CWLGC exhibits high genetic and morphological variations [77,81]. For instance, microsatellite marker-based genetic diversity has been investigated in the wild soybean at CWLGC that originated from the southern islands of Korea. Five hundred and thirty wild soybean accessions originating from China, Japan, and Korea included in the CWLGC have also been utilized for the successful identification of GmSALT3 haplotypes and the development of molecular markers related to salt tolerance [103].
Moreover, the CWLGC germplasms were studied for their natural variations in saponin contents and were used to help identify different group-A acetylsaponin-deficient mutants [32,104,105,106]. The wild soybean soyasaponin mutants from CWLGC have been used to identify and characterize the genes involved in the saponin biosynthesis pathway, e.g., Sg-1 locus (CWS2133), sg-5 locus (CWS5095), and Sg-6 locus [33,105,107,108,109]. In addition to saponins, wild soybean accessions at the CWLGC have also been screened for the presence of Kunitz trypsin inhibitor polymorphism, alpha-linolenic acid concentration, and soyisoflavone profile diversity [110,111,112,113]. Furthermore, the germplasms have been used to functionally characterize two seed-specific flavonoid glycosyltransferases and a β-amyrin synthase gene [109,114].
Another interesting discovery using the CWLGC collection is the identification of the W1 locus and the analysis of four new alleles at this locus associated with flower color in soybean [40]. A complete range of flower color has been observed in the CWLGC wild soybean collection and different flower color variants have been studied to help understand the genetic and molecular basis of flower color e.g., the pinkish-white flowers of accession CW13133 [39] (Figure 4).
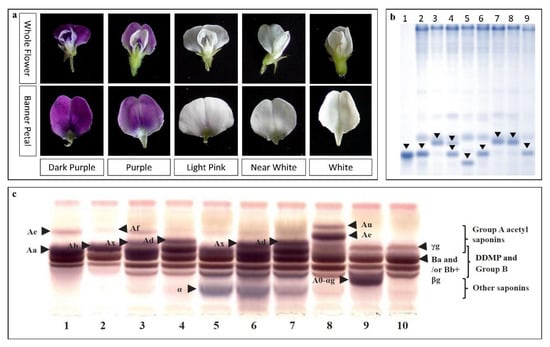
Figure 4.
Demonstrations of the diversity in the CWLGC. (a) Flower color variations [40]. (b) Using Kunitz trypsin inhibitor, different KTi forms were detected in Korean G. soja accessions by non-denaturing PAGE. Lanes 1–9 are: Tia protein standard (Sigma), Tia, Tib, Tia/Tib, Tibi7-1, Tia, Tibi5, Tib, and Tia, respectively [110]. (c) Comparison of saponin composition phenotypes in seed hypocotyls in the Korean G. soja collection by thin-layer chromatography (TLC). Lanes 1–7 are the common phenotypes: Aa, Ab, AaBc, AbBc, Aa+a, AaBc+a, and AbBc+a types, respectively. Lanes 8, 9, and 10 are mutant phenotypes: AuAeBc (CWS0115), A0Bc+ag (CWS2133), and A0Bc-S (CWS5095) types, respectively [32].
Being a major source of plant oils and proteins, the demand for soybean is escalating. Soybean yield has been significantly increased since the 1960s and is forecasted to reach 3.0 metric tonnes per hectare in 2028. It was observed that soybean yields in East Asian countries have been stagnant for some time. To meet the projected demands, it is important to consider breeding soybean cultivars with higher yield potentials and better tolerance to biotic and abiotic stresses. Wild soybean has been shown to provide an important resource for soybean breeding in terms of nutrition, biotic and abiotic stress tolerance, and yield-related traits (Table 1).
So far, only a few characteristics of wild soybean contained in the CWLGC have been explored. Phenotypic screening towards biotic and abiotic resistance under diverse climate conditions would reveal more relevant results to meet current needs. In this regard, the CWLGC G. soja collection has been initially explored for the diversity in root system architecture (Figure 3b). Regarding biotic stress tolerance, no screening has yet been done, leaving the opportunity open for researchers interested in disease and insect pest resistance. Soybean cyst nematode, soybean mosaic virus, bacterial blight and wilt, and a whole list of fungal diseases could be the initial screening targets. Furthermore, early emergence and vigor traits in wild soybean have not yet been examined. Wild soybean seeds can survive in extreme climatic conditions in their native habitats, making them a good target for studying dormancy as well as longevity traits. These traits are important in terms of soybean improvement as well as ex situ germplasm conservation. By analyzing the morphological variations in leaf shape in the CWLGC G. soja accessions, we can potentially unveil the process of domestication of this trait and enhance our understanding of the regulation of photosynthesis. A comparative and in-depth genome resequencing of G. soja chloroplast genomes could enhance our understanding of the domestication process of G. max, as well as of the traits regulated by the plastid genome. Up until now, only limited knowledge of these aspects is available [115,116].
7. Conclusions
It is now well understood that during the process of domestication of soybean, many genes/alleles related to abiotic and biotic stress tolerance and agronomic traits may have been lost due to bottleneck and human selection. The narrow genetic background of cultivated soybean can be expanded by exploring wild soybean germplasms. The wild soybean present on the Korean peninsula is a valuable resource and has high genetic diversity. CWLGC maintains a comprehensive wild soybean collection and strives to preserve its valuable genetic diversity. Through this review, we introduce a genetic resource of wild soybean that could be beneficial to soybean researchers worldwide.
Author Contributions
G.C. led the collection, maintenance, and characterization of the germplasm bank. G.C. and H.M.L. coordinated the preparation and writing of this manuscript. All authors actively participated in conducting literature review, collecting relevant information, writing, preparing tables and figures, and proofreading. All authors have read and agreed to the published version of the manuscript.
Funding
The work is supported by the Cooperative Research Program for Agriculture Science and Technology Development (project no. PJ012942), Rural Development Administration, Korea, to G.C., and the Hong Kong RGC AoE Scheme (AoE/M-403/16), to H.M.L. G.C. thanks all graduate and post-graduate students, collaborators, and friends who participated in germplasm collection. Jeeyan Chu copy-edited this manuscript.
Conflicts of Interest
No conflicts of interest are to be declared.
Availability of Germplasm
The germplasm and genetic resource described in this review is currently available to the international soybean breeding community under the jurisdiction of international as well as Korean laws. Germplasm requests and plans for collaborations should be sent to G.C. (chung@chonnam.ac.kr).
References
- Kofsky, J.; Zhang, H.; Song, B.-H. The untapped genetic reservoir: the past, current, and future applications of the wild soybean (Glycine soja). Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef]
- Nawaz, M.A.; Rehman, H.M.; Imtiaz, M.; Baloch, F.S.; Lee, J.D.; Yang, S.H.; Lee, S.I.; Chung, G. Systems Identification and Characterization of Cell Wall Reassembly and Degradation Related Genes in Glycine max (L.) Merill, a Bioenergy Legume. Sci. Rep. 2017, 7, 10862. [Google Scholar] [CrossRef]
- Lam, H.-M.; Remais, J.; Fung, M.-C.; Xu, L.; Sun, S.S.-M. Food supply and food safety issues in China. The Lancet 2013, 381, 2044–2053. [Google Scholar] [CrossRef]
- Considine, M.J.; Siddique, K.H.; Foyer, C.H. Nature’s pulse power: Legumes, food security and climate change. J. Exp. Bot. 2017, 68, 1815–1818. [Google Scholar] [CrossRef] [PubMed]
- Allender, C. The Second Report on the State of the World’s Plant. Genetic Resources for Food and Agriculture; FAO Commission on Genetic Resources for Food and Agriculture: Rome, Italy, 2010.
- Lin, B.B. Resilience in agriculture through crop diversification: adaptive management for environmental change. BioScience 2011, 61, 183–193. [Google Scholar] [CrossRef]
- Hyten, D.L.; Song, Q.; Zhu, Y.; Choi, I.-Y.; Nelson, R.L.; Costa, J.M.; Specht, J.E.; Shoemaker, R.C.; Cregan, P.B. Impacts of genetic bottlenecks on soybean genome diversity. Proc. Natl. Acad. Sci. USA 2006, 103, 16666–16671. [Google Scholar] [CrossRef]
- Cowling, W.; Li, L.; Siddique, K.; Henryon, M.; Berg, P.; Banks, R.; Kinghorn, B. Evolving gene banks: Improving diverse populations of crop and exotic germplasm with optimal contribution selection. J. Exp. Bot. 2016, 68, 1927–1939. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Lam, H.-M.; Nguyen, H.T.; Siddique, K.H.; Varshney, R.K.; Colmer, T.D.; Cowling, W.; Bramley, H.; Mori, T.A.; Hodgson, J.M. Neglecting legumes has compromised human health and sustainable food production. Nat. Plant. 2016, 2, 16112. [Google Scholar] [CrossRef]
- Muñoz, N.; Liu, A.; Kan, L.; Li, M.-W.; Lam, H.-M. Potential uses of wild germplasms of grain legumes for crop improvement. Int. J. Mol. Sci. 2017, 18, 328. [Google Scholar] [CrossRef]
- Chung, G.; Singh, R.J. Broadening the genetic base of soybean: A multidisciplinary approach. Crit. Rev. Plant Sci. 2008, 27, 295–341. [Google Scholar] [CrossRef]
- Ladizinsky, G.; Newell, C.; Hymowitz, T. Wide crosses in soybeans: Prospects and limitations. Euphytica 1979, 28, 421–423. [Google Scholar] [CrossRef]
- Singh, R.; Nelson, R.L. Methodology for creating alloplasmic soybean lines by using Glycine tomentella as a maternal parent. Plant Breed. 2014, 133, 624–631. [Google Scholar] [CrossRef]
- Singh, R.J. Cytogenetics and genetic introgression from wild relatives in soybean. Nucleus 2019, 62, 3–14. [Google Scholar] [CrossRef]
- Singh, R.; Hymowitz, T. The genomic relationship between Glycine max (L.) Merr. and G. soja Sieb. and Zucc. as revealed by pachytene chromosome analysis. Theor. Appl. Genet. 1988, 76, 705–711. [Google Scholar] [CrossRef]
- He, S.-L.; Wang, Y.-S.; Li, D.-Z.; Yi, T.-S. Environmental and historical determinants of patterns of genetic differentiation in wild soybean (Glycine soja Sieb. et Zucc). Sci. Rep. 2016, 6, 22795. [Google Scholar] [CrossRef]
- Zhou, Z.; Jiang, Y.; Wang, Z.; Gou, Z.; Lyu, J.; Li, W.; Yu, Y.; Shu, L.; Zhao, Y.; Ma, Y. Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat. Biotechnol. 2015, 33, 408–414. [Google Scholar] [CrossRef]
- Wang, L.-x.; Lin, F.-y.; LI, L.-h.; Wei, L.; Zhe, Y.; LUAN, W.-j.; PIAO, R.-h.; Yuan, G.; NING, X.-c.; Li, Z. Genetic diversity center of cultivated soybean (Glycine max) in China–New insight and evidence for the diversity center of Chinese cultivated soybean. J. Integr. Agric. 2016, 15, 2481–2487. [Google Scholar] [CrossRef]
- Leamy, L.J.; Lee, C.R.; Song, Q.; Mujacic, I.; Luo, Y.; Chen, C.Y.; Li, C.; Kjemtrup, S.; Song, B.H. Environmental versus geographical effects on genomic variation in wild soybean (Glycine soja) across its native range in northeast Asia. Ecol. Evolut. 2016, 6, 6332–6344. [Google Scholar] [CrossRef]
- Hay, F.R.; Probert, R.J. Advances in seed conservation of wild plant species: A review of recent research. Conserv. Physiol. 2013, 1. [Google Scholar] [CrossRef]
- Castañeda-Álvarez, N.P.; Khoury, C.K.; Achicanoy, H.A.; Bernau, V.; Dempewolf, H.; Eastwood, R.J.; Guarino, L.; Harker, R.H.; Jarvis, A.; Struik, P.C.; et al. Global conservation priorities for crop wild relatives. Nat. Plant. 2016, 2, 16022. [Google Scholar] [CrossRef]
- Lam, H.-M.; Xu, X.; Liu, X.; Chen, W.; Yang, G.; Wong, F.-L.; Li, M.-W.; He, W.; Qin, N.; Wang, B. Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat. Genet. 2010, 42, 1053–1059. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Chung, C.Y.-L.; Li, M.-W.; Wong, F.-L.; Wang, X.; Liu, A.; Wang, Z.; Leung, A.K.-Y.; Wong, T.-H.; Tong, S.-W. A reference-grade wild soybean genome. Nat. Commun. 2019, 10, 1216. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Li, M.-W.; Xie, M.; Liu, X.; Ni, M.; Shao, G.; Song, C.; Yim, A.K.-Y.; Tao, Y.; Wong, F.-L. Identification of a novel salt tolerance gene in wild soybean by whole-genome sequencing. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef]
- Kim, M.; Hyten, D.L.; Niblack, T.L.; Diers, B.W. Stacking resistance alleles from wild and domestic soybean sources improves soybean cyst nematode resistance. Crop Sci. 2011, 51, 934–943. [Google Scholar] [CrossRef]
- Guo, B.; Sleper, D.A.; Nguyen, H.T.; Arelli, P.R.; Shannon, J.G. Quantitative trait loci underlying resistance to three soybean cyst nematode populations in soybean PI 404198A. Crop Sci. 2006, 46, 224–233. [Google Scholar] [CrossRef]
- Zhang, J.; Wen, Z.; Li, W.; Zhang, Y.; Zhang, L.; Dai, H.; Wang, D.; Xu, R. Genome-wide association study for soybean cyst nematode resistance in Chinese elite soybean cultivars. Mol. Breed. 2017, 37, 60. [Google Scholar] [CrossRef]
- Luo, X.; Bai, X.; Sun, X.; Zhu, D.; Liu, B.; Ji, W.; Cai, H.; Cao, L.; Wu, J.; Hu, M. Expression of wild soybean WRKY20 in Arabidopsis enhances drought tolerance and regulates ABA signalling. J. Exp. Bot. 2013, 64, 2155–2169. [Google Scholar] [CrossRef]
- Lu, X.; Xiong, Q.; Cheng, T.; Li, Q.-T.; Liu, X.-L.; Bi, Y.-D.; Li, W.; Zhang, W.-K.; Ma, B.; Lai, Y.-C. A PP2C-1 allele underlying a quantitative trait locus enhances soybean 100-seed weight. Mol. Plant 2017, 10, 670–684. [Google Scholar] [CrossRef]
- Leamy, L.J.; Zhang, H.; Li, C.; Chen, C.Y.; Song, B.-H. A genome-wide association study of seed composition traits in wild soybean (Glycine soja). BMC Genomics 2017, 18, 18. [Google Scholar] [CrossRef]
- Bi, Y.; Li, W.; Xiao, J.; Lin, H.; Liu, M.; Liu, M.; Luan, X.; Zhang, B.; Xie, X.; Guo, D. Heterosis and combining ability estimates in isoflavone content using different parental soybean accessions: Wild soybean, a valuable germplasm for soybean breeding. PLoS ONE 2015, 10, e0114827. [Google Scholar] [CrossRef]
- Panneerselvam, K.; Tsukamoto, C.; Honda, N.; Kikuchi, A.; Lee, J.D.; Yang, S.H.; Chung, G. Saponin polymorphism in the Korean wild soybean (Glycine soja Sieb. and Zucc.). Plant Breed. 2013, 132, 121–126. [Google Scholar] [CrossRef]
- Rehman, H.M.; Nawaz, M.A.; Shah, Z.H.; Yang, S.H.; Chung, G. Functional characterization of naturally occurring wild soybean mutant (sg-5) lacking astringent saponins using whole genome sequencing approach. Plant Sci. 2017. [Google Scholar] [CrossRef] [PubMed]
- Yano, R.; Takagi, K.; Takada, Y.; Mukaiyama, K.; Tsukamoto, C.; Sayama, T.; Kaga, A.; Anai, T.; Sawai, S.; Ohyama, K. Metabolic switching of astringent and beneficial triterpenoid saponins in soybean is achieved by a loss-of-function mutation in cytochrome P450 72A69. Plant J. 2017, 89, 527–539. [Google Scholar] [CrossRef]
- Kim, M.Y.; Lee, S.; Van, K.; Kim, T.-H.; Jeong, S.-C.; Choi, I.-Y.; Kim, D.-S.; Lee, Y.-S.; Park, D.; Ma, J. Whole-genome sequencing and intensive analysis of the undomesticated soybean (Glycine soja Sieb. and Zucc.) genome. Proc. Natl. Acad. Sci. USA 2010, 107, 22032–22037. [Google Scholar] [CrossRef]
- Chung, W.-H.; Jeong, N.; Kim, J.; Lee, W.K.; Lee, Y.-G.; Lee, S.-H.; Yoon, W.; Kim, J.-H.; Choi, I.-Y.; Choi, H.-K. Population structure and domestication revealed by high-depth resequencing of Korean cultivated and wild soybean genomes. DNA Res. 2013, 21, 153–167. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Wang, Y.; Wu, S.; Wang, Y.-Y.; Ye, C.-Y.; Bai, X.; Li, Z.; Yan, C.; Wang, W.; Wang, Z. Genome re-sequencing of semi-wild soybean reveals a complex Soja population structure and deep introgression. PLoS ONE 2014, 9, e108479. [Google Scholar] [CrossRef]
- Li, Y.-h.; Zhou, G.; Ma, J.; Jiang, W.; Jin, L.-g.; Zhang, Z.; Guo, Y.; Zhang, J.; Sui, Y.; Zheng, L. De novo assembly of soybean wild relatives for pan-genome analysis of diversity and agronomic traits. Nat. Biotechnol. 2014, 32, 1045–1052. [Google Scholar] [CrossRef]
- Park, G.T.; Sundaramoorthy, J.; Lee, S.; Lee, J.-D.; Kim, J.H.; Park, S.-K.; Seo, H.S.; Chung, G.; Song, J.T. Color Variation in a Novel Glycine soja Mutant W4-S1 with Pinkish-White Flowers Is Controlled by a Single Recessive Allele at the W4 Locus. Crop Sci. 2017, 57, 3112–3121. [Google Scholar] [CrossRef]
- Sundaramoorthy, J.; Park, G.T.; Chang, J.H.; Lee, J.-D.; Kim, J.H.; Seo, H.S.; Chung, G.; Song, J.T. Identification and molecular analysis of four new alleles at the W1 locus associated with flower color in soybean. PLoS ONE 2016, 11, e0159865. [Google Scholar] [CrossRef]
- Park, G.T.; Sundaramoorthy, J.; Lee, S.; Lee, J.-D.; Kim, J.H.; Park, S.-K.; Seo, H.S.; Chung, G.; Song, J.T. Color Variation in a Novel Mutant with Pinkish-White Flowers Is Controlled by a Single Recessive Allele at the Locus. Crop Sci. 2017, 57, 3112–3121. [Google Scholar] [CrossRef]
- Asekova, S.; Kulkarni, K.P.; Patil, G.; Kim, M.; Song, J.T.; Nguyen, H.T.; Shannon, J.G.; Lee, J.-D. Genetic analysis of shoot fresh weight in a cross of wild (G. soja). Mol. Breed. 2016, 36, 1–15. [Google Scholar] [CrossRef]
- Muñoz, N.; Qi, X.; Li, M.; Xie, M.; Gao, Y.; Cheung, M.; Wong, F.; Lam, H. Improvement in nitrogen fixation capacity could be part of the domestication process in soybean. Heredity 2016, 117, 84–93. [Google Scholar] [CrossRef] [PubMed]
- Fehr, W.; Cianzio, S.; Welke, G. Registration of’SS202’soybean. Crop Sci. 1990, 30. [Google Scholar] [CrossRef]
- Concibido, V.; La Vallee, B.; Mclaird, P.; Pineda, N.; Meyer, J.; Hummel, L.; Yang, J.; Wu, K.; Delannay, X. Introgression of a quantitative trait locus for yield from Glycine soja into commercial soybean cultivars. Theor. Appl. Genet. 2003, 106, 575–582. [Google Scholar] [CrossRef]
- Wang, D.; Graef, G.; Procopiuk, A.; Diers, B. Identification of putative QTL that underlie yield in interspecific soybean backcross populations. Theor. Appl. Genet. 2004, 108, 458–467. [Google Scholar] [CrossRef]
- Li, D.; Pfeiffer, T.; Cornelius, P. Soybean QTL for yield and yield components associated with alleles. Crop Sci. 2008, 48, 571–581. [Google Scholar] [CrossRef]
- Wang, W.; Li, X.; Chen, S.; Song, S.; Gai, J.; Zhao, T. Using presence/absence variation markers to identify the QTL/allele system that confers the small seed trait in wild soybean (Glycine soja Sieb. & Zucc.). Euphytica 2016, 208, 101–111. [Google Scholar]
- Kulkarni, K.P.; Asekova, S.; Lee, D.-H.; Bilyeu, K.; Song, J.T.; Lee, J.-D. Mapping QTLs for 100-seed weight in an interspecific soybean cross of Williams 82 (Glycine max) and PI 366121 (Glycine soja). Crop Pasture Sci. 2017, 68, 148–155. [Google Scholar] [CrossRef]
- Zhang, H.; Li, C.; Davis, E.L.; Wang, J.; Griffin, J.D.; Kofsky, J.; Song, B.-H. Genome-wide association study of resistance to soybean cyst nematode (Heterodera glycines) HG Type 2.5. 7 in wild soybean (Glycine soja). Front. Plant Sci. 2016, 7, 1214. [Google Scholar]
- Winter, S.M.; Shelp, B.J.; Anderson, T.R.; Welacky, T.W.; Rajcan, I. QTL associated with horizontal resistance to soybean cyst nematode in Glycine soja PI464925B. Theor. Appl. Genet. 2007, 114, 461–472. [Google Scholar] [CrossRef]
- Yu, N.; Diers, B.W. Fine mapping of the SCN resistance QTL cqSCN-006 and cqSCN-007 from Glycine soja PI 468916. Euphytica 2017, 213, 54. [Google Scholar] [CrossRef]
- Wang, D.; Diers, B.W.; Arelli, P.; Shoemaker, R. Loci underlying resistance to race 3 of soybean cyst nematode in Glycine soja plant introduction 468916. Theor. Appl. Genet. 2001, 103, 561–566. [Google Scholar] [CrossRef]
- Carter, T.E.; Huei, E.; Burton, J.; Farmer, F.; Gizlice, Z. Registration of ‘Pearl’soybean. Crop Sci. 1995, 35, 1713. [Google Scholar] [CrossRef]
- Lee, J.S.; Yoo, M.-h.; Jung, J.K.; Bilyeu, K.D.; Lee, J.-D.; Kang, S. Detection of novel QTLs for foxglove aphid resistance in soybean. Theor. Appl. Genet. 2015, 128, 1481–1488. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhang, Z.; Wen, Z.; Gu, C.; An, Y.-Q.C.; Bales, C.; DiFonzo, C.; Song, Q.; Wang, D. Fine mapping of the soybean aphid-resistance genes Rag6 and Rag3c from Glycine soja 85-32. Theor. Appl. Genet. 2017, 130, 2601–2615. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, Z.; Bales, C.; Gu, C.; DiFonzo, C.; Li, M.; Song, Q.; Cregan, P.; Yang, Z.; Wang, D. Mapping novel aphid resistance QTL from wild soybean, Glycine soja 85-32. Theor. Appl. Genet. 2017, 130, 1941–1952. [Google Scholar] [CrossRef]
- Hill, C.B.; Li, Y.; Hartman, G.L. Resistance of Glycine species and various cultivated legumes to the soybean aphid (Homoptera: Aphididae). J. Econ. Entomol. 2004, 97, 1071–1077. [Google Scholar] [CrossRef]
- Iquira, E.; Humira, S.; François, B. Association mapping of QTLs for sclerotinia stem rot resistance in a collection of soybean plant introductions using a genotyping by sequencing (GBS) approach. BMC Plant Biol. 2015, 15, 5. [Google Scholar] [CrossRef]
- Zhang, H.; Song, B.-H. RNA-seq data comparisons of wild soybean genotypes in response to soybean cyst nematode (Heterodera glycines). Genomics Data 2017, 14, 36–39. [Google Scholar] [CrossRef]
- Zhang, H.; Song, Q.; Griffin, J.D.; Song, B.-H. Genetic architecture of wild soybean (Glycine soja) response to soybean cyst nematode (Heterodera glycines). Mol. Genet. Genomics 2017, 292, 1257–1265. [Google Scholar] [CrossRef]
- Rani, A.; Kumar, V.; Gill, B.; Shukla, S.; Rathi, P.; Singh, R. Mapping of duplicate dominant genes for Mungbean yellow mosaic India virus resistance in Glycine soja. Crop Sci. 2018, 58, 1566–1574. [Google Scholar] [CrossRef]
- Tucker, D.; Maroof, S.; Mideros, S.; Skoneczka, J.; Nabati, D.; Buss, G.; Hoeschele, I.; Tyler, B.; St Martin, S.; Dorrance, A. Mapping quantitative trait loci for partial resistance to Phytophthora sojae in a soybean interspecific cross. Crop Sci. 2010, 50, 628–635. [Google Scholar] [CrossRef]
- Luo, Q.; Yu, B.; Liu, Y. Differential sensitivity to chloride and sodium ions in seedlings of Glycine max and G. soja under NaCl stress. J. Plant Physiol. 2005, 162, 1003–1012. [Google Scholar] [CrossRef]
- Lee, J.-D.; Shannon, J.G.; Vuong, T.D.; Nguyen, H.T. Inheritance of salt tolerance in wild soybean (Glycine soja Sieb. and Zucc.) accession PI483463. J. Hered. 2009, 100, 798–801. [Google Scholar] [CrossRef]
- Tuyen, D.; Lal, S.; Xu, D. Identification of a major QTL allele from wild soybean (Glycine soja Sieb. & Zucc.) for increasing alkaline salt tolerance in soybean. Theor. Appl. Genet. 2010, 121, 229–236. [Google Scholar] [PubMed]
- Ha, B.-K.; Vuong, T.D.; Velusamy, V.; Nguyen, H.T.; Shannon, J.G.; Lee, J.-D. Genetic mapping of quantitative trait loci conditioning salt tolerance in wild soybean (Glycine soja) PI 483463. Euphytica 2013, 193, 79–88. [Google Scholar] [CrossRef]
- Kilen, T.C.; He, G. Identification and inheritance of metribuzin tolerance in wild soybean. Crop Sci. 1992, 32, 684–685. [Google Scholar] [CrossRef]
- Manavalan, L.P.; Prince, S.J.; Musket, T.A.; Chaky, J.; Deshmukh, R.; Vuong, T.D.; Song, L.; Cregan, P.B.; Nelson, J.C.; Shannon, J.G. Identification of novel QTL governing root architectural traits in an interspecific soybean population. PLoS ONE 2015, 10, e0120490. [Google Scholar] [CrossRef]
- Prince, S.J.; Song, L.; Qiu, D.; dos Santos, J.V.M.; Chai, C.; Joshi, T.; Patil, G.; Valliyodan, B.; Vuong, T.D.; Murphy, M. Genetic variants in root architecture-related genes in a Glycine soja accession, a potential resource to improve cultivated soybean. BMC Genomics 2015, 16, 132. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, J.; Jiang, W.; Liu, J.; Yang, S.; Gai, J.; Li, Y. Identification and analysis of NaHCO3 stress responsive genes in wild soybean (Glycine soja) roots by RNA-seq. Front. Plant Sci. 2016, 7, 1842. [Google Scholar] [CrossRef]
- Ning, W.; Zhai, H.; Yu, J.; Liang, S.; Yang, X.; Xing, X.; Huo, J.; Pang, T.; Yang, Y.; Bai, X. Overexpression of Glycine soja WRKY20 enhances drought tolerance and improves plant yields under drought stress in transgenic soybean. Mol. Breed. 2017, 37, 19. [Google Scholar] [CrossRef]
- Diers, B.W.; Keim, P.; Fehr, W.; Shoemaker, R. RFLP analysis of soybean seed protein and oil content. Theor. Appl. Genet. 1992, 83, 608–612. [Google Scholar] [CrossRef]
- Pantalone, V.; Rebetzke, G.; Burton, J.; Wilson, R. Genetic regulation of linolenic acid concentration in wild soybean Glycine soja accessions. J. Am. Oil Chem. Soc. 1997, 74, 159–163. [Google Scholar] [CrossRef]
- Park, J.; Kim, J.H.; Krishnamurthy, P.; Tsukamoto, C.; Song, J.T.; Chung, G.; Shannon, J.G.; Lee, J.-D. Characterization of a New Allele of the Saponin-Synthesizing Gene in Soybean. Crop Sci. 2016, 56, 385–391. [Google Scholar] [CrossRef]
- Li, M.-W.; Muñoz, N.B.; Wong, C.-F.; Wong, F.-L.; Wong, K.-S.; Wong, J.W.-H.; Qi, X.; Li, K.-P.; Ng, M.-S.; Lam, H.-M. QTLs regulating the contents of antioxidants, phenolics, and flavonoids in soybean seeds share a common genomic region. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.D.; Shannon, J.; Vuong, T.; Moon, H.; Nguyen, H.; Tsukamoto, C.; Chung, G. Genetic diversity in wild soybean (Glycine soja Sieb. and Zucc.) accessions from southern islands of Korean peninsula. Plant Breed. 2010, 129, 257–263. [Google Scholar] [CrossRef]
- Nawaz, M.A.; Yang, S.H.; Rehman, H.M.; Baloch, F.S.; Lee, J.D.; Park, J.H.; Chung, G. Genetic diversity and population structure of Korean wild soybean (Glycine soja Sieb. and Zucc.) inferred from microsatellite markers. Biochem. Syst. Ecol. 2017, 71, 87–96. [Google Scholar] [CrossRef]
- Carter, T.; Nelson, R.; Sneller, C.; Cui, Z. Genetic diversity in soybean. Soybean Monogr. Am. Soc. Agron. Madison Wis. USA 2004. [Google Scholar]
- Li, M.-W.; Wang, Z.; Jiang, B.; Kaga, A.; Wong, F.-L.; Zhang, G.; Han, T.; Chung, G.; Nguyen, H.; Lam, H.-M. Impacts of genomic research on soybean improvement in East Asia. Theor. Appl. Genet. 2019, 1–24. [Google Scholar] [CrossRef]
- Lee, G.-A.; Crawford, G.W.; Liu, L.; Sasaki, Y.; Chen, X. Archaeological soybean (Glycine max) in East Asia: Does size matter? PLoS ONE 2011, 6, e26720. [Google Scholar] [CrossRef]
- Crawford, G.W.; Lee, G.-A. Agricultural origins in the Korean Peninsula. Antiquity 2003, 77, 87. [Google Scholar] [CrossRef]
- Barnes, G.L. Archaeology of East. Asia: The Rise of Civilization in China, Korea and Japan; Oxbow Books: Oxford, UK; Casemate Publishing: Pennsylvania, PA, USA, 2015. [Google Scholar]
- Fujita, R.; Ohara, M.; Okazaki, K.; Shimamoto, Y. The extent of natural cross-pollination in wild soybean (Glycine soja). J. Hered. 1997, 88, 124–128. [Google Scholar] [CrossRef]
- Kuroda, Y.; Kaga, A.; Tomooka, N.; Vaughan, D.A. Gene flow and genetic structure of wild soybean (Glycine soja) in Japan. Crop Sci. 2008, 48, 1071–1079. [Google Scholar] [CrossRef]
- Wang, Y.; Shahid, M.Q.; BALOCH, F.S. Phylogeographical studies of Glycine soja: Implicating the refugium during the quaternary glacial period and large-scale expansion after the last glacial maximum. Turk. J. Agric. For. 2016, 40, 825–838. [Google Scholar] [CrossRef]
- Lu, H.; Yi, S.; Xu, Z.; Zhou, Y.; Zeng, L.; Zhu, F.; Feng, H.; Dong, L.; Zhuo, H.; Yu, K. Chinese deserts and sand fields in Last Glacial Maximum and Holocene Optimum. Chin. Sci. Bull. 2013, 58, 2775–2783. [Google Scholar] [CrossRef]
- Park, H.; Hur, S. Growth habit and protein content of various wild soybean strains. Korean J. Bot. 1979. [Google Scholar]
- Yu, H.; Kiang, Y.-T. Genetic variation in South Korean natural populations of wild soybean (Glycine soja). Euphytica 1993, 68, 213–221. [Google Scholar] [CrossRef]
- Kim, K.-U.; Gang, T.-D.; Lee, J.-H.; Lee, I.-J.; Shin, D.-H.; Hwang, Y.-H.; Kim, S.-U.; Kim, H.-M. Physio-ecological characteristics of wild soybeans (Glycine soja) collected throughout Korea and their response to glyphosate. Korean J. Weed Sci. 2003, 23, 153–159. [Google Scholar]
- Kim, K.C.; Park, E.-H. Variation of protein, oil contents and fatty acid composition of Korean wild soybean (Glycine soja Sieb. & Zucc.) seeds. Korean J. Crop Sci. 2005, 50, 118–122. [Google Scholar]
- Lee, J.-D.; Yoon, Y.-H.; Chung, I.-K.; Park, S.-K.; Hwang, Y.-H. A new Glycine soja germplasm accession with green seed-coat color. Breed. Sci. 2005, 55, 21–25. [Google Scholar] [CrossRef][Green Version]
- Yun, H.-T.; Seo, M.-J.; Kim, S.-L.; An, S.-O.; Kim, S.-J. Variation of seed component contents in wild soybean (Glycine soja Sieb. & Zucc.). Korean J. Crop Sci. 2005, 50, 108–111. [Google Scholar]
- Choi, I.-Y.; Kang, J.-H.; Song, H.-S.; Kim, N.-S. Genetic diversity measured by simple sequence repeat variations among the wild soybean, Glycine soja, collected along the riverside of five major rivers in Korea. Genes Genet. Syst. 1999, 74, 169–177. [Google Scholar] [CrossRef]
- Cho, Y.; Yoon, M.; Lee, J.; Baek, H.; Kim, C.; Kim, T.; Cho, E.; Lee, H. Diversity and geographical relationships by SSR marker in subgenus soja originated from Korea. Korean J. Crop Sci. 2006, 51, 239–247. [Google Scholar]
- Lee, J.-D.; Yu, J.-K.; Hwang, Y.-H.; Blake, S.; So, Y.-S.; Lee, G.-J.; Nguyen, H.; Shannon, J.G. Genetic Diversity of Wild Soybean (Glycine soja Sieb. and Zucc.) Accessions from South Korea and Other Countries. Crop Sci. 2008, 48, 606–616. [Google Scholar] [CrossRef]
- He, S.; Wang, Y.; Volis, S.; Li, D.; Yi, T. Genetic diversity and population structure: Implications for conservation of wild soybean (Glycine soja Sieb. et Zucc) based on nuclear and chloroplast microsatellite variation. Int. J. Mol. Sci. 2012, 13, 12608–12628. [Google Scholar] [CrossRef]
- Frankham, R. Relationship of genetic variation to population size in wildlife. Conserv. Biol. 1996, 10, 1500–1508. [Google Scholar] [CrossRef]
- Frankham, R. Do island populations have less genetic variation than mainland populations? Heredity 1997, 78, 311–327. [Google Scholar] [CrossRef]
- Frankham, R. Inbreeding and extinction: Island populations. Conserv. Boil. 1998, 12, 665–675. [Google Scholar] [CrossRef]
- Maxted, N.; Kell, S. Establishment of a global network for the in situ conservation of crop wild relatives: Status and needs. FAO Comm. Genet. Resour. Food Agric. Rome 2009, 266, 509. [Google Scholar]
- Kim, M.Y.; Van, K.; Kang, Y.J.; Kim, K.H.; Lee, S.-H. Tracing soybean domestication history: From nucleotide to genome. Breed. Sci. 2012, 61, 445–452. [Google Scholar] [CrossRef]
- Lee, S.; Kim, J.-H.; Sundaramoorthy, J.; Park, G.T.; Lee, J.-D.; Kim, J.H.; Chung, G.; Seo, H.S.; Song, J.T. Identification of GmSALT3 haplotypes and development of molecular markers based on their diversity associated with salt tolerance in soybean. Mol. Breed. 2018, 38, 86. [Google Scholar] [CrossRef]
- Krishnamurthy, P.; Lee, J.M.; Tsukamoto, C.; Takahashi, Y.; Singh, R.J.; Lee, J.D.; Chung, G. Evaluation of genetic structure of Korean wild soybean (Glycine soja) based on saponin allele polymorphism. Genet. Res. Crop Evol. 2014, 61, 1121–1130. [Google Scholar] [CrossRef]
- Krishnamurthy, P.; Tsukamoto, C.; Singh, R.J.; Lee, J.-D.; Kim, H.-S.; Yang, S.-H.; Chung, G. The Sg-6 saponins, new components in wild soybean (Glycine soja Sieb. and Zucc.): polymorphism, geographical distribution and inheritance. Euphytica 2014, 198, 413–424. [Google Scholar] [CrossRef]
- Krishnamurthy, P.; Tsukamoto, C.; Takahashi, Y.; Hongo, Y.; Singh, R.J.; Lee, J.D.; Chung, G. Comparison of saponin composition and content in wild soybean (Glycine soja Sieb. and Zucc.) before and after germination. Biosci. Biotechnol. Biochem. 2014, 78, 1988–1996. [Google Scholar] [CrossRef] [PubMed]
- Sundaramoorthy, J.; Park, G.T.; Mukaiyama, K.; Tsukamoto, C.; Chang, J.H.; Lee, J.-D.; Kim, J.H.; Seo, H.S.; Song, J.T. Molecular elucidation of a new allelic variation at the Sg-5 gene associated with the absence of group A saponins in wild soybean. PLoS ONE 2018, 13, e0192150. [Google Scholar] [CrossRef]
- Krishnamurthy, P.; Lee, J.D.; Ha, B.K.; Chae, J.H.; Song, J.T.; Tsukamoto, C.; Singh, R.J.; Chung, G. Genetic characterization of group A acetylsaponin-deficient mutants from wild soybean (Glycine soja Sieb. and Zucc.). Plant Breed. 2015, 134, 316–321. [Google Scholar] [CrossRef]
- Rehman, H.M.; Nawaz, M.A.; Shah, Z.H.; Chung, G.; Yang, S.H. Molecular Elucidation of Two Novel Seed Specific Flavonoid Glycosyltransferases in Soybean. J. Plant Biol. 2018, 61, 320–329. [Google Scholar] [CrossRef]
- Krishnamurthy, P.; Singh, R.J.; Tsukamoto, C.; Park, J.H.; Lee, J.D.; Chung, G. Kunitz trypsin inhibitor polymorphism in the Korean wild soybean (Glycine soja Sieb. & Zucc.). Plant Breed. 2013, 132, 311–316. [Google Scholar]
- Nawaz, M.A.; Golokhvast, K.S.; Rehman, H.M.; Tsukamoto, C.; Kim, H.-S.; Yang, S.H.; Chung, G. Soyisoflavone diversity in wild soybeans (Glycine soja Sieb. & Zucc.) from the main centres of diversity. Biochem. Syst. Ecol. 2018, 77, 16–21. [Google Scholar]
- Tsukamoto, C.; Nawaz, M.A.; Kurosaka, A.; Le, B.; Lee, J.D.; Son, E.; Yang, S.H.; Kurt, C.; BALOCH, F.S.; Chung, G. Isoflavone profile diversity in Korean wild soybeans (Glycine soja Sieb. & Zucc.). Turk. J. Agric. For. 2018, 42, 248–261. [Google Scholar]
- Chae, J.-H.; Ha, B.-K.; Chung, G.; Park, J.-E.; Park, E.; Ko, J.-M.; Shannon, J.G.; Song, J.T.; Lee, J.-D. Identification of environmentally stable wild soybean genotypes with high alpha-linolenic acid concentration. Crop Sci. 2015, 55, 1629–1636. [Google Scholar] [CrossRef]
- Ali, M.; Krishnamurthy, P.; El-Hadary, M.; Kim, J.; Nawaz, M.; Yang, S.; Chung, G. Identification and expression profiling of a new β-amyrin synthase gene (GmBAS3) from soybean. Russ. J. Plant physiol. 2016, 63, 383–390. [Google Scholar] [CrossRef]
- Fang, C.; Ma, Y.; Yuan, L.; Wang, Z.; Yang, R.; Zhou, Z.; Liu, T.; Tian, Z. Chloroplast DNA Underwent Independent Selection from Nuclear Genes during Soybean Domestication and Improvement. J. Genet. Genomics = Yi Chuan Xue Bao 2016, 43, 217. [Google Scholar] [CrossRef] [PubMed]
- Asaf, S.; Khan, A.L.; Khan, M.A.; Imran, Q.M.; Kang, S.-M.; Al-Hosni, K.; Jeong, E.J.; Lee, K.E.; Lee, I.-J. Comparative analysis of complete plastid genomes from wild soybean (Glycine soja) and nine other Glycine species. PLoS ONE 2017, 12, e0182281. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).