Selection of the Root Endophyte Pseudomonas brassicacearum CDVBN10 as Plant Growth Promoter for Brassica napus L. Crops
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation and Identification of Bacterial Isolates
2.2. In Vitro Analyses of Plant Growth-Promoting Mechanisms and Biosynthesis of Polysaccharides
2.3. Effects of Bacterial Isolates on Rapeseed Seedlings
2.4. Draft Genome Sequencing and Annotation
2.5. Field Experiment
2.6. Amplicon Sequencing and Sequence Analysis
2.7. Statistical Analysis of Plant Parameters
3. Results
3.1. Bacterial Culturome Shows the High Diversity of B. Napus Associated Endophytic Bacteria
3.2. In Vitro Analyses of Plant Growth-Promoting Mechanisms
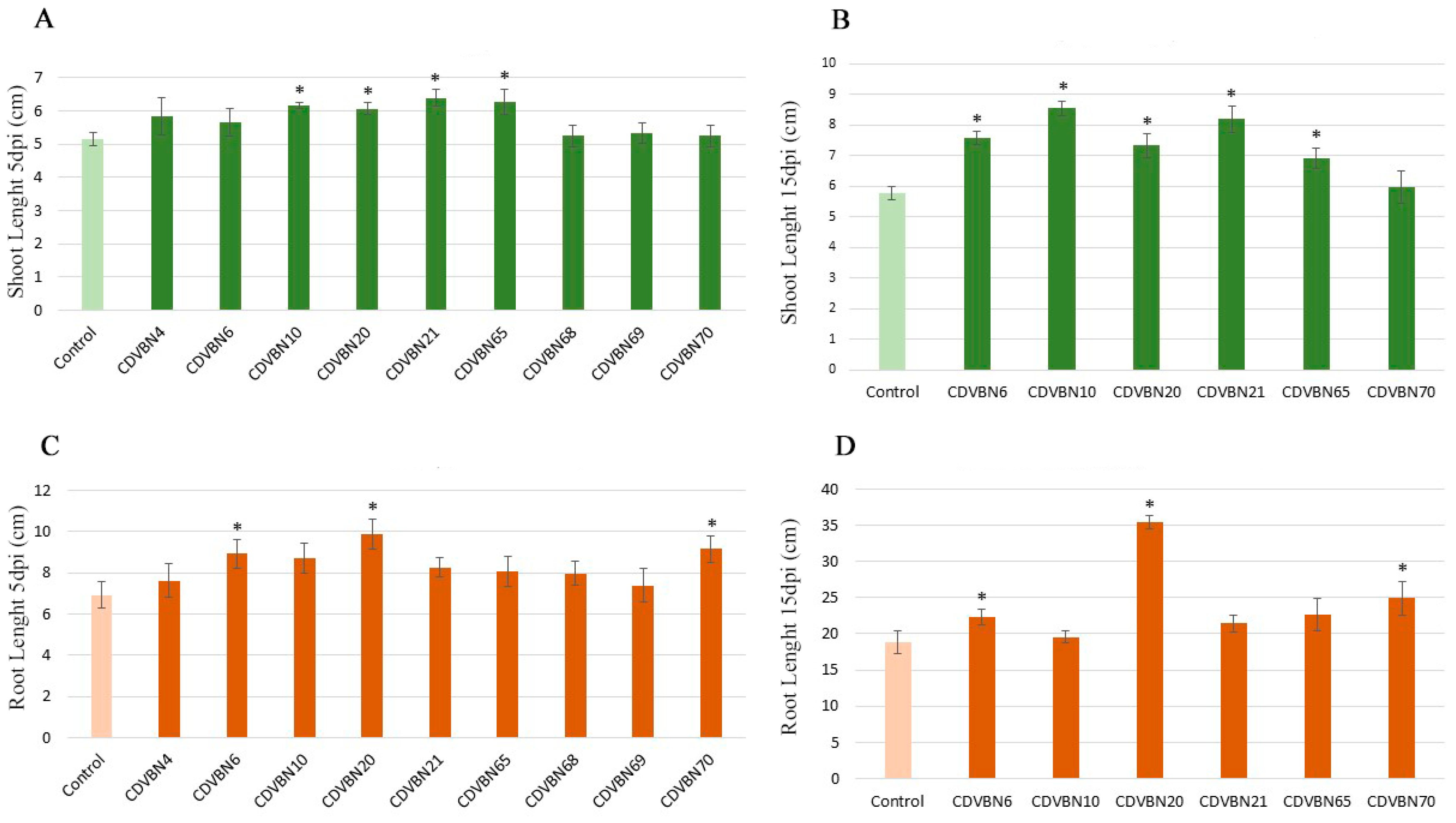
3.3. Plant Growth Promotion in Rapeseed Seedlings under Controlled Conditions and Additional PGP Traits
3.4. Taxonomic Affiliation of the Best Performing Strains
3.5. Genome in Silico Analysis of Plant Growth-Promoting and Putative Colonization Related Mechanisms
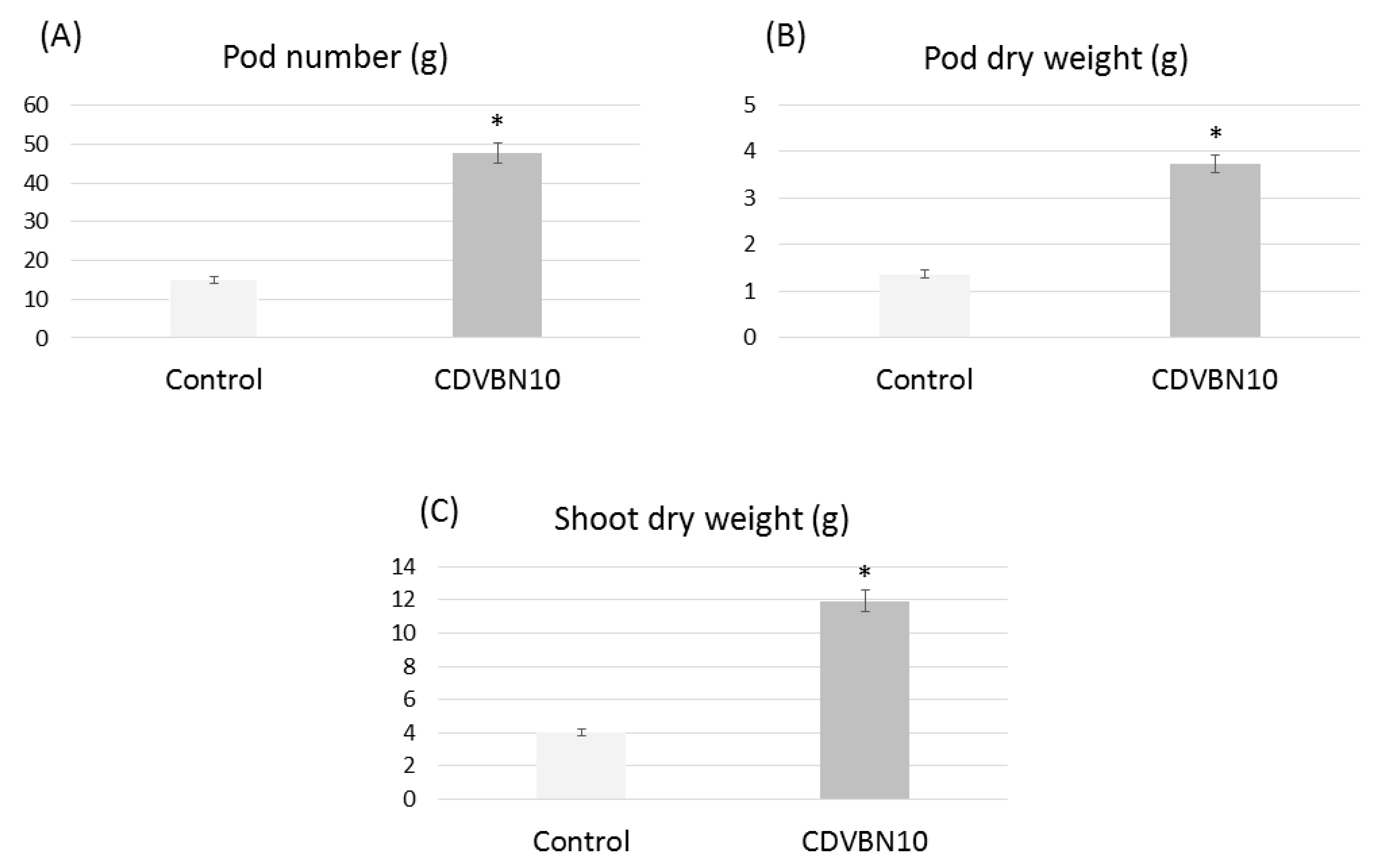
3.6. Pseudomonas brassicacearum CDVBN10 Displays Beneficial Effects in Rapeseed Plants Cultivated in the Field
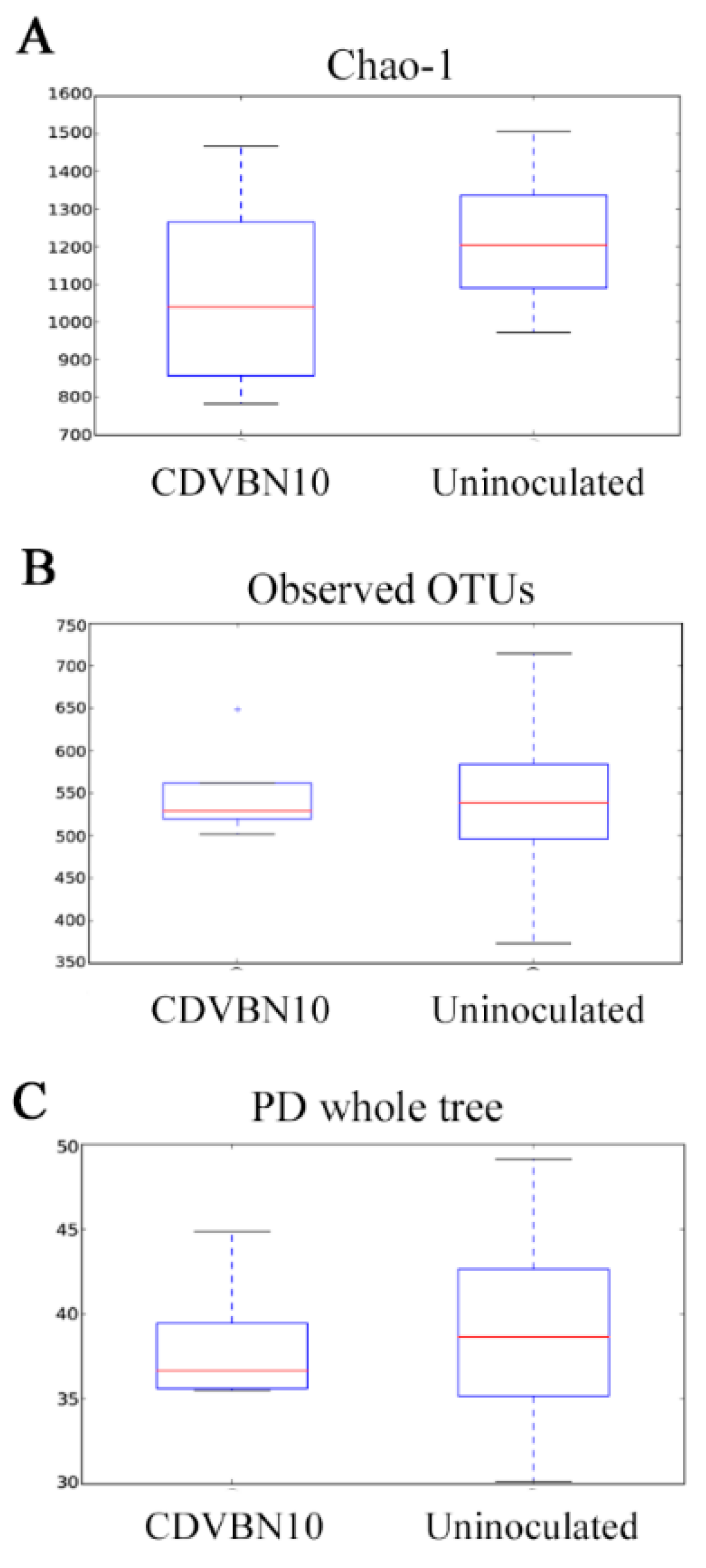
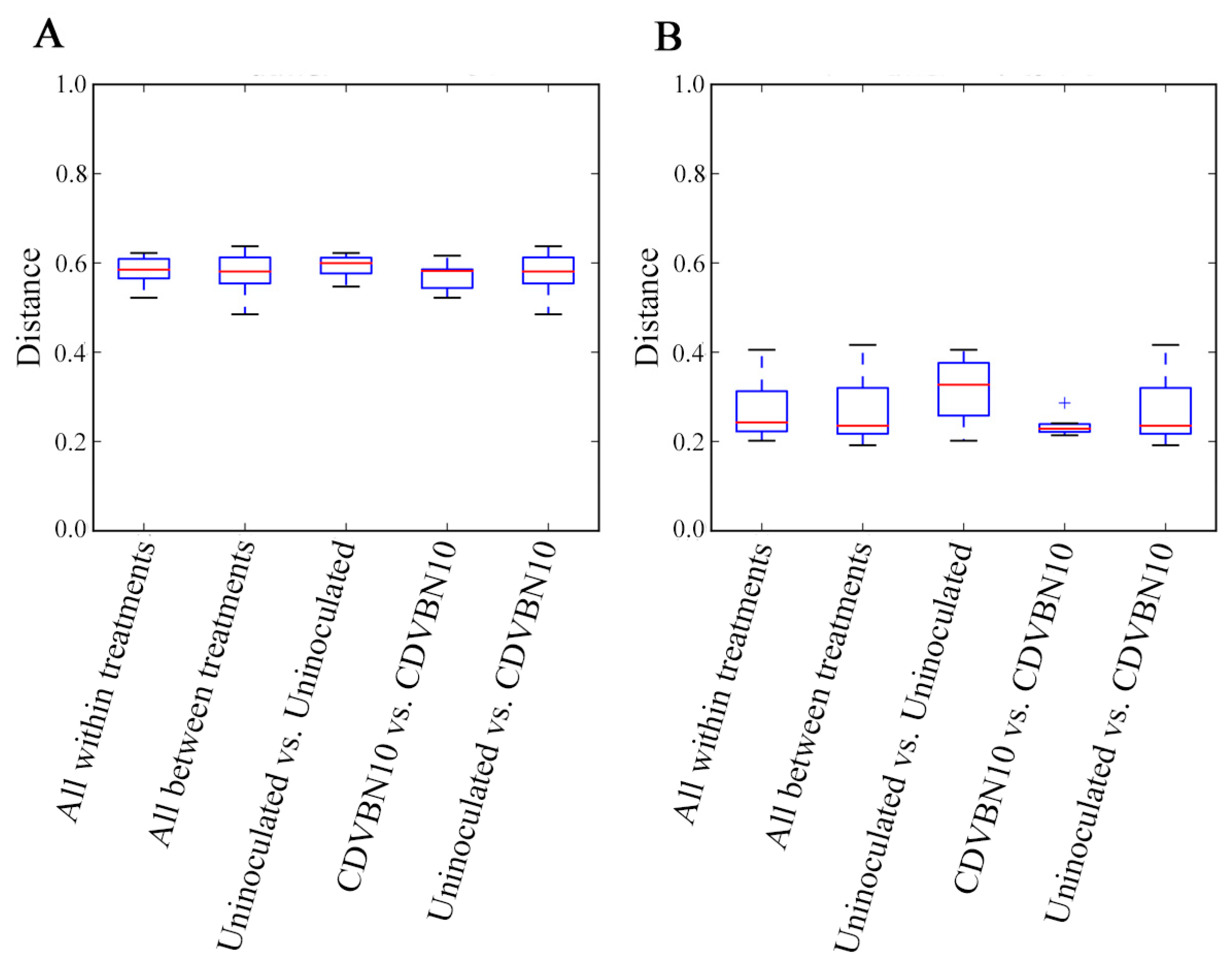
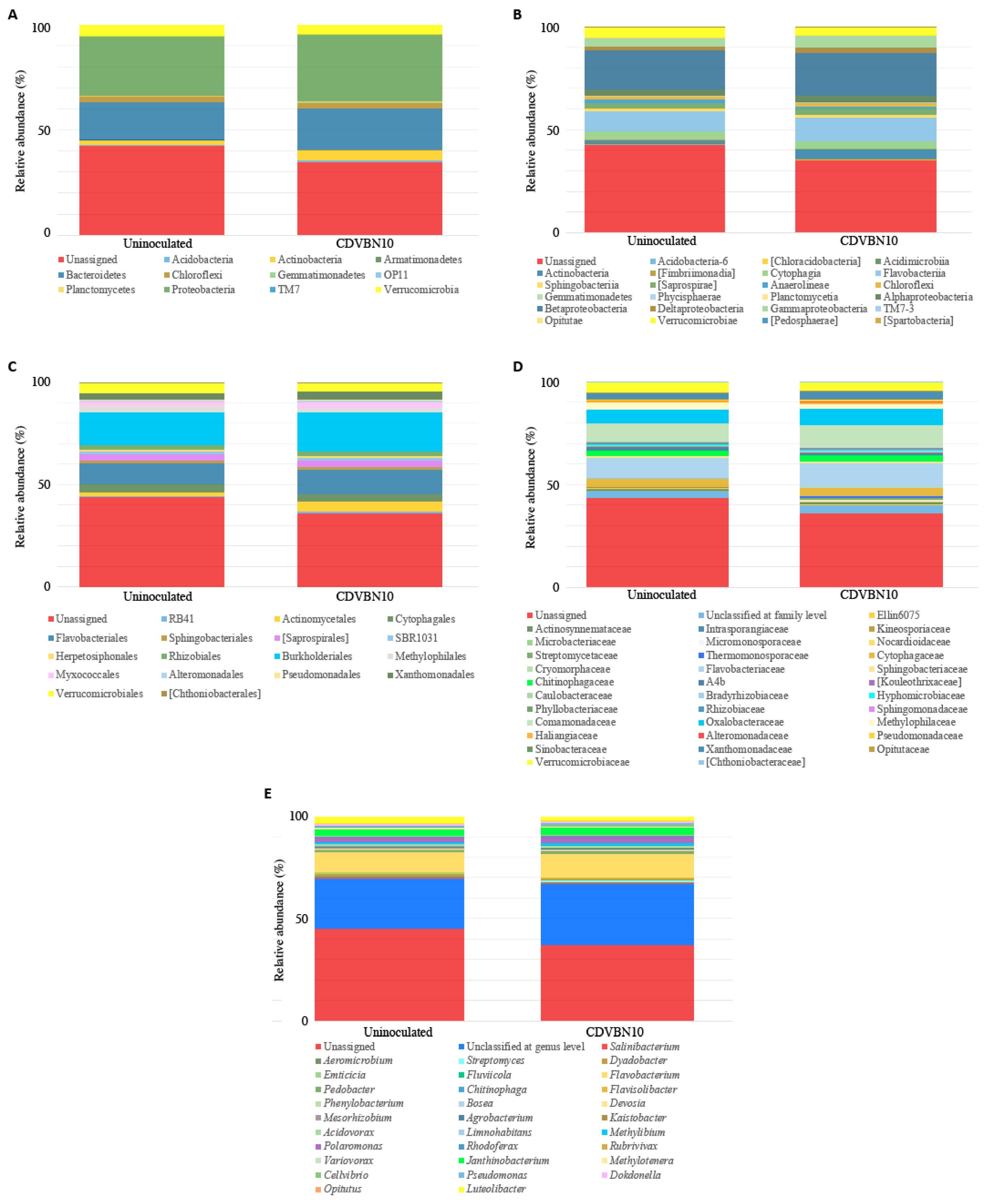
3.7. CDVBN10 Inoculation Does Not Significantly Alter Bacterial Diversity in Rapeseed Roots Grown in the Field Trial
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wu, W.; Ma, B. Integrated nutrient management (INM) for sustaining crop productivity and reducing environmental impact: A review. Sci. Total Environ. 2015, 512, 415–427. [Google Scholar] [CrossRef]
- Adesemoye, A.O.; Torbert, H.A.; Kloepper, J.W. Plant Growth-Promoting Rhizobacteria Allow Reduced Application Rates of Chemical Fertilizers. Microb. Ecol. 2009, 58, 921–929. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, D.; Ansari, M.W.; Sahoo, R.K.; Tuteja, N. Biofertilizers function as key player in sustainable agriculture by improving soil fertility, plant tolerance and crop productivity. Microb. Cell Fact. 2014, 13, 66. [Google Scholar] [CrossRef] [PubMed]
- Mahanty, T.; Bhattacharjee, S.; Goswami, M.; Bhattacharyya, P.; Das, B.; Ghosh, A. Biofertilizers: A potential approach for sustainable agriculture development. Environ. Sci. Pollut. Res. 2017, 24, 3315–3335. [Google Scholar] [CrossRef] [PubMed]
- Olanrewaju, O.S.; Glick, B.R.; Babalola, O.O. Mechanisms of action of plant growth promoting bacteria. World J. Microbiol. Biotechnol. 2017, 33, 197. [Google Scholar] [CrossRef] [PubMed]
- Menéndez, E.; Garcia-Fraile, P. Plant probiotic bacteria: Solutions to feed the world. AIMS Microbiol. 2017, 3, 502–524. [Google Scholar] [CrossRef] [PubMed]
- Gaiero, J.R.; McCall, C.A.; Thompson, K.A.; Day, N.J.; Best, A.S.; Dunfield, K.E. Inside the root microbiome: Bacterial root endophytes and plant growth promotion. Am. J. Bot. 2013, 100, 1738–1750. [Google Scholar] [CrossRef]
- Santoyo, G.; Moreno-Hagelsieb, G.; del Carmen Orozco-Mosqueda, M.; Glick, B.R. Plant growth-promoting bacterial endophytes. Microbiol. Res. 2016, 183, 92–99. [Google Scholar] [CrossRef]
- Gopal, M.; Gupta, A. Microbiome selection could spur next-generation plant breeding strategies. Front. Microbiol. 2016, 7, 1971. [Google Scholar] [CrossRef]
- Velázquez, E.; García-Fraile, P.; Ramírez-Bahena, M.H.; Rivas, R.; Martínez-Molina, E. Bacteria Involved in Nitrogen-Fixing Legume Symbiosis: Current Taxonomic Perspective. In Microbes for Legume Improvement; Springer: Vienna, Austria, 2010; pp. 1–5. [Google Scholar]
- Brundrett, M.C.; Tedersoo, L. Evolutionary history of mycorrhizal symbioses and global host plant diversity. New Phytol. 2018, 220, 1108–1115. [Google Scholar] [CrossRef]
- Clear, M.R.; Hom, E.F. The evolution of symbiotic plant-microbe signaling. Ann. Plant. Rev. Online 2019, 2, 1–52. [Google Scholar] [CrossRef]
- Vorholt, J.A.; Vogel, C.; Carlström, C.I.; Müller, D.B. Establishing causality: Opportunities of synthetic communities for plant microbiome research. Cell Host Microb. 2017, 22, 142–155. [Google Scholar] [CrossRef] [PubMed]
- Etesami, H.; Alikhani, H.A. Rhizosphere and endorhiza of oilseed rape (Brassica napus L.) plant harbor bacteria with multifaceted beneficial effects. Biol. Cont. 2016, 94, 11–24. [Google Scholar] [CrossRef]
- Card, S.D.; Hume, D.E.; Roodi, D.; McGill, C.R.; Millner, J.P.; Johnson, R.D. Beneficial endophytic microorganisms of Brassica–A review. Biol. Control. 2015, 90, 102–112. [Google Scholar] [CrossRef]
- Rathore, R.; Germaine, K.J.; Forristal, P.D.; Spink, J.; Dowling, D. Meta-Omics Approach to Unravel the Endophytic Bacterial Communities of Brassica napus and Other Agronomically Important Crops. In Endophytes for a Growing World; Hodkinson, T.R., Ed.; Cambridge University Press: Cambridge, UK, 2019; p. 232. [Google Scholar]
- Farina, R.; Beneduzi, A.; Ambrosini, A.; de Campos, S.B.; Lisboa, B.B.; Wendisch, V. Diversity of plant growth-promoting rhizobacteria communities associated with the stages of canola growth. Appl. Soil Ecol. 2012, 55, 44–52. [Google Scholar] [CrossRef]
- Bertrand, H.; Nalin, R.; Bally, R.; Cleyet-Marel, J.C. Isolation and identification of the most efficient plant growth-promoting bacteria associated with canola (Brassica napus). Biol. Fertil. Soils 2001, 33, 152–156. [Google Scholar] [CrossRef]
- Sheng, X.F.; Xia, J.J.; Jiang, C.Y.; He, L.Y.; Qian, M. Characterization of heavy metal-resistant endophytic bacteria from rape (Brassica napus) roots and their potential in promoting the growth and lead accumulation of rape. Environ. Pollut. 2008, 156, 1164–1170. [Google Scholar] [CrossRef]
- Etesami, H.; Emami, S.; Alikhani, H.A. Potassium solubilizing bacteria (KSB): Mechanisms, promotion of plant growth and future prospects A review. J. Soil Sci. Plant. Nutr. 2017, 17, 897–911. [Google Scholar] [CrossRef]
- Puri, A.; Padda, K.P.; Chanway, C.P. Evidence of nitrogen fixation and growth promotion in canola (Brassica napus L.) by an endophytic diazotroph Paenibacillus polymyxa P2b-2R. Biol. Fertil. Soils 2016, 52, 119–125. [Google Scholar] [CrossRef]
- Lally, R.D.; Galbally, P.; Moreira, A.S.; Spink, J.; Ryan, D.; Germaine, K.J. Application of endophytic Pseudomonas fluorescens and a bacterial consortium to Brassica napus can increase plant height and biomass under greenhouse and field conditions. Front. Plant. Sci. 2017, 8, 2193. [Google Scholar] [CrossRef]
- Petrova, S.N.; Andronov, E.E.; Belimov, A.A.; Beregovaya, Y.V.; Denshchikov, V.A.; Minakov, D.L. Prokaryotic Community Structure in the Rapeseed (Brassica napus L.) Rhizosphere Depending on Addition of 1-Aminocyclopropane-1-Carboxylate-Utilizing Bacteria. Microbiology 2020, 89, 115–121. [Google Scholar] [CrossRef]
- Bruto, M.; Prigent-Combaret, C.; Muller, D.; Moënne-Loccoz, Y. Analysis of genes contributing to plant-beneficial functions in plant growth-promoting rhizobacteria and related Proteobacteria. Sci. Rep. 2014, 4, 6261. [Google Scholar] [CrossRef] [PubMed]
- López-Mondéjar, R.; Kostovčík, M.; Lladó, S.; Carro, L.; García-Fraile, P. Exploring the Plant Microbiome through Multi-Omics Approaches. In Probiotics in Agroecosystem; Springer: Singapore, 2017; pp. 233–268. [Google Scholar]
- Igual, J.M.; Valverde, A.; Rivas, R.; Mateos, P.F.; Rodríguez-Barrueco, C.; Martínez-Molina, E. Genomic Fingerprinting of Frankia Strains by PCR-Based Techniques. Assessment of a Primer Based on the Sequence of 16S rRNA Gene of Escherichia coli. In Frankia Symbiosis; Normand, P., Pawlowski, K., Dawson, J.I., Eds.; Springer: Dordrecht, The Netherland, 2003; pp. 115–123. [Google Scholar]
- Rivas, R.; Garcia-Fraile, P.; Peix, A.; Mateos, P.F.; Martínez-Molina, E.; Velazquez, E. Alcanivorax balearicus sp. nov., isolated from Lake Martel. Int. J. Syst. Evol. Microbiol. 2007, 57, 1331–1335. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Poveda, J.; Jiménez-Gómez, A.; Saati-Santamaría, Z.; Usategui-Martín, R.; Rivas, R.; García-Fraile, P. Mealworm frass as a potential biofertilizer and abiotic stress tolerance-inductor in plants. Appl. Soil Ecol. 2019, 142, 110–122. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Chun, J.; Lee, J.H.; Jung, Y.; Kim, M.; Kim, S.; Kim, B.K. EzTaxon: A web-based tool for the Identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int. J. Syst. Evol. Microbiol. 2007, 57, 2259–2261. [Google Scholar] [CrossRef]
- Jiménez-Gómez, A.; Saati-Santamaría, Z.; Igual, J.M.; Rivas, R.; Mateos, P.F.; García-Fraile, P. Genome Insights into the Novel Species Microvirga brassicacearum, a Rapeseed Endophyte with Biotechnological Potential. Microorganisms 2019, 7, 354. [Google Scholar] [CrossRef]
- Schwyn, B.; Neilands, J.B. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Alexander, D.B.; Zuberer, D.A. Use of Chrome Azurol S reagents to evaluate siderophore production by rhizosphere bacteria. Biol. Fertil. Soils 1991, 12, 39–45. [Google Scholar] [CrossRef]
- Pikovskaya, R.I. Mobilization of phosphorus in soil connection with the vital activity of some microbial species. Microbiologiya 1948, 17, 362–370. [Google Scholar]
- Robledo, M.; Rivera, L.; Jiménez-Zurdo, J.I.; Rivas, R.; Dazzo, F.; Velázquez, E.; Mateos, P.F. Role of Rhizobium endoglucanase CelC2 in cellulose biosynthesis and biofilm formation on plant roots and abiotic surfaces. Microb. Cell Fact. 2012, 11, 125. [Google Scholar] [CrossRef] [PubMed]
- Khalid, A.; Arshad, M.; Zahir, Z.A. Screening plant growth-promoting rhizobacteria for improving growth and yield of wheat. J. Appl. Microbiol. 2004, 96, 473–480. [Google Scholar] [CrossRef] [PubMed]
- Saati-Santamaría, Z.; López-Mondéjar, R.; Jiménez-Gómez, A.; Díez-Méndez, A.; Větrovský, T.; Igual, J.M.; Garcia-Fraile, P. Discovery of phloeophagus beetles as a source of Pseudomonas strains that produce potentially new bioactive substances and description of Pseudomonas bohemica sp. nov. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Zerbino, D.R.; Birney, E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008, 18, 821–829. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A. The RAST Server: Rapid annotations using subsystems technology. BMC Genomics 2008, 9, 75. [Google Scholar] [CrossRef]
- Dellavalle, N.B. Determination of Specific Conductance in Supernatant 1:2 soil:water solution. In Handbook on Reference Methods for Soil Analysis; Council, Inc.: Athens, GA, USA, 1992; pp. 44–50. [Google Scholar]
- Větrovský, T.; Baldrian, P.; Morais, D. SEED 2: A user-friendly platform for amplicon high-throughput sequencing data analyses. Bioinformatics 2018, 34, 2292–2294. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335. [Google Scholar] [CrossRef]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Kruskal, W.H.; Wallis, W.A. Use of ranks in one-criterion variance analysis. J. Am. Stat. Assoc. 1952, 47, 583–621. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Bonferroni, C.E. Teoria Statistica Delle Classi e Calcolo Delle Probabilità; Libreria Internazionale Seeber: Florence, Italy, 1936. [Google Scholar]
- Landau, S.; Rabe-Hesketh, S. Software review: StatView for windows, version 5.0. Stat. Methods Med. Res. 1999, 8, 337–341. [Google Scholar] [CrossRef] [PubMed]
- Santos-Beneit, F. The Pho regulon: A huge regulatory network in bacteria. Front. Microbiol. 2015, 6, 402. [Google Scholar] [CrossRef] [PubMed]
- Epstein, W. The roles and regulation of potassium in bacteria. Prog. Nucleic Acid Res. 2003, 75, 293–320. [Google Scholar]
- Meyer, J.M. Pyoverdine Siderophores as Taxonomic and Phylogenic Markers. Pseudomonas 2010, 201–233. [Google Scholar] [CrossRef]
- Rivas, R.; Peix, A.; Mateos, P.F.; Trujillo, M.E.; Martínez-Molina, E.; Velázquez, E. Biodiversity of populations of phosphate solubilizing rhizobia that nodulates chickpea in different Spanish soils. Plant. Soil 2006, 287, 23–33. [Google Scholar] [CrossRef]
- Germida, J.J.; Siciliano, S.D.; Renato de Freitas, J.; Seib, A.M. Diversity of root-associated bacteria associated with field-grown canola (Brassica napus L.) and wheat (Triticum aestivum L.). FEMS Microbiol. Ecol. 1998, 26, 43–50. [Google Scholar] [CrossRef]
- Dunfield, K.E.; Germida, J.J. Diversity of bacterial communities in the rhizosphere and root interior of field-grown genetically modified Brassica napus. FEMS Microbiol. Ecol. 2001, 38, 1–9. [Google Scholar] [CrossRef]
- Granér, G.; Persson, P.; Meijer, J.; Alström, S. A study on microbial diversity in different cultivars of Brassica napus in relation to its wilt pathogen, Verticillium longisporum. FEMS Microbiol. Lett. 2003, 224, 269–276. [Google Scholar] [CrossRef]
- Zhang, Y.F.; He, L.Y.; Chen, Z.J.; Wang, Q.Y.; Qian, M.; Sheng, X.F. Characterization of ACC deaminase-producing endophytic bacteria isolated from copper-tolerant plants and their potential in promoting the growth and copper accumulation of Brassica napus. Chemosphere 2011, 83, 57–62. [Google Scholar] [CrossRef]
- Croes, S.; Weyens, N.; Janssen, J.; Vercampt, H.; Colpaert, J.V.; Carleer, R. Bacterial communities associated with Brassica napus L. grown on trace element-contaminated and non-contaminated fields: A genotypic and phenotypic comparison. Microb. Biotechnol. 2013, 6, 371–384. [Google Scholar] [CrossRef] [PubMed]
- Montalbán, B.; Croes, S.; Weyens, N.; Lobo, M.C.; Pérez-Sanz, A.; Vangronsveld, J. Characterization of bacterial communities associated with Brassica napus L. growing on a Zn-contaminated soil and their effects on root growth. Int. J. Phytoremediat. 2016, 18, 985–993. [Google Scholar] [CrossRef]
- Gkarmiri, K.; Mahmood, S.; Ekblad, A.; Alström, S.; Högberg, N.; Finlay, R. Identifying the active microbiome associated with roots and rhizosphere soil of oilseed rape. Appl. Environ. Microbiol. 2017, 83, e01938-17. [Google Scholar] [CrossRef] [PubMed]
- Larcher, M.; Rapior, S.; Cleyet-Marel, J.C. Bacteria from the rhizosphere and roots of Brassica napus influence its root growth promotion by Phyllobacterium brassicacearum. Acta Bot. Gallica 2008, 155, 355–366. [Google Scholar] [CrossRef]
- Lay, C.Y.; Bell, T.H.; Hamel, C.; Harker, K.N.; Mohr, R.; Greer, C.W. Canola Root–Associated Microbiomes in the Canadian Prairies. Front. Microbiol. 2018, 9, 1188. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; He, L.Y.; Wang, Q.; Sheng, X.F. Synergistic effects of plant growth-promoting Neorhizobium huautlense T1-17 and immobilizers on the growth and heavy metal accumulation of edible tissues of hot pepper. J. Hazard. Mater. 2016, 312, 123–131. [Google Scholar] [CrossRef]
- Msaddak, A.; Rejili, M.; Durán, D.; Rey, L.; Imperial, J.; Palacios, J.M. Members of Microvirga and Bradyrhizobium genera are native endosymbiotic bacteria nodulating Lupinus luteus in Northern Tunisian soils. FEMS Microbiol. Ecol. 2017, 93. [Google Scholar] [CrossRef] [PubMed]
- Dall’Asta, P.; Velho, A.C.; Pereira, T.P.; Stadnik, M.J.; Arisi, A.C.M. Herbaspirillum seropedicae promotes maize growth but fails to control the maize leaf anthracnose. Physiol. Mol. Biol. Plant. 2019, 25, 167–176. [Google Scholar] [CrossRef]
- Mouazen, A.M.; Kuang, B. On-line visible and near infrared spectroscopy for in-field phosphorus management. Soil Tillage Res. 2016, 155, 471–477. [Google Scholar] [CrossRef]
- Beneduzi, A.; Ambrosini, A.; Passaglia, L.M.P. Plant growth-promoting rhizobacteria (PGPR): Their potential as antagonists and biocontrol agents. Genet. Mol. Biol. 2012, 35, 1044–1051. [Google Scholar] [CrossRef]
- Jiménez-Gómez, A.; Flores-Félix, J.D.; García-Fraile, P.; Mateos, P.F.; Menéndez, E.; Velázquez, E.; Rivas, R. Probiotic activities of Rhizobium laguerreae on growth and quality of spinach. Sci. Rep. 2018, 8, 295. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.X.; Li, M.A.; Ming-Hui, C.; Jia-Qin, X.I.; Feng, H.E.; Chang-Qun, D. Phosphate solubilizing ability and phylogenetic diversity of bacteria from P-rich soils around Dianchi Lake drainage area of China. Pedosphere 2012, 22, 707–716. [Google Scholar] [CrossRef]
- Li, X.J.; Tang, H.Y.; Duan, J.L.; Gao, J.M.; Xue, Q.H. Bioactive alkaloids produced by Pseudomonas brassicacearum subsp. neoaurantiaca, an endophytic bacterium from Salvia miltiorrhiza. Nat. Prod. Res. 2013, 27, 496–499. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Mavrodi, D.V.; Ke, L.; Mavrodi, O.V.; Yang, M.; Thomashow, L.S. Biocontrol and plant growth-promoting activity of rhizobacteria from Chinese fields with contaminated soils. Microb. Biotechnol. 2015, 8, 404–418. [Google Scholar] [CrossRef] [PubMed]
- Matthijs, S.; Brandt, N.; Ongena, M.; Achouak, W.; Meyer, J.M.; Budzikiewicz, H. Pyoverdine and histicorrugatin-mediated iron acquisition in Pseudomonas thivervalensis. Biometals 2016, 29, 467–485. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, B.N.; Allay, S.; Chakraborty, A.P.; Chakraborty, U. PGPR in managing root rot disease and enhancing growth in mandarin (Citrus reticulata Blanco.) seedlings. J. Hortic. Sci. 2017, 11, 104–115. [Google Scholar]
- Ortiz-Ojeda, P.; Ogata-Gutiérrez, K.; Zúñiga-Dávila, D. Evaluation of plant growth promoting activity and heavy metal tolerance of psychrotrophic bacteria associated with maca (Lepidium meyenii Walp.) rhizosphere. AIMS Microbiol. 2017, 3, 279–292. [Google Scholar] [CrossRef]
- Park, Y.G.; Mun, B.G.; Kang, S.M.; Hussain, A.; Shahzad, R.; Seo, C.W. Bacillus aryabhattai SRB02 tolerates oxidative and nitrosative stress and promotes the growth of soybean by modulating the production of phytohormones. PLoS ONE 2017, 12, e0173203. [Google Scholar] [CrossRef]
- Naili, F.; Neifar, M.; Elhidri, D.; Cherif, H.; Bejaoui, B.; Aroua, M. Optimization of the effect of PGPR–based biofertlizer on wheat growth and yield. Biom. Biostat. Int. J. 2018, 7, 226–232. [Google Scholar] [CrossRef]
- Cézard, C.; Farvacques, N.; Sonnet, P. Chemistry and biology of pyoverdines, Pseudomonas primary siderophores. Curr. Med. Chem. 2015, 22, 165–186. [Google Scholar] [CrossRef]
- Trapet, P.; Avoscan, L.; Klinguer, A.; Pateyron, S.; Citerne, S.; Chervin, C. The Pseudomonas fluorescens siderophore pyoverdine weakens Arabidopsis thaliana defense in favor of growth in iron-deficient conditions. Plant. Physiol. 2016, 171, 675–693. [Google Scholar] [CrossRef] [PubMed]
- Kuzmanović, N.; Eltlbany, N.; Ding, G.; Baklawa, M.; Min, L.; Wei, L. Analysis of the genome sequence of plant beneficial strain Pseudomonas sp. RU47. J. Biotechnol. 2018, 281, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.K.; Singh, P.P.; Gupta, A.; Singh, A.K.; Keshri, J. Tolerance of Heavy Metal Toxicity Using PGPR Strains of Pseudomonas Species. In PGPR Amelioration in Sustainable Agriculture; Singh, K., Ed.; Woodhead Publishing: Sawston, UK, 2019; pp. 239–252. [Google Scholar]
- Nascimento, F.X.; Rossi, M.J.; Glick, B.R. Ethylene and 1-Aminocyclopropane-1-carboxylate (ACC) in plant–bacterial interactions. Front. Plant. Sci. 2018, 9, 114. [Google Scholar] [CrossRef] [PubMed]
- Glick, B.R.; Todorovic, B.; Czarny, J.; Cheng, Z.; Duan, J.; McConkey, B. Promotion of plant growth by bacterial ACC deaminase. Crit. Rev. Plant Sci. 2007, 26, 227–242. [Google Scholar] [CrossRef]
- Ciocchini, A.E.; Roset, M.S.; Briones, G.; de Iannino, N.I.; Ugalde, R.A. Identification of active site residues of the inverting glycosyltransferase Cgs required for the synthesis of cyclic β-1, 2-glucan, a Brucella abortus virulence factor. Glycobiology 2006, 16, 679–691. [Google Scholar] [CrossRef] [PubMed]
- Glick, B.R. Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol. Res. 2014, 169, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Skorupska, A.; Janczarek, M.; Marczak, M.; Mazur, A.; Król, J. Rhizobial exopolysaccharides: Genetic control and symbiotic functions. Microb. Cell Fact. 2006, 5, 7. [Google Scholar] [CrossRef]
- Henrissat, B.; Sulzenbacher, G.; Bourne, Y. Glycosyltransferases, glycoside hydrolases: Surprise. Curr. Opin. Struct. Biol. 2008, 18, 527–533. [Google Scholar] [CrossRef]
- Rodríguez-Navarro, D.N.; Dardanelli, M.S.; Ruíz-Saínz, J.E. Attachment of bacteria to the roots of higher plants. FEMS Microbiol. Lett. 2007, 272, 127–136. [Google Scholar] [CrossRef]
- Bogino, P.; Oliva, M.; Sorroche, F.; Giordano, W. The role of bacterial biofilms and surface components in plant-bacterial associations. Int. J. Mol. Sci. 2013, 14, 15838–15859. [Google Scholar] [CrossRef]
- Yang, J.; Tauschek, M.; Robins-Browne, R.M. Control of bacterial virulence by AraC-like regulators that respond to chemical signals. Trends Microbiol. 2011, 19, 128–135. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Fraile, P.; Seaman, J.C.; Karunakaran, R.; Edwards, A.; Poole, P.S.; Downie, J.A. Arabinose and protocatechuate catabolism genes are important for growth of Rhizobium leguminosarum biovar viciae in the pea rhizosphere. Plant Soil 2015, 390, 251–264. [Google Scholar] [CrossRef]
- Achouak, W.; Sutra, L.; Heulin, T.; Meyer, J.M.; Fromin, N.; Degraeve, S. Pseudomonas brassicacearum sp. nov. and Pseudomonas thivervalensis sp. nov., two root-associated bacteria isolated from Brassica napus and Arabidopsis thaliana. Int. J. Syst. Evol. Microbiol. 2000, 50, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Chung, B.S.; Aslam, Z.; Kim, S.W.; Kim, G.G.; Kang, H.S.; Ahn, J.W. A bacterial endophyte, Pseudomonas brassicacearum YC5480, isolated from the root of Artemisia sp. producing antifungal and phytotoxic compounds. Plant Pathol. J. 2008, 24, 461–468. [Google Scholar] [CrossRef]
- Pereira, S.I.A.; Monteiro, C.; Vega, A.L.; Castro, P.M. Endophytic culturable bacteria colonizing Lavandula dentata L. plants: Isolation, characterization and evaluation of their plant growth-promoting activities. Ecol. Eng. 2016, 87, 91–97. [Google Scholar] [CrossRef]
- Deng, Z.S.; Zhao, L.F.; Kong, Z.Y.; Yang, W.Q.; Lindström, K.; Wang, E.T. Diversity of endophytic bacteria within nodules of the Sphaerophysa salsula in different regions of Loess Plateau in China. FEMS Microbiol. Ecol. 2011, 76, 463–475. [Google Scholar] [CrossRef] [PubMed]
- Safronova, V.I.; Stepanok, V.V.; Engqvist, G.L.; Alekseyev, Y.V.; Belimov, A.A. Root-associated bacteria containing 1-aminocyclopropane-1-carboxylate deaminase improve growth and nutrient uptake by pea genotypes cultivated in cadmium supplemented soil. Biol. Fertil. Soil 2006, 42, 267–272. [Google Scholar] [CrossRef]
- Long, H.H.; Schmidt, D.D.; Baldwin, I.T. Native bacterial endophytes promote host growth in a species-specific manner; phytohormone manipulations do not result in common growth responses. PLoS ONE 2008, 3, e2702. [Google Scholar] [CrossRef]
- Kong, Z.; Deng, Z.; Glick, B.R.; Wei, G.; Chou, M. A nodule endophytic plant growth-promoting Pseudomonas and its effects on growth, nodulation and metal uptake in Medicago lupulina under copper stress. Ann. Microbiol. 2017, 67, 49–58. [Google Scholar] [CrossRef]
- Zachow, C.; Müller, H.; Monk, J.; Berg, G. Complete genome sequence of Pseudomonas brassicacearum strain L13-6-12, a biological control agent from the rhizosphere of potato. Stand. Genomic Sci. 2017, 12, 6. [Google Scholar] [CrossRef]
- Zengerer, V.; Schmid, M.; Bieri, M.; Müller, D.C.; Remus-Emsermann, M.N.; Ahrens, C.H. Pseudomonas orientalis F9: A potent antagonist against phytopathogens with phytotoxic effect in the apple flower. Front. Microbiol. 2018, 9, 145. [Google Scholar] [CrossRef] [PubMed]
- Madani, H.; Malboobi, M.A.; Bakhshkelarestaghi, K.; Stoklosa, A. Biological and Chemical Phosphorus Fertilizers Effect on Yield and P Accumulation in Rapeseed (Brassica napus L.). Not. Bot. Hortic. Agrobot. Cluj-Napoca 2012, 40, 210–214. [Google Scholar] [CrossRef]
- Mohammadi, K.; Rokhzadi, A. An integrated fertilization system of canola (Brassica napus L.) production under different crop rotations. Ind. Crops Prod. 2012, 37, 264–269. [Google Scholar] [CrossRef]
- Valetti, L.; Iriarte, L.; Fabra, A. Growth promotion of rapeseed (Brassica napus) associated with the inoculation of phosphate solubilizing bacteria. Appl. Soil Ecol. 2018, 132, 1–10. [Google Scholar] [CrossRef]
- Qiao, J.; Yu, X.; Liang, X.; Liu, Y.; Borriss, R.; Liu, Y. Addition of plant-growth-promoting Bacillus subtilis PTS-394 on tomato rhizosphere has no durable impact on composition of root microbiome. BMC Microbiol. 2017, 17, 131. [Google Scholar] [CrossRef] [PubMed]
- Gadhave, K.R.; Devlin, P.F.; Ebertz, A.; Ross, A.; Gange, A.C. Soil inoculation with Bacillus spp. modifies root endophytic bacterial diversity, evenness, and community composition in a context-specific manner. Microb. Ecol. 2018, 76, 741–750. [Google Scholar] [CrossRef] [PubMed]
- Compant, S.; Samad, A.; Faist, H.; Sessitsch, A. A review on the plant microbiome: Ecology, functions and emerging trends in microbial application. J. Adv. Res. 2019, 19, 29–37. [Google Scholar] [CrossRef]
- Van der Heijden, M.G.; Hartmann, M. Networking in the plant microbiome. PLoS Biol. 2016, 14, e1002378. [Google Scholar] [CrossRef]
- Cordero, J.; de Freitas, J.R.; Germida, J.J. Bacterial microbiome associated with the rhizosphere and root interior of crops in Saskatchewan, Canada. Can. J. Microbiol. 2020, 66, 71–85. [Google Scholar] [CrossRef]
- Berg, G.; Smalla, K. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol. Ecol. 2009, 68, 1–13. [Google Scholar] [CrossRef]
- Hartmman, K.; Tringe, S.G. Interactions between plants and soil shaping the root microbiome under abiotic stress. Biochem. J. 2019, 476, 2705–2724. [Google Scholar] [CrossRef] [PubMed]
- Naylor, D.; DeGraaf, S.; Purdom, E.; Coleman-Derr, D. Drought and host selection influence bacterial community dynamics in the grass root microbiome. ISME J. 2017, 11, 2691–2704. [Google Scholar] [CrossRef] [PubMed]

| Strain | Bacterial Growth Medium | 879F * | Most Closely Related Type Strain Based on the 16S rRNA Gene | % Similarity with the Most Closely Related Type Strain (16S rRNA) | Taxonomy | Siderophores | Cellulose | P Solub |
|---|---|---|---|---|---|---|---|---|
| CDVBN92A | 869 1/10 | I | Pseudarthrobacter oxydans ATCC 14358T | - | Actinobacteria, Actinobacteria, Micrococcales, Micrococcaceae | |||
| CDVBN98 * | 869 1/10 | I | Pseudarthrobacter oxydans ATCC 14358T | 99.58 | Actinobacteria, Actinobacteria, Micrococcales, Micrococcaceae | |||
| CDVBN100 * | 869 1/10 | II | Isoptericola nanjingensis H17T | 97.42 | Actinobacteria, Actinobacteria, Micrococcales, Promicromonosporaceae | |||
| PDABN24A * | YMA | IV | Dermacoccus nishinomiyaensis DSM 20448T | 99.35 | Actinobacteria, Actinobacteria, Micrococcales, Dermacoccaceae | |||
| CDVBN92B * | 869 1/10 | V | Agromyces ramosus DSM 43045T | 99.45 | Actinobacteria, Actinobacteria, Micrococcales, Microbacteriaceae | |||
| CDVBN29 * | YMA | VI | Clavibacter capsici LMG 29047T | 99.93 | Actinobacteria, Actinobacteria, Micrococcales, Microbacteriaceae | |||
| CDVBN34 | TSA | VI | Clavibacter capsici LMG 29047T | - | Actinobacteria, Actinobacteria, Micrococcales, Microbacteriaceae | |||
| CDVBN89* | 869 1/10 | VII | Microbacterium yannicii DSM 23203T | 98.95 | Actinobacteria, Actinobacteria, Micrococcales, Microbacteriaceae | |||
| CDVBN50 * | 869 1/10 | VIII | Microbacterium yannicii G72T | 100 | Actinobacteria, Actinobacteria, Micrococcales, Microbacteriaceae | |||
| CDVBN46A | 869 1/10 | IX | Arthrobacter humícola KV-653T | - | Actinobacteria, Actinobacteria, Micrococcales, Micrococcaceae | |||
| CDVBN60 * | 869 1/10 | IX | Arthrobacter humícola KV-653T | 99.71 | Actinobacteria, Actinobacteria, Micrococcales, Micrococcaceae | |||
| CDVBN84 * | TSA | X | Arthrobacter pascens DSM 20545T | 98.73 | Actinobacteria, Actinobacteria, Micrococcales, Micrococcaceae | |||
| PDABN28 * | 869 1/10 | XI | Micrococcus yunnanensis YIM 65004T | 99.57 | Actinobacteria, Actinobacteria, Micrococcales, Micrococcaceae | |||
| CDVBN49 * | 869 1/10 | XII | Pseudarthrobacter oxydans ATCC 14358T | 99.58 | Actinobacteria, Actinobacteria, Micrococcales, Micrococcaceae | |||
| CDVBN42 * | 869 1/10 | XIII | Pseudarthrobacter oxydans ATCC 14358T | 99.58 | Actinobacteria, Actinobacteria, Micrococcales, Micrococcaceae | |||
| CDVBN43 | 869 1/10 | XIII | Pseudarthrobacter oxydans ATCC 14358T | - | Actinobacteria, Actinobacteria, Micrococcales, Micrococcaceae | |||
| CDVBN44 | 869 1/10 | XIII | Pseudarthrobacter oxydans ATCC 14358T | - | Actinobacteria, Actinobacteria, Micrococcales, Micrococcaceae | |||
| CDVBN53 * | 869 1/10 | XIV | Pseudarthrobacter oxydans ATCC 14358T | 99.58 | Actinobacteria, Actinobacteria, Micrococcales, Micrococcaceae | |||
| CDVBN73 | 869 1/10 | XIV | Pseudarthrobacter oxydans ATCC 14358T | - | Actinobacteria, Actinobacteria, Micrococcales, Micrococcaceae | |||
| CDVBN57 * | 869 1/10 | XV | Pseudarthrobacter oxydans ATCC 14358T | 99.58 | Actinobacteria, Actinobacteria, Micrococcales, Micrococcaceae | |||
| CDVBN61 | 869 1/10 | XV | Pseudarthrobacter oxydans ATCC 14358T | - | Actinobacteria, Actinobacteria, Micrococcales, Micrococcaceae | |||
| CDVBN51 * | 869 1/10 | XVI | Pseudarthrobacter oxydans ATCC 14358T | 99.58 | Actinobacteria, Actinobacteria, Micrococcales, Micrococcaceae | |||
| CDVBN33 * | TSA | XVII | Pseudarthrobacter siccitolerans LMG 27359T | 99.44 | Actinobacteria, Actinobacteria, Micrococcales, Micrococcaceae | |||
| CDVBN72 * | 869 1/10 | XVIII | Nocardioides cavernae YIM A1136T | 99.36 | Actinobacteria, Actinobacteria, Propionibacteriales, Nocardioidaceae | |||
| CDVBN90 * | 869 1/10 | XIX | Nocardioides cavernae YIM A1136T | 99.36 | Actinobacteria, Actinobacteria, Propionibacteriales, Nocardioidaceae | |||
| CDVBN101 | 869 1/10 | XIX | Nocardioides cavernae YIM A1136T | - | Actinobacteria, Actinobacteria, Propionibacteriales, Nocardioidaceae | |||
| CDVBN102 * | 869 1/10 | XX | Micromonospora coxensis DSM 45161T | 99.86 | Actinobacteria; Actinobacteria; Micromonosporales; Micromonosporaceae | |||
| PDABN18 * | 869 1/10 | XXI | Flavobacterium pectinovorum DSM6368T | 99.09 | Bacteroidetes, Bacteroidetes, Flavobacteriia, Flavobacteriales, Flavobacteriaceae | |||
| PDABN27 * | 869 | XXII | Staphylococcus cohnii subsp. cohnii ATCC 29974T | 100 | Firmicutes, Bacilli, Bacillales, Staphylococcaceae | |||
| CDVBN19 * | 869 | XXIII | Staphylococcus cohnii subsp. cohnii ATCC 29974T | 99.93 | Firmicutes, Bacilli, Bacillales, Staphylococcaceae | |||
| CDVBN54 | 869 1/10 | XXIV | Bacillus aryabhattai JCM 13839T | - | Firmicutes, Bacilli, Bacillales, Bacillaceae | |||
| CDVBN55 | 869 1/10 | XXIV | Bacillus aryabhattai JCM 13839T | - | Firmicutes, Bacilli, Bacillales, Bacillaceae | |||
| CDVBN58 | 869 1/10 | XXIV | Bacillus aryabhattai JCM 13839T | - | Firmicutes, Bacilli, Bacillales, Bacillaceae | |||
| CDVBN68 * | YMA | XXIV | Bacillus aryabhattai JCM 13839T | 99.86 | Firmicutes, Bacilli, Bacillales, Bacillaceae | |||
| CDVBN9 * | 869 1/10 | XXV | Bacillus megaterium NBRC 15308T | 100 | Firmicutes, Bacilli, Bacillales, Bacillaceae | |||
| CDVBN91 * | 869 1/10 | XXVI | Bacillus niacini IFO 15566T | 99.38 | Firmicutes, Bacilli, Bacillales, Bacillaceae | |||
| PDABN29 * | 869 1/10 | XXVII | Bacillus safensis FO-36BT | 99.93 | Firmicutes, Bacilli, Bacillales, Bacillaceae | |||
| PDABN11 | TSA | XXVIII | Bacillus siamensis PD-A10T | - | Firmicutes, Bacilli, Bacillales, Bacillaceae | |||
| PDABN19B * | TSA | XXVIII | Bacillus siamensis PD-A10T | 99.86 | Firmicutes, Bacilli, Bacillales, Bacillaceae | |||
| CDVBN6 * | 869 | III | Bacillus simplex LMG 25856T | 99.93 | Firmicutes, Bacilli, Bacillales, Bacillaceae | |||
| CDVBN18 * | 869 | XXIX | Pseudomonas baetica A390T | 99.79 | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae | |||
| CDVBN66 * | YMA | XXX | Pseudomonas baetica A390T | 99.79 | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae | |||
| CDVBN28 * | YMA | XXXI | Pseudomonas baetica A390T | 99.79 | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae | |||
| CDVBN2 | YMA | XXXII | Pseudomonas baetica A390T | - | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae | |||
| CDVBN4 * | YMA | XXXII | Pseudomonas baetica A390T | 99.79 | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae | |||
| CDVBN8 * | 869 | XXXIII | Pseudomonas baetica A390T | 99.79 | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae | |||
| CDVBN41 * | 869 1/10 | XXXIV | Pseudomonas baetica A390T | 99.79 | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae | |||
| CDVBN45 | 869 1/10 | XXXIV | Pseudomonas baetica A390T | - | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae | |||
| CDVBN38 * | 869 1/10 | XXXV | Pseudomonas baetica A390T | 99.79 | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae | |||
| CDVBN39 | 869 1/10 | XXXV | Pseudomonas baetica A390T | - | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae | |||
| CDVBN37 | 869 1/10 | XXXV | Pseudomonas baetica A390T | - | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae | |||
| CDVBN22 * | YMA | XXXVI | Pseudomonas baetica A390T | 99.79 | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae | |||
| CDVBN23 * | YMA | XXXVII | Pseudomonas baetica A390T | 99.79 | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae | |||
| CDVBN70 | YMA | XXXVIII | Pseudomonas baetica A390T | - | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae | |||
| CDVBN71 * | YMA | XXXVIII | Pseudomonas baetica A390T | 99.79 | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae | |||
| CDVBN13 * | TSA | XXXIX | Pseudomonas brassicacearum subsp. brassicacearum DBK11T | 99.72 | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae | |||
| CDVBN14 | TSA | XXXIX | Pseudomonas brassicacearum subsp. brassicacearum DBK11T | - | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae | |||
| CDVBN62 * | YMA | XL | Pseudomonas brassicacearum subsp. brassicacearum DBK11T | 99.79 | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae | |||
| CDVBN47 * | 869 1/10 | XLI | Pseudomonas brassicacearum subsp. brassicacearum DBK11T | 99.79 | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae | |||
| CDVBN25 * | YMA | XLII | Pseudomonas brassicacearum subsp. brassicacearum DBK11T | 99.79 | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae | |||
| CDVBN27 | YMA | XLII | Pseudomonas brassicacearum subsp. brassicacearum DBK11T | - | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae | |||
| CDVBN52 * | 869 1/10 | XLIII | Pseudomonas brassicacearum subsp. neoaurantiaca ATCC 49054T | 99.79 | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae | |||
| CDVBN64 * | YMA | XLIV | Pseudomonas brassicacearum subsp. neoaurantiaca ATCC 49054T | 99.79 | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae | |||
| CDVBN26 * | YMA | XLV | Pseudomonas brassicacearum subsp. neoaurantiaca ATCC 49054T | 99.79 | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae | |||
| CDVBN108 | TSA | XLVI | Pseudomonas brassicacearum subsp. neoaurantiaca ATCC 49054T | - | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae | |||
| CDVBN21 * | TSA | XLVI | Pseudomonas brassicacearum subsp. neoaurantiaca ATCC 49054T | 99.86 | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae | |||
| CDVBN10 * | 869 | XLVII | Pseudomonas brassicacearum subsp. neoaurantiaca ATCC 49054T | 99.86 | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae | |||
| CDVBN17 | 869 | XLVII | Pseudomonas brassicacearum subsp. neoaurantiaca ATCC 49054T | - | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae | |||
| CDVBN24 | YMA | XLVII | Pseudomonas brassicacearum subsp. neoaurantiaca ATCC 49054T | - | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae | |||
| CDVBN11 | TSA | XLVII | Pseudomonas brassicacearum subsp. neoaurantiaca ATCC 49054T | - | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae | |||
| CDVBN15 | TSA | XLVII | Pseudomonas brassicacearum subsp. neoaurantiaca ATCC 49054T | - | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae | |||
| CDVBN1 * | YMA | XLVIII | Pseudomonas brassicacearum subsp. neoaurantiaca ATCC 49054T | 99.86 | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae | |||
| CDVBN69 * | YMA | XLIX | Pseudomonas brassicacearum subsp. neoaurantiaca ATCC 49054T | 99.86 | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae | |||
| CDVBN65 * | YMA | L | Pseudomonas orientalis CFML96-170T | 99.86 | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae | |||
| CDVBN3 * | YMA | LI | Pseudomonas orientalis CFML96-170T | 99.65 | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae | |||
| CDVBN20 * | 869 | LII | Pseudomonas orientalis CFML96-170T | 99.79 | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae | |||
| PDABN1 * | TSA | LIII | Pseudomonas poae DSM 14936T | 100 | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae | |||
| PDABN14 * | YMA | LIV | Pseudomonas poae DSM 14936T | 100 | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae | |||
| PDABN5 * | 869 | LV | Pseudomonas thivervalensis DSM 13194T | 99.86 | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae | |||
| PDABN12 | YMA | LV | Pseudomonas thivervalensis DSM 13194T | - | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae | |||
| PDABN3 * | 869 1/10 | LVI | Pseudomonas thivervalensis DSM 13194T | 99.86 | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae | |||
| PDABN4 | 869 | LVI | Pseudomonas thivervalensis DSM 13194T | - | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae | |||
| PDABN6 | YMA | LVI | Pseudomonas thivervalensis DSM 13194T | - | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae | |||
| PDABN7 | YMA | LVI | Pseudomonas thivervalensis DSM 13194T | - | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae | |||
| PDABN8 | YMA | LVI | Pseudomonas thivervalensis DSM 13194T | - | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae | |||
| PDABN13 | YMA | LVI | Pseudomonas thivervalensis DSM 13194T | - | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae | |||
| PDABN15 | YMA | LVI | Pseudomonas thivervalensis DSM 13194T | - | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae | |||
| PDABN2 | YMA | LVI | Pseudomonas thivervalensis DSM 13194T | - | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae | |||
| CDVBN16 * | 869 1/10 | LVII | Pseudomonas thivervalensis DSM 13194T | 99.86 | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae | |||
| PDABN26 * | YMA | LVIII | Bosea lathyri DSM 26656T | 99.22 | Proteobacteria, Alphaproteobacteria, Rhizobiales, Bradyrhizobiaceae | |||
| CDVBN78 * | 869 1/10 | LIX | Devosia psychrophila Cr7-05T | 99.09 | Proteobacteria, Alphaproteobacteria, Rhizobiales, Hyphomicrobiaceae | |||
| CDVBN77 * | 869 1/10 | LX | Microvirga aerophila KACC 12743T | 97.64 | Proteobacteria, Alphaproteobacteria, Rhizobiales, Methylobacteriaceae | |||
| PDABN20 * | YMA | LXI | Neorhizobium alkalisoli CCBAU 01393T | 99.76 | Proteobacteria, Alphaproteobacteria, Rhizobiales, Rhizobiaceae | |||
| PDABN21 | YMA | LXI | Neorhizobium alkalisoli CCBAU 01393T | - | Proteobacteria, Alphaproteobacteria, Rhizobiales, Rhizobiaceae | |||
| PDABN21B * | 869 1/10 | LXII | Agrobacterium nepotum 39/7T | 100 | Proteobacteria, Alphaproteobacteria, Rhizobiales, Rhizobiaceae | |||
| PDABN22B * | 869 1/10 | LXIII | Agrobacterium nepotum 39/7T | 100 | Proteobacteria, Alphaproteobacteria, Rhizobiales, Rhizobiaceae | |||
| PDABN19A * | 869 | LXIV | Shinella kummerowiae CCBAU 25048T | 98.53 | Proteobacteria, Alphaproteobacteria, Rhizobiales, Rhizobiaceae | |||
| PDABN23 * | 869 1/10 | LXV | Shinella kummerowiae CCBAU 25048T | 98.76 | Proteobacteria, Alphaproteobacteria, Rhizobiales, Rhizobiaceae | |||
| PDABN24B | YMA | LXV | Shinella kummerowiae CCBAU 25048T | - | Proteobacteria, Alphaproteobacteria, Rhizobiales, Rhizobiaceae | |||
| PDABN32 * | TSA | LXVI | Shinella kummerowiae CCBAU 25048T | 99.76 | Proteobacteria, Alphaproteobacteria, Rhizobiales, Rhizobiaceae | |||
| PDABN23A * | TSA | LXVII | Shinella kummerowiae CCBAU 25048T | 99.76 | Proteobacteria, Alphaproteobacteria, Rhizobiales, Rhizobiaceae | |||
| CDVBN83 * | YMA | LXVIII | Sphingomonas faeni DSM 14747T | 99.78 | Proteobacteria, Alphaproteobacteria, Sphingomonadales, Sphingomonadaceae | |||
| CDVBN46B * | 869 1/10 | LXIX | Massilia suwonensis 5414S-25T | 99.01 | Proteobacteria, Betaproteobacteria, Burkholderiales, Oxalobacteraceae | |||
| CDVBN40 * | 869 1/10 | LXX | Massilia yuzhufengensis ZD1-4T | 98.59 | Proteobacteria, Betaproteobacteria, Burkholderiales, Oxalobacteraceae | |||
| PDABN9 * | YMA | LXXI | Acidovorax radicis N35T | 99.65 | Proteobacteria, Betaproteobacteria, Burkholderiales, Comamonadaceae | |||
| CDVBN31 * | TSA | LXXII | Variovorax paradoxus NBRC 15149T | 99.52 | Proteobacteria, Betaproteobacteria, Burkholderiales, Comamonadaceae | |||
| CDVBN59 * | 869 1/10 | LXXIII | Herbaspirillum lusitanum LMG 21710T | 100 | Proteobacteria, Betaproteobacteria, Burkholderiales, Oxalobacteraceae | |||
| CDVBN63 | YMA | LXXIII | Herbaspirillum lusitanum LMG 21710T | - | Proteobacteria, Betaproteobacteria, Burkholderiales, Oxalobacteraceae | |||
| CDVBN67 | YMA | LXXIII | Herbaspirillum lusitanum LMG 21710T | - | Proteobacteria, Betaproteobacteria, Burkholderiales, Oxalobacteraceae | |||
| CDVBN32 * | TSA | LXXIV | Herbaspirillum lusitanum LMG 21710T | 99.45 | Proteobacteria, Betaproteobacteria, Burkholderiales, Oxalobacteraceae | |||
| PDABN25 * | YMA | LXXV | Shigella flexneri ATCC 29903T | 99.58 | Proteobacteria, Gammaproteobacteria, Enterobacterales, Enterobacteriaceae | |||
| CDVBN81 * | TSA | LXXVI | Acinetobacter johnsonii ATCC 17909T | 99.51 | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Moraxellaceae |
| Strain | IAA-like Molecules (µg·mL-1) | P Solubilization (Ca3(PO4)2) | N Fixation | Siderophores | Cellulose | P Solubilization (CaHPO4) |
|---|---|---|---|---|---|---|
| CDVBN4 | 24.53 | + | - | +++ | ++ | +++ |
| CDVBN6 | 5.34 | - | + | +++ | ++ | +++ |
| CDVBN10 * | 8.18 | + | - | +++ | ++ | +++ |
| CDVBN20 * | 75.19 | w | - | +++ | + | +++ |
| CDVBN21 | 13.72 | + | - | +++ | ++ | +++ |
| CDVBN65 | 5.14 | + | - | +++ | - | +++ |
| CDVBN68 | 10.07 | + | - | +++ | ++ | +++ |
| CDVBN69 | 8.45 | + | - | +++ | + | +++ |
| CDVBN70 | 0.00 | + | - | +++ | ++ | ++ |
| Attributes | CDVBN10 | CDVBN20 |
|---|---|---|
| Genome size (bp) | 6,180,897 | 5,666,760 |
| GC Content (%) | 60.8 | 60.6 |
| N50 value | 128,213 | 49,053 |
| L50 value | 15 | 34 |
| Number of contigs (with PEGs) | 85 | 271 |
| Number of subsystems | 403 | 393 |
| Number of coding sequences | 5773 | 5199 |
| Number of RNAs | 61 | 37 |
| Number of Genes Related to: | CDVBN10 | CDVBN20 |
|---|---|---|
| Cofactors, vitamins, prosthetic groups, pigments | 219 | 232 |
| Cell wall and capsule | 49 | 49 |
| Virulence, disease and defense | 58 | 60 |
| Potassium metabolism | 11 | 9 |
| Miscellaneous | 37 | 39 |
| Phages, prophages, transposable elements, plasmids | 8 | 3 |
| Membrane transport | 194 | 151 |
| Iron acquisition and metabolism | 19 | 52 |
| RNA metabolism | 50 | 52 |
| Nucleosides and nucleotides | 96 | 101 |
| Protein metabolism | 230 | 212 |
| Motility and Chemotaxis | 68 | 74 |
| Regulation and cell signalling | 55 | 61 |
| Secondary metabolism | 4 | 4 |
| DNA metabolism | 101 | 95 |
| Fatty acids, lipids and isoprenoids | 155 | 138 |
| Nitrogen metabolism | 55 | 19 |
| Dormancy and sporulation | 4 | 1 |
| Respiration | 133 | 111 |
| Stress response | 106 | 102 |
| Metabolism of aromatic compounds | 94 | 71 |
| Amino acids and derivatives | 548 | 481 |
| Sulfur metabolism | 24 | 14 |
| Phosphorus metabolism | 35 | 49 |
| Carbohydrates | 316 | 261 |
| Treatment | N (g/100g) | C (g/100g) | Fe (mg/kg) | K (g/100g) | P (g/100g) |
|---|---|---|---|---|---|
| Control | 3.56 ± 0.05 | 53.69 ± 0.49 | 67.34 ± 2.52 | 1.04 ± 0.01 | 0.58 ± 0.02 |
| CDVBN10 | 3.82 ± 0.07 * | 54.89 ± 0.19 * | 59.60 ± 1.50 * | 0.99 ± 0.03 | 0.65 ± 0.03 * |
| Samples | Raw Reads | Reads after Processing * | Observed OTUs | PD whole Tree | Chao-1 | Shannon | Simpson | Good´s Coverage | |
|---|---|---|---|---|---|---|---|---|---|
| CDVBN10 | A1 | 40297 | 15884 | 540 | 69.68 | 2465.58 | 5.56 | 0.75 | 0.95 |
| A2 | 49926 | 15190 | 714 | 71.17 | 2416.30 | 8.35 | 0.97 | 0.96 | |
| A3 | 50086 | 7353 | 537 | 44.89 | 1369.97 | 7.46 | 0.97 | 0.95 | |
| A4 | 30213 | 14870 | 373 | 53.98 | 2083.64 | 3.95 | 0.61 | 0.96 | |
| Uninoculated | B1 | 47285 | 2381 | 532 | 31.42 | 782.39 | 7.51 | 0.97 | 0.90 |
| B2 | 61430 | 4491 | 525 | 35.55 | 1041.56 | 7.86 | 0.99 | 0.93 | |
| B3 | 44494 | 14480 | 501 | 64.73 | 2201.01 | 5.51 | 0.77 | 0.95 | |
| B4 | 52639 | 21274 | 648 | 81.05 | 2083.64 | 7.48 | 0.94 | 0.96 | |
| Total | 376370 | 96105 | |||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiménez-Gómez, A.; Saati-Santamaría, Z.; Kostovcik, M.; Rivas, R.; Velázquez, E.; Mateos, P.F.; Menéndez, E.; García-Fraile, P. Selection of the Root Endophyte Pseudomonas brassicacearum CDVBN10 as Plant Growth Promoter for Brassica napus L. Crops. Agronomy 2020, 10, 1788. https://doi.org/10.3390/agronomy10111788
Jiménez-Gómez A, Saati-Santamaría Z, Kostovcik M, Rivas R, Velázquez E, Mateos PF, Menéndez E, García-Fraile P. Selection of the Root Endophyte Pseudomonas brassicacearum CDVBN10 as Plant Growth Promoter for Brassica napus L. Crops. Agronomy. 2020; 10(11):1788. https://doi.org/10.3390/agronomy10111788
Chicago/Turabian StyleJiménez-Gómez, Alejandro, Zaki Saati-Santamaría, Martin Kostovcik, Raúl Rivas, Encarna Velázquez, Pedro F. Mateos, Esther Menéndez, and Paula García-Fraile. 2020. "Selection of the Root Endophyte Pseudomonas brassicacearum CDVBN10 as Plant Growth Promoter for Brassica napus L. Crops" Agronomy 10, no. 11: 1788. https://doi.org/10.3390/agronomy10111788
APA StyleJiménez-Gómez, A., Saati-Santamaría, Z., Kostovcik, M., Rivas, R., Velázquez, E., Mateos, P. F., Menéndez, E., & García-Fraile, P. (2020). Selection of the Root Endophyte Pseudomonas brassicacearum CDVBN10 as Plant Growth Promoter for Brassica napus L. Crops. Agronomy, 10(11), 1788. https://doi.org/10.3390/agronomy10111788







