Phenotypic Divergence Analysis in Pigeonpea [Cajanus cajan (L.) Millspaugh] Germplasm Accessions
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Study Sites
2.3. Experimental Design and Data Collection
2.4. Statistical Analysis
3. Results
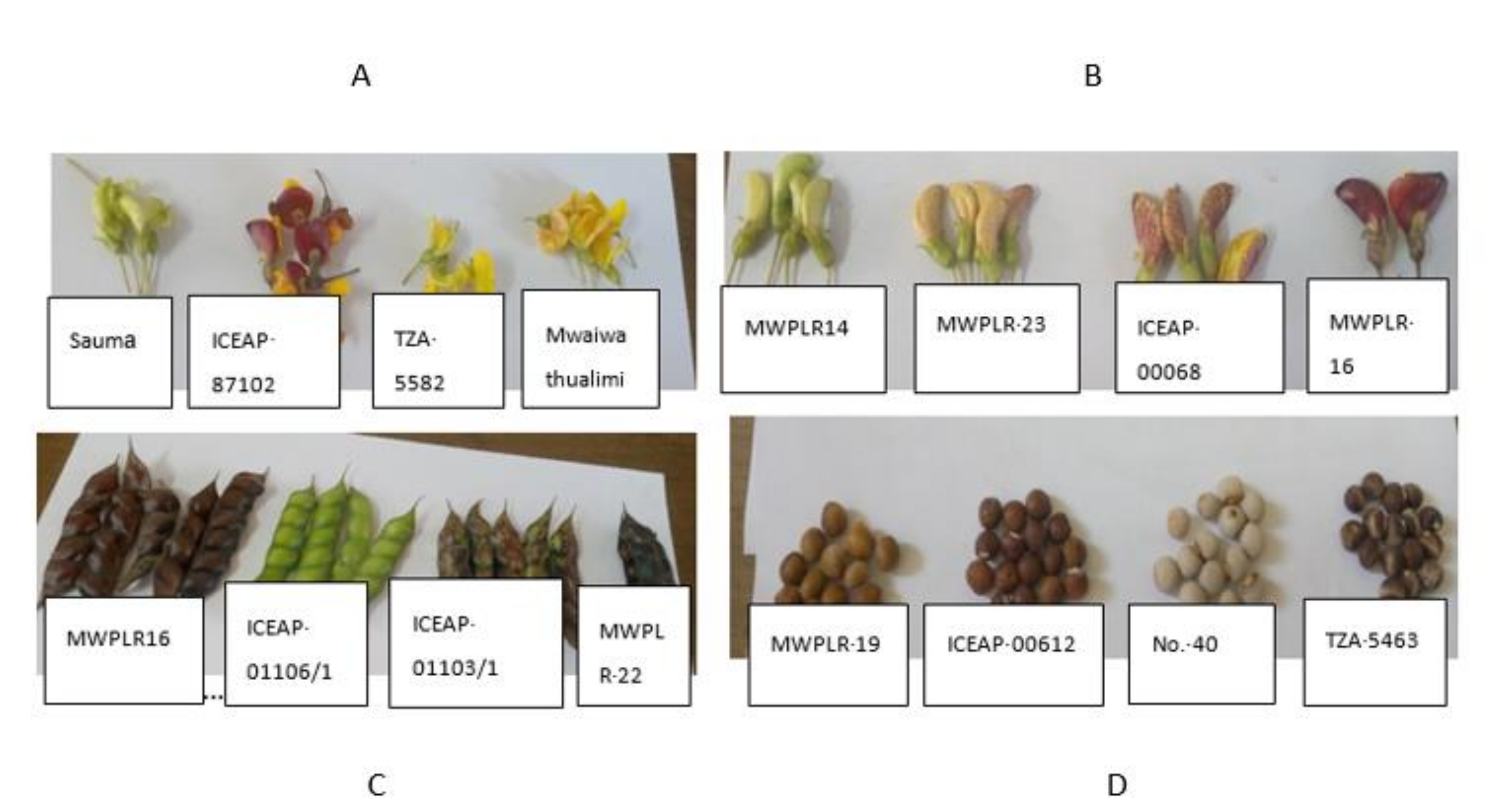
3.1. Genotype Variation Based on Qualitative Traits
3.2. Genotype and Environment Variances for Quantitative Traits
3.3. Mean Performance of Pigeonpea Genotypes across the Test Environments
3.4. Correlation Analysis among Phenotypic Traits
3.5. Nonlinear Principal Component (PC) and Cluster Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ade-Omowaye, B.; Tucker, G.; Smetanska, I. Nutritional potential of nine underexploited legumes in Southwest Nigeria. Int. Food. Res. J. 2015, 22, 798. [Google Scholar]
- Subbarao, G.; Chauhan, Y.; Johansen, C. Patterns of osmotic adjustment in pigeonpea—Its importance as a mechanism of drought resistance. Eur. J. Agron. 2000, 12, 239–249. [Google Scholar] [CrossRef]
- Giller, K.E. Nitrogen Fixation in Tropical Cropping Systems; Cabi: Wallingford, UK, 2001. [Google Scholar]
- Kwena, K.; Karuku, G.; Ayuke, F.; Esilaba, A. Nitrogen Deficiency in Semi-Arid Kenya: Can Pigeonpea fix it? East Afr. Agric For. J 2019, 83, 322–340. [Google Scholar] [CrossRef]
- Mula, M.; Saxena, K. Lifting the Level of Awareness on Pigeonpea—A Global Perspective; International Crops Research Institute for the Semi-Arid Tropics: Patancheru, India, 2010. [Google Scholar]
- Food and Agriculture Organisation of the United Nations. FAOSTAT. Crop Statistics. 2017. Available online: www.fao.org/faostat/en/#data/QC (accessed on 20 January 2019).
- Kananji, G.; Yohane, E.; Mviha, P.J.Z.; Siambi, M.; Silim, S. A Guide to Pigeonpea Production in Malawi; Department of Agricultural Research Services: Lilongwe, Malawi, 2016. [Google Scholar]
- Bohra, A.; Dubey, A.; Saxena, R.K.; Penmetsa, R.V.; Poornima, K.; Kumar, N.; Farmer, A.D.; Srivani, G.; Upadhyaya, H.D.; Gothalwal, R. Analysis of BAC-end sequences (BESs) and development of BES-SSR markers for genetic mapping and hybrid purity assessment in pigeonpea (Cajanus spp.). Biomed. Cent. BMC Plant Bio. 2011, 11, 56. [Google Scholar] [CrossRef] [PubMed]
- Saxena, R.K.; Von Wettberg, E.; Upadhyaya, H.D.; Sanchez, V.; Songok, S.; Saxena, K.; Kimurto, P.; Varshney, R.K. Genetic diversity and demographic history of Cajanus spp. illustrated from genome-wide SNPs. PLoS ONE 2014, 9, e88568. [Google Scholar] [CrossRef]
- Upadhyaya, H.D.; Reddy, K.N.; Sharma, S.; Dwivedi, S.L.; Ramachandran, S. Enhancing the value of genetic resources for use in pigeonpea improvement. Legume Persp. 2016, 11, 13–16. [Google Scholar]
- IBPGR; ICRISAT. Descriptors for Pigeonpea [Cajanus cajan (L.) Millsp.]; International Board of Plant Genetic Resources: Rome, Italy; International Crops Research Institute for Semi-Arid Tropics: Patancheru, India, 1993. [Google Scholar]
- George, D.; Mallery, P. IBM SPSS Statistics 26 Step by Step: A Simple Guide and Reference; Routledge: Abbingdon, UK, 2019. [Google Scholar]
- Payne, R.; Murray, D.; Harding, S.; Baired, D.; Soultar, D. Genstat Windows, 17th ed.; VSN International: Hemel Hempstead, UK, 2017. [Google Scholar]
- Shimelis, H.; Shiringani, R. Variance components and heritabilities of yield and agronomic traits among cowpea genotypes. Euphytica 2010, 176, 383–389. [Google Scholar] [CrossRef]
- Meulman, J.J.; Van der Kooij, A.J.; Heiser, W.J. Principal components analysis with nonlinear optimal scaling transformations for ordinal and nominal data. Sage Handb. Quant. Methodol. Soc. Sci. 2004, 3, 49–72. [Google Scholar]
- Upadhyaya, H.; Reddy, K.; Gowda, C.; Singh, S. Phenotypic diversity in the pigeonpea (Cajanus cajan) core collection. Genet. Res. Crop Evol 2007, 54, 1167–1184. [Google Scholar] [CrossRef]
- Manyasa, E.; Silim, S.; Christiansen, J. Variability patterns in Ugandan pigeonpea landraces. J. Sat. Agric. Res. 2009, 7, 1–9. [Google Scholar]
- Ayenan, M.A.T.; Ofori, K.; Ahoton, L.E.; Danquah, A. Pigeonpea [(Cajanus cajan (L.) Millsp.)] production system, farmers’ preferred traits and implications for variety development and introduction in Benin. Agric. Food Sec. 2017, 6, 48. [Google Scholar] [CrossRef]
- Grausgruber, H.; Sailer, C.; Ghambashidze, G.; Bolyos, L.; Ruckenbauer, P. Genetic variation for plant breeding. In Proceedings of the 17th EUCARPIA General Congress, Tulln, Austria, 8–11 September 2004. [Google Scholar]
- Bustos-Korts, D.; Romagosa, I.; Borràs-Gelonch, G.; Casas, A.; Slafer, G.; Van Eeuwijk, F. Genotype by Environment Interaction and Adaptation; In Encyclopedia of Sustainability Science and Technology; Springer: New York, NY, 2018; pp. 29–71. [Google Scholar]
- Vales, M.; Srivastava, R.; Sultana, R.; Singh, S.; Singh, I.; Singh, G.; Patil, S.; Saxena, K. Breeding for earliness in pigeonpea: Development of new determinate and nondeterminate lines. Crop Sci. 2012, 52, 2507–2516. [Google Scholar] [CrossRef]
- Kimaro, D. Genetic Improvement of Pigeonpea (Cajanus cajan (L.) Millsp.) for Fusarium wilt resistance in Tanzania. Ph.D. Thesis, University of Kwazulu-Natal, Pitermaritzburg, South Africa, December 2016. [Google Scholar]
- Gerrano, A.S.; Van Rensburg, W.S.J.; Mathew, I.; Shayanowako, A.I.T.; Bairu, M.W.; Venter, S.L.; Swart, W.; Mofokeng, A.; Mellem, J.J.; Labuschagne, M. Genotype and genotype 3 environment interaction effects on the grain yield performance of cowpea genotypes in dryland farming system in South Africa. Euphytica 2020, 216, 80. [Google Scholar] [CrossRef]
- Rekha, R.; Prasanthi, L.; Sekhar, M.; Priya, M. Studies on selection indices in pigeonpea [Cajanus cajan (L.) Millsp]. Int. J. Appl. Biol. Pharm. 2013, 4, 291–294. [Google Scholar]
- Saroj, S.; Singh, M.; Kumar, R.; Singh, T.; Singh, M. Genetic variability, correlation and path analysis for yield attributes in pigeonpea. Bioscan 2013, 8, 941–944. [Google Scholar]
- Sreelakshmi, C.; Shivani, D.; Kumar, C. Genetic divergence and stability analysis in Pigeonpea (Cajanus cajan L.). Electr. J. Plant Breed 2010, 1, 530–535. [Google Scholar]
- Hemavathy, A.T.; Bapu, J.; Priyadharshini, C. Principal component analysis in pigeonpea (Cajanus cajan (L.) millsp.). Electr. J. Plant Breed 2017, 8, 1133–1139. [Google Scholar] [CrossRef]
- Narayanan, S.L.; Manivannan, N.; Mahalingam, A. Correlation and Path Analyses of Yield and its Component Traits in Pigeonpea [Cajanus cajan(L.) Millsp.]. Int. J. Curr. Microbiol. App. Sci. 2018, 7, 614–618. [Google Scholar] [CrossRef]
- Choudhary, A.K.; Sultana, R.; Ontagodi, T.; Singh, I.; Bhatt, B. Recent advances in breeding pigeonpea [Cajanus cajan (L.) Millsp.]. In Proceedings of the National Conference on Global Research Initiatives for Sustainable Agriculture & Allied Sciences (GRISASS-2015), Gwalior, MP, India, 12–13 December 2015. [Google Scholar]
- Yang, S.; Pang, W.; Ash, G.; Harper, J.; Carling, J.; Wenzl, P.; Huttner, E.; Zong, X.; Kilian, A. Low level of genetic diversity in cultivated pigeonpea compared to its wild relatives is revealed by diversity arrays technology. Appl. Gen. 2006, 113, 585–595. [Google Scholar] [CrossRef] [PubMed]


| Code | Genotype Designation | Description | Source | Origin | Code | Genotype Designation | Description | Source | Origin |
|---|---|---|---|---|---|---|---|---|---|
| G1 | ICEAP 0673/1 | Breeding line | ICRISAT | Kenya | G42 | ICEAP 87105 | Cultivar | ICRISAT | Kenya |
| G2 | ICEAP 00554 | Breeding line | ICRISAT | Kenya | G43 | MWPLR 16 | Landrace | GENEBANK | Malawi |
| G3 | ICEAP 01164/1 | Breeding line | ICRISAT | Kenya | G44 | TZA 2496 | Landrace | TARI | Tanzania |
| G4 | MWPLR 19 | Landrace | GENEBANK | Malawi | G45 | TZA 5582 | Landrace | TARI | Tanzania |
| G5 | MWPLR 22 | Landrace | GENEBANK | Malawi | G46 | TZA 5596 | Landrace | TARI | Tanzania |
| G6 | ICEAP 01170 | Breeding line | ICRISAT | Kenya | G47 | Chitedze Pigeonpea 2 | Cultivar | DARS | Malawi |
| G7 | ICEAP 01169 | Breeding line | ICRISAT | Tanzania | G48 | MWPLR 7 | Landrace | GENEBANK | Malawi |
| G8 | TZA 2439 | Landrace | TARI | Tanzania | G49 | Babati | Landrace | TARI | Tanzania |
| G9 | MWPLR 9 | Landrace | GENEBANK | Malawi | G50 | TZA 5557 | Landrace | TARI | Tanzania |
| G10 | MWPLR 6 | Landrace | GENEBANK | Malawi | G51 | MWPLR 14 | Landrace | ICRISAT | Kenya |
| G11 | MWPLR 17 | Landrace | GENEBANK | Malawi | G52 | ICEAP 01101/1 | Breeding line | ICRISAT | Kenya |
| G12 | TZA 253 | Landrace | TARI | Tanzania | G53 | TZA 2456 | Landrace | TARI | Tanzania |
| G13 | MWPLR 1 | Landrace | GENEBANK | Malawi | G54 | TZA 5464 | Landrace | TARI | Tanzania |
| G14 | MWPLR 18 | Landrace | GENEBANK | Malawi | G55 | ICEAP 01101/2 | Breeding line | ICRISAT | Kenya |
| G15 | TZA 2464 | Landrace | TARI | Tanzania | G56 | ICEAP 01285 | Breeding line | ICRISAT | Kenya |
| G16 | ICEAP 00604 | Breeding line | ICRISAT | Kenya | G57 | MWPLR 25 | Landrace | GENEBANK | Malawi |
| G17 | TZA 2509 | Landrace | GENEBANK | Malawi | G58 | ICEAP 87091 | Breeding line | ICRISAT | Kenya |
| G18 | ICEAP 01146/1 | Breeding line | ICRISAT | Kenya | G59 | TZA 2692 | Landrace | TARI | Tanzania |
| G19 | MWPLR 11 | Landrace | GENEBANK | Malawi | G60 | TZA 2807 | Landrace | TARI | Tanzania |
| G20 | TZA 5555 | Landrace | TARI | Tanzania | G61 | ICEAP 00068 | Breeding line | ICRISAT | Kenya |
| G21 | No. 40 | Landrace | TARI | Tanzania | G62 | TZA 2785 | Landrace | TARI | Tanzania |
| G22 | ICEAP 01150 | Breeding line | ICRISAT | Kenya | G63 | MWPLR 10 | Landrace | GENEBANK | Malawi |
| G23 | MZ2/9 | Breeding line | TARI | Tanzania | G64 | ICEAP 00612 | Breeding line | ICRISAT | Kenya |
| G24 | ICEAP 01172/1 | Breeding line | ICRISAT | Kenya | G65 | MWPLR 21 | Landrace | GENEBANK | Malawi |
| G25 | ICEAP 01103/1 | Breeding line | ICRISAT | Kenya | G66 | TZA 2514 | Landrace | TARI | Tanzania |
| G26 | MWPLR 24 | Landrace | GENEBANK | Malawi | G67 | TZA 2466 | Landrace | TARI | Tanzania |
| G27 | ICEAP 01155 | Breeding line | ICRISAT | Kenya | G68 | ICEAP 01179 | Breeding line | ICRISAT | Kenya |
| G28 | ICEAP 01180/2 | Breeding line | ICRISAT | Malawi | G69 | MWPLR 13 | Landrace | GENEBANK | Malawi |
| G29 | MWPLR 4 | Landrace | GENEBANK | Malawi | G70 | MWPLR 2 | Landrace | GENEBANK | Malawi |
| G30 | Kachangu | Cultivar | DARS | Malawi | G71 | TZA 250 | Landrace | DARS | Malawi |
| G31 | Mwayiwathualimi | Cultivar | DARS | Kenya | G72 | MWPLR 3 | Landrace | GENEBANK | Malawi |
| G32 | MWPLR 8 | Landrace | ICRISAT | Malawi | G73 | TZA 5541 | Landrace | TARI | Tanzania |
| G33 | ICEAP 01154/2 | Breeding line | ICRISAT | Kenya | G74 | MWPLR 23 | Landrace | GENEBANK | Malawi |
| G34 | Chitedze Pigeonpea 1 | Cultivar | DARS | Malawi | G75 | ICEAP 00979/1 | Breeding line | ICRISAT | Kenya |
| G35 | ICEAP 01164 | Breeding line | ICRISAT | Kenya | G76 | TZA 197 | Landrace | TARI | Tanzania |
| G36 | Bangili | Landrace | TARI | Tanzania | G77 | MWPLR 20 | Landrace | GENEBANK | Malawi |
| G37 | ICEAP 00053 | Breeding line | ICRISAT | Kenya | G78 | HOMBOLO | Landrace | TARI | Tanzania |
| G38 | MWPLR 12 | Landrace | GENEBANK | Malawi | G79 | ICEAP 86012 | Breeding line | ICRISAT | Kenya |
| G39 | TZA5463 | Landrace | TARI | Tanzania | G80 | ICEAP 01106/1 | Breeding line | ICRISAT | Kenya |
| G40 | MWPLR 5 | Landrace | GENEBANK | Malawi | G81 | Sauma | Cultivar | DARS | Malawi |
| G41 | MWPLR 15 | Landrace | GENEBANK | Malawi |
| Site | Latitude | Longitude | Altitude (Masl) | Soil Texture | Rainfall (mm) | Min Temp (°C) | Max Temp (°C) | |||
|---|---|---|---|---|---|---|---|---|---|---|
| 2017/18 | 2018/19 | 2017/18 | 2018/19 | 2017/18 | 2018/19 | |||||
| Bvumbwe | 15°55′ S | 35°04′ E | 1228 | Sandy clay loam | 975.2 | 1442 | 16.2 | 17.9 | 22.6 | 24.9 |
| Chitedze | 13°59′ S | 33°38′ E | 1146 | Sandy clay | 929.8 | 693.4 | 18.5 | 20.2 | 24.7 | 29.4 |
| Makoka | 15°32′ S | 35°11′ E | 1029 | Sandy clay loam | 566.6 | 1184.8 | 16.3 | 15.6 | 23.2 | 28.2 |
| Traits | Code | Description |
|---|---|---|
| Qualitative Traits | ||
| Plant habit | PH | 1 = Compact (erect), 2 = semi-spreading (semi-erect) or 3 = spreading |
| Flower streak pattern | FSP | 0 = no streaks, 1 = sparse, 2 = medium and 3 = dense streaks, 4 = uniform coverage of second color |
| Flower base/main color | FBC | 1 = ivory (green white), 2 = light yellow, 3 = yellow, 4 = orange, 5 = red, 6 = purple |
| Leaf shape | LS | 1 = ovate, 2 = triangular, 3 = trullate |
| Leaf hairiness | LH | 1 = hairy, 2 = non-hairy |
| Pod form | PF | 1 = flat, 2 = cylindrical |
| Pod color | PC | 1 = green, 2 = purple, 3 = mixed (green +purple) and 4 = dark purple |
| Seed color pattern | SCP | 1 = plain, 2 = mottled, 3 = speckled, 4 = mottled and speckled, 5 = ringed |
| Seed main color | SMC | 1 = white (yellow white), 2 = cream (gray white), 3 = orange, 4 = brown, 5 = grey, 6 = purple, 7 = black |
| Seed eye color | SEC | 1 = purple, 2 = light brown, 3 = reddish brown, 4 = gray/dark, 5 = cream/white |
| Seed shape | SSH | 1 = Oval, 2 = pea-shape, 3 = square/angular, 4 = elongate |
| Quantitative Traits | ||
| Plant height | PH | Measured in cm from plant base to the tip of the main stem |
| Days to 50% flowering | DTF | Number of days from sowing until when 50% of the plants have at least one open flower |
| Primary branches | PBR | The average number of primary branches of 10 randomly selected and tagged plants |
| Secondary branches | NSB | The average number of secondary branches of 10 randomly selected and tagged plants |
| Days to 75% maturity | DTM | Number of days from sowing until when 75% of the pods in a plot turn brown |
| Number of seeds per pod | NSP | The average number of pods per plant from 10 randomly selected and tagged pods |
| Number of pods per plant | NPP | The average number of pods from 10 randomly selected and tagged plants |
| Number of racemes per plant | NRP | The average number of racemes from 10 randomly selected and tagged plants |
| Grain yield (t/ha) | GYD | Weight of the grain harvested in a plot extrapolated to t/ha |
| 100 seed weight (g) | HSWT | Weight of a random sample of 100 grain |
| Trait | Description | Frequency (%) | DF | Chi-Square | Genotype Code a |
|---|---|---|---|---|---|
| Growth habit | Compact | 11.5 | 160 | 304.52 ** | G53, G2, G1, G27, G26 |
| Semi-spreading | 61.9 | G63, G50, G28, G70, G76, G80, G51, G78, G49, G32, G62, G39, G67, G5, G8, G13, G72, G24, G74, G3,32, G22, G4, G40, G30, G52, G56, G48, G79, G36, G23,G16, G77, G7, G71, G44, G67, G46, G69, G33, G54, G20, G43, G42, G71, G62, G65,G39, G69, G17, G18, G59 | |||
| Spreading | 26.6 | G45, G41, G29, G49, G56, G64, G37, G60, G15, G11, G65, G75, G81, G44, G67, G11, G46 | |||
| Flower color | Ivory | 13.6 | 240 | 910.08 *** | G78, G40, G36, G27, G33, G80, G51 |
| Light yellow | 7.4 | G13, G5, G31 | |||
| Yellow | 64.9 | G50, G45, G70, G53, G76, G72, G24, G74, G3, G22, G4, G58, G68, G18, G19, G17, G9, G62, G29, G32, G65, G21, G52, G1, G56, G37, G48, G79. G23, G16, G61, G77, G7, G71, G44, G15, G67, G11, G69, G65, G75, G20, G43, G26, G71, G44, G15, G67, G62, G11, G46, G65 | |||
| Purple | 16.8 | G63, G28, G41, G56, G60, G25, G46, G54, G26, G42 | |||
| Flower streak pattern | No streaks | 60.5 | 320 | 589.69 *** | G17, G53, G36, G12, G15, G37, G20, G60, G9, G54, G11, G66, G55, G80, G81, G71, G73, G23, G1, G65, G21, G18, G7, G13, G51, G62, G48, G49, G58, G14, G32, G16, G2, G27, G22, G6, G57, G10, G31, G8, G39, G30 |
| Sparse streaks | 8.1 | G49, G69, G42, G33, G28, G5, G70 | |||
| Medium sparse | 1.9 | G72, G74 | |||
| Dense streaks | 14.5 | G47, G61, G29, G60, G34, G40, G45, G67, G45, G68, G63, G77, G19 | |||
| Uniform coverage | 15 | G79, G50, G76, G59, G25, G46, G78, G38, G51, G75, G26, G35, G52, G56, G41, G43 | |||
| Pod color | Green | 48.7 | 240 | 647.43 *** | G73, G42, G1, G24, G74, G75, G52, G16, G65, G21, G18, G7, G13, G62, G17, G47, G61, G15, G20, G29, G44, G72, G60, G64, G9, G11, G66, G55, G80, G71, G58, G14, G27, G6, G57, G10, G8, G19 |
| Purple | 7.1 | G76, G45, G67, G38 | |||
| Mixed (green + purple) | 33.9 | G81, G70, G53, G36, G61, G43, G37, G34, G54, G79, G50, G40, G25, G33, G46, G42, G51, G4, G68, G26, G49, G3, G35, G32, G69, G2, G63, G22, G56, G77, G41, G30 | |||
| Dark purple | 10.3 | G31, G28, G39, G48, G59, G43 | |||
| Seed color pattern | Plain | 56.6 | 240 | 841.57 *** | G59, G80, G5, G18, G6, G53, G65, G62, G35, G34, G67, GG4, G60, G66, G21, G70, G36, G42, G40, G14, G50, G66, G20, G79, G49, G2, G3, G69, G56, G81, G47, G72, G15, G44 |
| Mottled | 15.3 | G41, G25, G34, G48, G28, G78, G23, G31, G9, G37, G57 | |||
| Speckled | 22.2 | G75, G68, G43, G38, G10, G19, G52, G58, G51, G73, G59, G76, G16, G29, G13, GG3, G17, G8, G54, G1, G24, G7, G71, G27, G12, G22, G55, G77 | |||
| Mottled + speckled | 5.9 | G46, G33, G30, 632, G39, G45, G26 | |||
| Seed main color | Cream | 76.8 | 320 | 1049.31 *** | G75, G68, G59, G43, G5, G18, G6, G38, G10, G53, G65, G63, G35, G19, G34, 52, G72, G15, G44, G22, G55, G57, G77, G60, G58, G78, G32, G73, G51, G70, G36, G16, G29, G42, G40, G23, G14, G17, G8, G50, G66, G20, G49, G54, G2, G3, G69, G1, G24, G45, G7, G9, G71, G81, G12, G47 |
| Orange | 3 | G4, G46, G25 | |||
| Brown | 11 | G64, G76, G63, G30, G34, G48, G28, G31, G37, G26 | |||
| Gray | 6.2 | G80, G66, G67, G56 | |||
| Purple | 3 | G39, G33, G41 | |||
| Seed shape | Oval | 30.7 | 80 | 480.21 *** | G75, G22, G5, G25, G38, G53, G35, G34, G28, G73, G51, G70, G36, G29, G42, G40, G31, G8, G18, G49, G3, G45, G37, G28, G27, G12, G55, G57 |
| Square/angular | 69.3 | G15, G44, G22, G77, G68, G59, G43, G46, G80, G18, G33, G30, G41, G6, G10, G65, G62, G19, G34, G67, G4, G52, G48, G60, G58, G66, G32, G64, G76, G21, G16, G13, G23, G14, G63, G17, G39, G52, G66, G79, G54, G2, G69, G1, G24, G56, G7, G9, G71, G81 | |||
| Seed eye color | Purple | 20.7 | 240 | 848.32 *** | G68, G5, G34, G25, G60, G78, G51, G64, G76, G21, G16, G29, G42, G40, G31, G50, G49, G2, G69, G24, G81, G55, G57 |
| Light brown | 70.2 | G75, G59, G43, G46, G18, G33, G30, G41, G6, G10, G53, G65, G62, G35, G19, G34, G67, G52, G48, G58, G28, G66, G32, G73, G36, G23, G14, G17, G39, G74, G20, G79, G54, G1, G46, G45, G9, G71, G37, G27, G12, G47, G15, G44, G22 | |||
| Gray/dark | 1.2 | G25 | |||
| Cream | 7.5 | G80, G38, G63, G8, G7, G26 |
| Source of Variation | DF | DTF | DTM | PH | NPB | NSB | NRP | NPP | NSP | GYD | HSWT |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Location | 2 | 9024.2 *** | 8735.4 *** | 54,965 *** | 114.4 *** | 93.7 * | 226.9 *** | 3236 ** | 22.5 *** | 5,968,860 *** | 1008.1 *** |
| Replication (Rep) | 1 | 701.9 ns | 289 ns | 118 ns | 1.2 ns | 105.4 * | 14,646 ns | 9810 * | 0.45 ns | 1,663,232 * | 9.5 ns |
| Block (Rep) | 8 | 3168.5 *** | 5703.4 *** | 7710.9 ns | 52.9 * | 93.7 * | 9099 * | 6433.6 ** | 2.4 * | 16,534,356.5 *** | 72.2 ** |
| Genotype (G) | 80 | 879.2 *** | 1234.9 *** | 2137 *** | 12.5 * | 30.9 * | 5004.9 * | 1990.3 * | 0.8 ns | 351,745.3 * | 16.8 * |
| Season (S) | 1 | 3370.5 ** | 2945.3 * | 447 ns | 409.6 *** | 650.1 *** | 2,023,492 *** | 437.5 *** | 31.5 *** | 30,308,789 *** | 50.2 * |
| G × L | 160 | 243 * | 361.9 * | 1106 * | 18 * | 35.6 * | 6150.9 * | 1916.1 * | 0.9 * | 360,816.9 * | 20.7 ** |
| G × S | 80 | 3610.3 ns | 606.9 ns | 1198 ns | 17.9 * | 34.7 * | 4642.7 ns | 1060.3 * | 0.9 ns | 400,468.2 * | 14.9 ns |
| G × L × S | 160 | 330.6 ns | 484.9 ns | 744 ns | 15.2* | 34.5 * | 6110.9 ns | 1502.8 * | 0.7 ns | 919,105.3 ns | 16.2 ns |
| Residual | 469 | 345.4 | 585.8 | 1243.1 | 14.5 | 11.8 | 5822.9 | 5667.2 | 0.8 | 313,554 | 15.4 |
| Source of Variation | DF | DTF | DTM | PH | NPB | NSB | NRP | NPP | NSP | GYD | HSWT |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Site | 2 | 9167 *** | 8020 *** | 55,114 *** | 108.6 *** | 93 * | 1,309,332 *** | 118,174 *** | 22.54 *** | 1.658 *** | 80.16 *** |
| Rep | 1 | 717 | 328 | 111 | 2.7 | 109 | 14184 | 9836 | 0.45 | 1.651 | 0.26 |
| Rep (Block) | 16 | 123 | 82 | 9769 | 33.3 | 71 | 14908 | 21,509 *** | 1.35 | 0.206 | 11.78 |
| Season | 1 | 3797 ** | 3625 * | 407 | 433.3 *** | 4672 *** | 2,043,617 *** | 440,237 *** | 31.51 *** | 3.092 *** | 0.04 |
| Type | 2 | 1629 * | 4725 ** | 44,433 ** | 18.5 | 14 | 4686 | 5891 * | 40.2 *** | 2.087 *** | 20.09 |
| Site*Season | 2 | 2523 ** | 700 | 55,081 *** | 910.5 *** | 253 *** | 1,018,464 *** | 149,039 *** | 36.09 *** | 6.38 *** | 39.42 * |
| Site*Type | 4 | 114 | 4385 ** | 1257 | 17 | 29 | 988,914 * | 8167 *** | 0.41 | 0.078 | 45.83 * |
| Season*Type | 2 | 161 | 388 | 3023 | 9.9 | 2 | 642 | 380 | 1.94 | 2.006 *** | 6.44 |
| Site*Season*Type | 4 | 676 | 7883 *** | 65,810 *** | 3.1 | 44 | 1771 | 2032 | 30.17 * | 0.2 | 1.82 |
| Residual | 937 | 386 | 619 | 1177 | 15.5 | 31 | 5823 | 1682 | 0.79 | 0.149 | 12.45 |
| Genotype | DTF | DTM | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Y1 | YII | Mean | YI | YII | Mean | |||||||||
| S1 | S2 | S3 | S1 | S2 | S3 | S1 | S2 | S3 | S1 | S2 | S3 | |||
| Top Ten Genotypes | ||||||||||||||
| 21 | 129 | 131 | 141 | 124 | 131 | 132 | 131 | 173 | 191 | 211 | 158 | 176 | 176 | 181 |
| 43 | 125 | 105 | 119 | 117 | 105 | 105 | 113 | 177 | 166 | 172 | 156 | 161 | 154 | 164 |
| 51 | 63 | 65 | 64 | 87 | 67 | 98 | 74 | 95 | 105 | 102 | 127 | 116 | 132 | 113 |
| 30 | 100 | 97 | 118 | 128 | 116 | 118 | 113 | 133 | 150 | 164 | 159 | 159 | 164 | 155 |
| 45 | 107 | 96 | 91 | 128 | 101 | 124 | 108 | 143 | 158 | 146 | 170 | 153 | 165 | 156 |
| 81 | 163 | 127 | 155 | 132 | 165 | 130 | 145 | 215 | 201 | 254 | 171 | 211 | 178 | 205 |
| 17 | 147 | 120 | 125 | 109 | 120 | 106 | 121 | 182 | 167 | 174 | 156 | 160 | 147 | 164 |
| 66 | 120 | 95 | 115 | 116 | 108 | 116 | 111 | 155 | 151 | 170 | 154 | 158 | 161 | 158 |
| 74 | 118 | 78 | 123 | 113 | 115 | 118 | 110 | 163 | 145 | 166 | 153 | 165 | 163 | 159 |
| 20 | 116 | 120 | 129 | 122 | 120 | 127 | 122 | 143 | 163 | 175 | 156 | 160 | 172 | 161 |
| Bottom Five Genotypes | ||||||||||||||
| 39 | 113 | 90 | 131 | 85 | 90 | 88 | 99 | 149 | 144 | 195 | 127 | 150 | 122 | 147 |
| 13 | 126 | 117 | 109 | 116 | 107 | 115 | 115 | 167 | 166 | 153 | 145 | 154 | 155 | 156 |
| 50 | 117 | 77 | 107 | 116 | 77 | 115 | 101 | 141 | 136 | 156 | 155 | 137 | 149 | 145 |
| 42 | 114 | 102 | 127 | 120 | 102 | 120 | 114 | 145 | 154 | 172 | 164 | 166 | 162 | 160 |
| 79 | 124 | 101 | 122 | 117 | 127 | 119 | 118 | 168 | 153 | 165 | 152 | 179 | 161 | 163 |
| Mean | 117.8 | 102.8 | 115.5 | 110.6 | 106.1 | 113.1 | 110.6 | 154.7 | 156.5 | 163.2 | 148.7 | 155.7 | 154.3 | 155.3 |
| STD | 17.9 | 18.2 | 15.1 | 13.0 | 16.9 | 12.3 | 10.5 | 22.0 | 22.0 | 21.1 | 13.7 | 18.4 | 14.9 | 11.9 |
| SED± | 2.0 | 2.0 | 1.7 | 1.4 | 1.9 | 1.4 | 1.2 | 2.4 | 2.4 | 2.3 | 1.5 | 2.0 | 1.7 | 1.3 |
| CV (%) | 15.2 | 17.7 | 13.1 | 11.8 | 15.9 | 10.8 | 9.5 | 14.2 | 14.0 | 12.9 | 9.2 | 11.8 | 9.6 | 7.7 |
| Genotype | PH | NPB | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Y1 | Y11 | Mean | Y1 | Y11 | Mean | |||||||||
| S1 | S2 | S3 | S1 | S2 | S3 | S1 | S2 | S3 | S1 | S2 | S3 | |||
| Top Ten Genotypes | ||||||||||||||
| 21 | 166.5 | 220.0 | 193.0 | 160.0 | 212.8 | 193.0 | 190.9 | 19 | 19 | 17 | 14 | 18 | 12 | 16 |
| 43 | 113.5 | 147.5 | 127.5 | 96.5 | 146.7 | 148.0 | 163.7 | 14 | 15 | 17 | 14 | 17 | 11 | 15 |
| 51 | 151.5 | 109.0 | 158.0 | 234.5 | 209.4 | 149.0 | 168.6 | 13 | 12 | 14 | 18 | 13 | 11 | 13 |
| 30 | 229.5 | 188.5 | 204.0 | 170.0 | 218.5 | 204.0 | 202.4 | 15 | 13 | 18 | 15 | 16 | 15 | 15 |
| 45 | 139.5 | 144.5 | 173.0 | 161.5 | 169.4 | 197.5 | 164.2 | 15 | 13 | 22 | 15 | 17 | 14 | 16 |
| 81 | 163.0 | 222.0 | 191.0 | 160.5 | 168.1 | 194.5 | 183.2 | 13 | 17 | 19 | 18 | 12 | 14 | 15 |
| 17 | 163.5 | 164.0 | 163.5 | 100.0 | 152.1 | 156.0 | 149.9 | 15 | 14 | 21 | 17 | 16 | 13 | 16 |
| 66 | 181.5 | 177.5 | 164.0 | 161.5 | 156.8 | 149.5 | 165.1 | 12 | 13 | 13 | 14 | 16 | 12 | 13 |
| 74 | 156.0 | 195.0 | 185.5 | 124.5 | 178.7 | 164.0 | 167.3 | 15 | 18 | 17 | 20 | 18 | 12 | 17 |
| 20 | 152.5 | 163.0 | 168.5 | 138.5 | 247.5 | 166.5 | 172.8 | 10 | 12 | 20 | 12 | 18 | 11 | 14 |
| Bottom Five Genotypes | ||||||||||||||
| 39 | 203 | 154.5 | 174 | 157.5 | 200 | 151.5 | 173.4 | 16 | 18 | 17 | 15 | 12 | 12 | 15 |
| 13 | 169 | 171.5 | 134 | 134.5 | 203.3 | 156.5 | 161.5 | 18 | 12 | 18 | 15 | 10 | 15 | 14 |
| 50 | 119 | 101.5 | 149.5 | 130.5 | 218.5 | 166.5 | 147.6 | 18 | 13 | 14 | 15 | 17 | 13 | 15 |
| 42 | 140 | 153 | 175.5 | 104.5 | 207.7 | 120 | 125.3 | 14 | 9 | 16 | 14 | 13 | 13 | 13 |
| 79 | 174 | 165.5 | 167.5 | 120.5 | 201.4 | 148 | 162.8 | 11 | 18 | 23 | 13 | 14 | 13 | 15 |
| Mean | 168.0 | 166.7 | 166.2 | 143.4 | 195.5 | 166.1 | 167.3 | 14.6 | 13.6 | 18.0 | 14.9 | 14.6 | 12.8 | 14.5 |
| STD | 23.9 | 34.5 | 22.1 | 23.0 | 27.0 | 23.1 | 12.6 | 2.7 | 4.4 | 2.7 | 2.4 | 3.2 | 2.0 | 1.3 |
| SED± | 2.7 | 3.8 | 2.5 | 2.6 | 3.0 | 2.6 | 1.4 | 0.3 | 0.5 | 0.3 | 0.3 | 0.4 | 0.2 | 0.1 |
| CV (%) | 14.2 | 20.7 | 13.3 | 16.0 | 13.8 | 13.9 | 7.5 | 18.7 | 32.1 | 15.0 | 16.3 | 22.0 | 15.6 | 9.1 |
| Genotype | NRP | NPP | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Y1 | Y11 | Mean | Y1 | Y11 | Mean | |||||||||
| S1 | S2 | S3 | S1 | S2 | S3 | S1 | S2 | S3 | S1 | S2 | S3 | |||
| Top Ten Genotypes | ||||||||||||||
| 21 | 214 | 402 | 71 | 130 | 61 | 47 | 154 | 157 | 270 | 66 | 61 | 92 | 98 | 124 |
| 43 | 138 | 173 | 97 | 117 | 95 | 58 | 113 | 119 | 315 | 98 | 72 | 110 | 90 | 134 |
| 51 | 260 | 155 | 146 | 113 | 80 | 51 | 134 | 167 | 231 | 109 | 65 | 76 | 90 | 123 |
| 30 | 178 | 430 | 134 | 132 | 73 | 52 | 166 | 127 | 362 | 95 | 97 | 83 | 101 | 144 |
| 45 | 191 | 647 | 160 | 151 | 88 | 83 | 220 | 96 | 261 | 106 | 81 | 92 | 122 | 126 |
| 81 | 200 | 536 | 85 | 89 | 69 | 40 | 170 | 140 | 240 | 69 | 61 | 70 | 82 | 110 |
| 17 | 184 | 258 | 96 | 139 | 94 | 61 | 139 | 102 | 158 | 65 | 35 | 112 | 89 | 93 |
| 66 | 148 | 168 | 108 | 119 | 76 | 49 | 111 | 69 | 186 | 82 | 26 | 78 | 64 | 84 |
| 74 | 196 | 414 | 98 | 125 | 84 | 81 | 166 | 128 | 112 | 64 | 46 | 40 | 94 | 81 |
| 20 | 126 | 259 | 106 | 148 | 130 | 73 | 140 | 115 | 177 | 78 | 38 | 157 | 45 | 101 |
| Bottom Five Genotypes | ||||||||||||||
| 39 | 161 | 465 | 103 | 145 | 55 | 60 | 165 | 128 | 125 | 93 | 38 | 61 | 82 | 88 |
| 13 | 155 | 228 | 80 | 119 | 99 | 52 | 122 | 98 | 195 | 55 | 37 | 60 | 95 | 90 |
| 50 | 116 | 321 | 199 | 195 | 81 | 46 | 159 | 79 | 78 | 60 | 59 | 96 | 84 | 76 |
| 42 | 122 | 150 | 87 | 151 | 80 | 62 | 109 | 99 | 78 | 90 | 62 | 67 | 70 | 78 |
| 79 | 98 | 552 | 70 | 131 | 163 | 54 | 178 | 53 | 226 | 51 | 26 | 165 | 90 | 102 |
| Mean | 174.1 | 312.3 | 99.0 | 161.6 | 91.8 | 58.9 | 149.4 | 114.6 | 148.2 | 80.0 | 51.0 | 80.9 | 86.7 | 93.4 |
| STD | 43.9 | 146.5 | 27.7 | 39.8 | 30.0 | 12.1 | 26.2 | 30.5 | 56.7 | 22.1 | 16.1 | 33.4 | 19.7 | 14.1 |
| SED± | 4.9 | 16.3 | 3.1 | 4.4 | 3.3 | 1.3 | 2.9 | 3.4 | 6.3 | 2.5 | 1.8 | 3.7 | 2.2 | 1.6 |
| CV (%) | 25.2 | 46.9 | 28.0 | 24.7 | 32.7 | 20.6 | 17.5 | 26.6 | 38.2 | 27.7 | 31.5 | 41.3 | 22.8 | 15.1 |
| Genotype | GYD | HSWT | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Y1 | Y11 | Mean | Y1 | Y11 | Mean | |||||||||
| S1 | S2 | S3 | S1 | S2 | S3 | S1 | S2 | S3 | S1 | S2 | S3 | |||
| Top Ten Genotypes | ||||||||||||||
| 21 | 2.1 | 0.9 | 2.3 | 2.4 | 1.3 | 1.7 | 1.8 | 16.0 | 16.5 | 10.0 | 10.5 | 12.5 | 15.5 | 13.5 |
| 43 | 1.7 | 1.7 | 1.6 | 1.8 | 1.6 | 1.9 | 1.7 | 17.0 | 14.5 | 14.0 | 17.0 | 22.5 | 13.0 | 16.3 |
| 51 | 1.8 | 1.0 | 2.1 | 2.1 | 1.7 | 1.7 | 1.7 | 16.5 | 17.5 | 14.5 | 18.5 | 21.5 | 13.5 | 17.0 |
| 30 | 2.3 | 1.6 | 1.2 | 1.2 | 1.4 | 1.8 | 1.6 | 17.5 | 17.0 | 15.0 | 16.0 | 16.0 | 12.0 | 15.6 |
| 45 | 1.5 | 0.9 | 1.4 | 1.5 | 2.3 | 1.9 | 1.6 | 18.4 | 19.0 | 15.5 | 16.0 | 16.5 | 18.0 | 17.2 |
| 81 | 1.3 | 0.5 | 1.5 | 1.6 | 2.3 | 2.3 | 1.6 | 19.5 | 16.0 | 15.5 | 19.0 | 15.0 | 11.0 | 16.0 |
| 17 | 1.1 | 0.5 | 0.7 | 1.4 | 2.5 | 3.0 | 1.5 | 18.5 | 14.0 | 11.0 | 17.5 | 20.0 | 15.5 | 16.1 |
| 66 | 2.4 | 1.5 | 1.2 | 1.2 | 1.4 | 1.5 | 1.5 | 15.5 | 15.5 | 15.0 | 17.5 | 17.5 | 13.5 | 15.8 |
| 74 | 2.2 | 1.6 | 1.1 | 1.0 | 1.1 | 1.8 | 1.5 | 14.5 | 14.5 | 15.5 | 16.9 | 20.0 | 13.5 | 15.8 |
| 20 | 1.2 | 0.9 | 1.7 | 1.7 | 1.7 | 1.2 | 1.4 | 16.0 | 12.5 | 15.0 | 18.5 | 15.0 | 14.0 | 15.2 |
| Bottom Five Genotypes | ||||||||||||||
| 39 | 0.4 | 0.4 | 1.1 | 1.1 | 1.2 | 0.9 | 0.8 | 15.5 | 14.5 | 14.5 | 16 | 15 | 16 | 15.3 |
| 13 | 0.8 | 0.2 | 0.5 | 1.4 | 0.5 | 0.3 | 0.6 | 12.5 | 15 | 14 | 15 | 16.5 | 16 | 14.8 |
| 50 | 0.9 | 0.5 | 0.4 | 0.5 | 0.4 | 0.7 | 0.6 | 13 | 10.5 | 17.5 | 21 | 19 | 14.5 | 15.9 |
| 42 | 0.6 | 0.4 | 0.9 | 0.5 | 0.4 | 0.4 | 0.5 | 12 | 12.5 | 14 | 19 | 20 | 14.5 | 15.3 |
| 79 | 0.8 | 0.3 | 0.4 | 0.5 | 0.3 | 0.5 | 0.5 | 13 | 16.5 | 14.5 | 17.5 | 17.5 | 14 | 15.5 |
| Mean | 1.1 | 0.6 | 1.3 | 1.3 | 1.5 | 1.3 | 1.2 | 15.9 | 13.9 | 13.5 | 17.6 | 12.9 | 14.2 | 14.7 |
| STD | 0.4 | 0.3 | 0.4 | 0.4 | 0.5 | 0.4 | 0.2 | 2.4 | 3.2 | 2.4 | 2.3 | 4.5 | 2.5 | 1.3 |
| SED± | 0.0 | 0.0 | 0.0 | 0.0 | 0.1 | 0.0 | 0.0 | 0.3 | 0.4 | 0.3 | 0.3 | 0.5 | 0.3 | 0.1 |
| CV (%) | 37.3 | 43.3 | 32.8 | 32.1 | 31.3 | 33.5 | 20.5 | 15.1 | 22.9 | 18.0 | 13.2 | 35.1 | 17.5 | 8.9 |
| Trait | DTF | DTM | PH | NPB | NSB | NRP | NPP | NSP | GYD | HSWT |
|---|---|---|---|---|---|---|---|---|---|---|
| DTF | 1 | 0.787 ** | 0.442 ** | 0.069 | 0.006 | 0.063 | 0.121 | −0.134 | 0.232 * | −0.021 |
| DTM | 1 | 0.409 ** | 0.066 | 0.037 | 0.034 | 0.121 | −0.020 | 0.131 | 0.023 | |
| PH | 1 | 0.057 | 0.149 | 0.249 * | 0.190 | −0.123 | 0.123 | 0.021 | ||
| NPB | 1 | 0.044 | 0.261 * | 0.145 | 0.406 ** | 0.174 | 0.350 ** | |||
| NSB | 1 | 0.024 | 0.152 | −0.101 | 0.214 | 0.090 | ||||
| NRP | 1 | 0.191 | 0.262 * | 0.177 | 0.124 | |||||
| NPP | 1 | 0.099 | 0.354 ** | 0.307 ** | ||||||
| NSP | 1 | 0.051 | 0.173 | |||||||
| GYD | 1 | 0.498 ** | ||||||||
| HSWT | 1 |
| Trait | Dimension | ||
|---|---|---|---|
| 1 | 2 | 3 | |
| FMC | −0.026 | −0.435 | −0.269 |
| FP | 0.016 | −0.568 | 0.050 |
| FSC | −0.101 | −0.172 | 0.291 |
| FSP | 0.357 | −0.095 | −0.080 |
| GH | −0.077 | 0.227 | 0.151 |
| LH | 0.629 | −0.435 | −0.112 |
| LS | −0.386 | −0.626 | 0.143 |
| PC | 0.010 | 0.175 | −0.310 |
| PF | 0.203 | 0.616 | 0.043 |
| SCP | −0.050 | 0.327 | −0.235 |
| SEC | −0.038 | 0.236 | −0.345 |
| SMC | 0.020 | −0.144 | −0.060 |
| SSH | 0.082 | −0.134 | 0.186 |
| STC | −0.042 | 0.000 | 0.023 |
| DTF | 0.186 | −0.069 | 0.827 |
| DTM | 0.236 | −0.190 | 0.793 |
| PH | 0.294 | −0.357 | 0.118 |
| NPB | 0.037 | 0.208 | 0.239 |
| NSB | 0.160 | 0.398 | 0.204 |
| NPP | 0.626 | 0.086 | −0.073 |
| PL | 0.252 | 0.353 | 0.104 |
| NRP | 0.001 | 0.735 | −0.117 |
| NSP | 0.476 | 0.076 | −0.075 |
| HSWT | 0.508 | 0.219 | −0.084 |
| GYD | 0.863 | 0.146 | −0.109 |
| Eigen value | 3.404 | 2.967 | 2.163 |
| Variance % | 39 | 34 | 25 |
| Cumulative | 39 | 73 | 98 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nyirenda Yohane, E.; Shimelis, H.; Laing, M.; Mathew, I.; Shayanowako, A. Phenotypic Divergence Analysis in Pigeonpea [Cajanus cajan (L.) Millspaugh] Germplasm Accessions. Agronomy 2020, 10, 1682. https://doi.org/10.3390/agronomy10111682
Nyirenda Yohane E, Shimelis H, Laing M, Mathew I, Shayanowako A. Phenotypic Divergence Analysis in Pigeonpea [Cajanus cajan (L.) Millspaugh] Germplasm Accessions. Agronomy. 2020; 10(11):1682. https://doi.org/10.3390/agronomy10111682
Chicago/Turabian StyleNyirenda Yohane, Esnart, Hussein Shimelis, Mark Laing, Isack Mathew, and Admire Shayanowako. 2020. "Phenotypic Divergence Analysis in Pigeonpea [Cajanus cajan (L.) Millspaugh] Germplasm Accessions" Agronomy 10, no. 11: 1682. https://doi.org/10.3390/agronomy10111682
APA StyleNyirenda Yohane, E., Shimelis, H., Laing, M., Mathew, I., & Shayanowako, A. (2020). Phenotypic Divergence Analysis in Pigeonpea [Cajanus cajan (L.) Millspaugh] Germplasm Accessions. Agronomy, 10(11), 1682. https://doi.org/10.3390/agronomy10111682





