Abstract
During recent centuries, cultivated olive has evolved to one of the major tree crops in the Mediterranean Basin and lately expanded to America, Australia, and Asia producing an estimated global average value of over USD 18 billion. A long-term research effort has been established with the long-term goal to preserve biodiversity, characterize agronomic behavior, and ultimately utilize genotypes suitable for cultivation in areas of unfavorable environmental conditions. In the present study, a combination of 10 simple sequence repeat (SSR) markers with the classification binary tree (CBT) analysis was evaluated as a method for discriminating genotypes within cultivated olive trees, while Olea europaea subsp. cuspidata was also used as an outgroup. The 10 SSR loci employed in this study, were highly polymorphic and gave reproducible amplification patterns for all accessions analyzed. Genetic analysis indicated that the group of SSR loci employed was highly informative. A further analysis revealed that two sub populations and pairwise relatedness gave insight about synonymies. In conclusion, the CBT method which employed SSR allelic sizes proved to be a valuable tool in order to distinguish olive cultivars over the traditional unweighted pair group method with the arithmetic mean (UPGMA) algorithm. Further research which will combine phenotyping characterization of olive germplasm will have the potential to enable the utilization of existing, and breeding of new, superior cultivars.
1. Introduction
During recent centuries, cultivated olive has evolved to one of the major tree crops in the Mediterranean Basin, while recently it has expanded to many areas in America, Australia, and Asia producing an estimated global average value of over USD 18 billion (FAOSTAT 2018). The expansion of its cultivation, however, did not come at no cost. Olive cultivars employed for new plantations were not always suitable for the local climate, resulting in investment failure [1]. Additionally, even Mediterranean countries where olive trees have been growing for centuries have been impacted by climate change [2]. As a result, some olive cultivars, for example, produce poor fruit yields when flowering has been destroyed by high air temperature [3] and drought stress [4].
Olea europaea has extensively been studied after the discovery of DNA-based molecular markers in order to characterize and discriminate olive germplasm and to detect possible adulterations in olive oils [5,6,7]. A comprehensive review provided by Sebastiani and Busconi, 2017 states the fact that SSR markers have been used as the marker of choice for olive germplasm analysis for both Mediterranean and non-Mediterranean countries compared to AFLPS, RAPDS, and ISSR markers. Nowadays, next generation sequencing (NGS) gave the opportunity to identify SNPs in olive germplasm [7,8] but until now relatively few studies which used NGS for olive germplasm characterization and discrimination exist [7]. Recently, Belaj, De La Rosa, Lorite, Mariotti, Cultrera, Beuzón, González-Plaza, Muñoz-Mérida, Trelles and Baldoni [8] produced EST-SNP markers for olive germplasm characterization which were able to discriminate different accessions and exhibited transferability to wild olive genotypes. Although, as their significant advantages, Belaj et al., 2018 stated that EST-SNPs displayed lower levels of genetic diversity than SSRs, and that SSR markers are the most rapid method for cultivar identification when a small number of samples exist. Furthermore, another recent published research from Li et al. [9] designed SSRs based on trinucleotide repeat sequences and showed their high discriminating capacity for 53 olive accessions.
The Institute for Olive Tree, Subtropical Crops and Viticulture in Chania, Greece, harbors the National Germplasm Depository of Greece comprising over 100 cultivars from the main olive producing countries of the world. These cultivars are formally exchanged between the members of the Network of Olive Collections which is coordinated by the International Olive Oil Council (http://www.internationaloliveoil.org/). Among them, over 45 cultivars originate from Greece and represent over 90% of cultivated olive groves. The main aim of this collection is to preserve biodiversity, characterize agronomic behavior, and ultimately utilize selected genotypes suitable for establishment in areas with unfavorable environmental conditions. The Institute has a database of morphological descriptors of olive cultivars with features of the tree, leaves, flowers, fruit, and seeds as described in Barranco et al. [10]. A previously published work by Koubouris, Avramidou, Metzidakis, Petrakis, Sergentani and Doulis [6] revealed the rich differentiation of morphological characters of 41 olive cultivars obtained in the Institute.
Classification binary trees (CBTs) were firstly introduced in 1984 from Breiman et al. [11] and reflect two sides, covering the use of trees as a data analysis method, and in a more mathematical framework, proving some of their fundamental properties. The construction of a CBT involves the split of the original set of samples (root) into two parts on the basis of a criterion involving a few variables, usually a simple algebraic expression of one (often) or two (rarely). All variables involved in the construction of the CBT can guarantee the group affiliation of the existing or new samples. The existing samples are arranged into the “leaves” of the tree; in an ideal situation six site groups are produced. CBTs were then used by Petrakis et al. [12] for geographical characterization of Greek extra virgin olive oils from one variety (Koroneiki) from three regions by using chemical values. The CBT of metabolomics data based on NMR analysis of samples, are used for the detection of adulteration of olive oil along with the forward stepwise canonical discriminant analysis [13]. These authors used CBTs in order to estimate the effect of harvesting time, cultivar, and geographical origin in the composition of olive oils [14]. The main reason for this is the independence of the data from any assumption, lack of linearity, or commensurability. CBTs have the ability to use the algorithm in order to weight the resultant groups without taking into account the number of their members provided, in this way a proper splitting criterion where the ‘twoing criterion’ is used [11]. In contrary, the split of the parental group into two on the basis of the most important variable (allele in the current study) is a common feature for UPGMA [15,16]. CBTs perform the selection with the twoing criterion which avoids the bias introduced by selecting variables that have more missing values [16] and overfitting common variables which have a wide domain [17]. Overfitting is a common problem of data mining methods that refers to a modelling error that occurs when a function corresponds too closely to a particular set of data. CBTs exceed overfitting, due to the fact that they perform the classification of any new sample on the basis of a simple algorithm constructed in a simple way dictated by the classification tree [18], which is called ‘mobile’ [19]. Furthermore, the CBT methodology uses the ‘surrogate splits’ method [20] which was introduced by Breiman, Friedman, Stone and Olshen [11], where the missing values in the data are not computed by data imputation. According to this method, a surrogate value has a similar splitting behavior with the predictor variable having the missing value and in this way its value can be put in the place of a missing value. For these reasons, the CBT methodology is capable of visually testing the monophyly hypothesis of olive cultivars and examining them independently due to the fact that cultivars are man-made combinations and they lack previous genetic structure information.
This last property does not exist in the neighbor joining method which uses phylogenetically observable substitution models in the implementation phase [21]. On the other hand, UPGMA [22] is intuitively simple and highly used but it suffers from several shortcomings such as the construction of different tree topologies from the same data set [23] or the existence in the computer memory of the entire dissimilarity matrix [24]. The first time that SSR allelic sizes were used for constructing a CBT was in Aksehirli-Pakyurek et al. [25], where several Cretan cultivars were compared with two major Turkish ones and wild olive tree fruits from Crete in order to estimate the genetic diversity and relationships between them.
The aim of the present study was to evaluate CBT as a method for the characterization of olive germplasm, test its discriminating capacity, and provide an insight in the within-cultivar-variation of the reference plant material conserved in the National Olive Germplasm Bank of Greece. The CBT method based on allelic sizes from 10 SSR loci that are employed in the current study will provide a novel and accurate method in order to discriminate cultivars in regards to traditional phenotyping or only SSR discriminating capability. Classification trees constructed on the basis of SSR polymorphic markers are valuable in order to characterize the richness of olive germplasm without a priori knowledge.
2. Materials and Methods
2.1. Plant Material
For the SSR genotyping, a total of 90 genotypes were analyzed originating from 53 Olea europaea subsp. europaea cultivars and one accession used as an outgroup of Olea europaea subsp. cuspidata. Depending on local availability, the cultivar membership varied as follows: Twenty-five cultivars were represented by one genotype, twenty-three by two, four by three, and one by six (Table 1). Plant material is maintained in the National Olive Germplasm Bank of Greece located at Chania, Crete located at the Chrisopigi Monastery area near the Institute of Olive Tree, Subtropical Crops and Viticulture, Hellenic Agricultural Organization ELGO “DIMITRA” (Chania, Southern Greece). The mean air temperature in the area was 18 °C, relative humidity (RH) 64%, and annual rainfall 600–800 mm (ELGO-DIMITRA. meteorological station, Chania, Greece).

Table 1.
List of samples analyzed in the present study including the cultivar full name and number of independent genotypes per cultivar.
2.2. DNA Extraction and Microsatellite Analysis
Total genomic DNA was isolated from the leaf material using the DNeasy Plant Mini kit (Qiagen, Hilden, Germany cat. No. 69104) according to the manufacturer’s instructions. Initial grinding was conducted using the automated grinder TissueLyzer (Qiagen, Hilden, Germany) in the presence of liquid nitrogen. For DNA quantification, the Nanodrop 2000 (Thermo Scientific, Waltham, Massachusetts, USA) spectrophotometer was employed. For genotyping, 10 microsatellite loci (DCA3, DCA5, DCA9, DCA14, DCA16, DCA18, Gapu101, UDO043, EM090, GAPU71B) were selected in agreement with Baldoni et al. [26] on the basis of their informativeness. Polymerase chain reactions were carried out in a 20 μL reaction in a Perkin Elmer 9600 (Waltham, MA, USA) thermocycler including 25 ng of template DNA, 0.2 mM of each dNTP, 0.2 μM of each primer, 2.5 mM MgCl2, and 1 U of Kapa Taq Polymerase (Kapa Biosystems, Cape Town, South Africa). Thermal cycling included: Initial denaturation at 95 °C for 5 min, followed by 35 cycles of 95 °C for 30 s, the corresponding annealing temperature for 45 s, and 72 °C for 45 s, with a final extension at 72 °C for 10 min. One micro liter portion of the PCR product mixtures were multiplexed, and electrophoretically separated using an automated fluorescence sequencer [ABI Prism 3730xl Genetic Analyzer (Applied Biosystems, Waltham, MA, USA). SSR binning and scoring were conducted and the initial data matrix was produced employing the proprietary software GeneMapper v4.0 (Applied Biosystems, Waltham, MA, USA).
The number of alleles per locus (Na), effective number of alleles (Ne), observed (Ho), expected heterozygosity (He), probability of identity (PI), polymorphic information content (PIC), and null allele frequency F (null) were estimated using the Cervus software package [27].
Subsequently, the SSR data was analyzed using the software Structure 2.3.1 [28] as described in Marra et al. [29] to elucidate relationships between the olive genotypes and achieve the most reliable grouping among them. In brief, the ‘admixture’ model, forming one to ten populations (K), a burn-in length of 10,000, followed by 100,000 runs at each K, with 10 replicates for every K, were employed. To select the right number of populations (K), the Structure Harvester program was used which performed the validation of the most likely number of clusters K with the Structure Harvester [30].
Furthermore, pairwise relatedness was also used to calculate the allelic similarity for codominant data using GenAlEx 6.501 [31], LRM estimator by Lynch and Ritland [32].
2.3. Cluster Analysis by the Classification Binary Tree (CBT)
The data matrix for the CBT analysis consisted of the sizes of 90 genotypes from each of the 53 Olea europaea subsp. europaea cultivars and one accession of Olea europaea subsp. cuspidata by 10 SSR loci, each having two alleles. Similarly, to the genetic analyses, the CBT input dataset consisted of SSR alleles base pair sizes. The output of CBT, which is called a mobile, initially entails the split of the original sample set into two parts on the basis of a criterion involving one or two discriminatory loci in a simple algebraic expression. Subsequently, each one of the two clusters is split into two, on the basis of a criterion while the quality of the improvement gained by the splitting of the parental cluster is measured by an impurity function which in this analysis is the twoing criterion [11]. This was proposed by [11] since the usually employed Gini index is problematic when the domain of the target attribute is relatively wide; it coincides with the Gini index when the domain of the target attribute is binary [17]. This criterion at each split is expressed on the basis of an inequality involving one (here) or a few (elsewhere) alleles. Thus, the tree, or better a mobile, grows according to splits that produce maximally informative and ‘pure’ groups according to the ‘twoing’ impurity function [11]. The reduction of error in the entire classification is monitored by means of an overall proportional reduction in error function originally proposed by Breiman, Friedman, Stone and Olshen [11]. To avoid overfitting, we used the complexity parameter which is a measure of the degree of tree complexity and the way that the tree describes the data [33]. The CBT analysis was performed using routines and packages within the R environment (R Development Core Team, 2017) and used the package ‘rpart’ [34], R package (2017) version 3.4.3 (Boston, MA, USA) and the SYSTAT 13.0 software (San Jose, CA, USA, 2009).
Subsequently, and for comparison with the CBT cluster analysis, a genetic similarity tree was constructed employing the agglomerative unweighted pair group method with the arithmetic mean (UPGMA) algorithm [22] using the MEGA X software (Old Main, University Park, PA, USA) [35].
3. Results
3.1. Genetic Parameters from SSR Analysis
In the current study, by using 10 microsatellite markers a total of 126 SSR alleles were produced for all the 90 olive genotypes. The number of alleles per locus varied from eight (UDO043) to 19 (DCA16) with an average number of 12.6 loci per locus (Table 2). The mean expected heterozygosity (He) was 0.801 (ranging from 0.513 for GAPU101 to 0.916 for DCA09), while the mean observed heterozygosity (Ho) was 0.663 (varying from 0.043 for GAPU101 to 0.932 for DCA18) for all 90 accessions. When the calculation for polymorphic information content (PIC) was performed we found that it ranged from 0.489 for GAPU101 to 0.904 for DCA09 and presented a mean value of 0.778) (Table 2). Moreover, when we examined null allele frequencies, due to the fact that the null allele can decrease heterozygosity we found that two SSR loci (GAPU101 and UDO043) showed significantly high estimated probability of null allele (0.846 and 0.224) (Table 2). Furthermore, in three markers (DCA03, DCA09, and DCA18), Ho was higher than He. This result could indicate high genetic variability amongst the cultivars analyzed (Table 2).

Table 2.
For each locus the following are reported: Number of alleles detected (Na), effective number of alleles (Ne), observed (Ho) and expected (He) heterozygosity, probability of identity (PI), polymorphic information content (PIC), Shannon Information Index (I), probability of null allele (F null), and fixation index (F).
The calculation of probability of identity (PI) can provide significant information about the discrimination of genotypes. In the current study, PI was estimated as being between 0.015 for the SSR locus DCA09 and 0.087 for UDO043. When we estimated the value of the combined probability of identity for all the 10 SSR analyzed, the value was very low, 1.708 × 10−13 (Table 2). This result indicates that all genotypes examined can be distinguished effectively.
The genetic population structure was assessed through the Structure software (Pritchard et al., 2000) and Structure Harvester [30] in order to define the best K among the olive cultivars Structure analysis, with a K value equal to 2, revealing the existence of two admixed groups (gene pools) within the analyzed germplasm. Each group is depicted with a different color (green vs. red) in Figure 1 One pool (pictured in red color, Figure 1) included 19 genotypes and eight cultivars (‘Adramytini’, ‘Koroneiki’, ‘Kothreiki’, ‘Dafnelia’, ‘Dopia Zakynthou’, ‘Myrtolia’, ‘Koutsourelia’, and ‘Rahati’), while the second group included 71 genotypes (Supplementary Table S1).
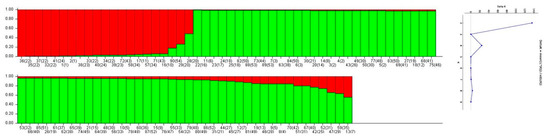
The LRM analysis displayed strong relationships for the cultivar ‘Chalkidiki’ and ‘Chondrolia Chalkidikis’ (LRM = 0.857) which are grown in the same geographic region and for ‘Frantoio’ and ‘Oblonga’ cultivars (LRM = 0.601) (Supplementary Table S2).
In the UPGMA similarity dendrogram (Supplementary Figure S1), it can be seen that all genotypes originating from the same cultivar were grouped together.
3.2. Classification Binary Trees
The produced CBT mobile is shown in Figure 2. The proportional reduction in error was 1 (100%) implying that the ability of the variables (allele sizes/loci) are the best descriptors of the classification of olive cultivars into terminal leaves (Table 3, Table S3).
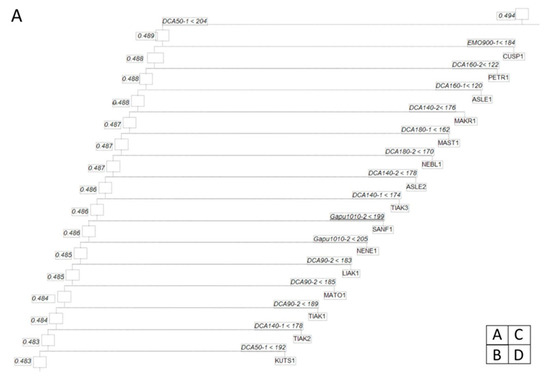

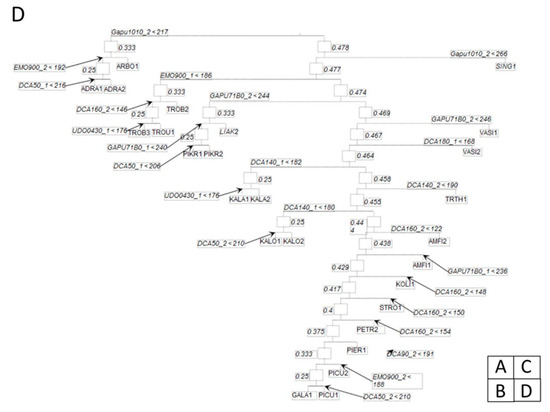
Figure 2.
Dendrogram (mobile) of olive cultivars based on simple sequence repeat (SSR) markers that entered in the classification binary tree (CBT) algorithm. The numbers inside the squares are the impurity at this node of the dendrogram and the number of olive cultivars. The inequality at the nodes corresponds to the responsible variables and this specific level of classification and the value of this variable. As a rule, the left branch corresponds to groups having smaller variable values and the right branch to larger values. Beneath each rectangle is the name of the olive cultivar. Due to CBT large mobile tree we divided the dendrogram to four figures (A), (B), (C) and (D) in order to illustrate the branches.

Table 3.
The number of splits in which the various loci (A) and alleles (B) participate.
The outgroup used in this CBT is Olea cuspidata. It is expectedly classified early in the tree (Figure 2A) on the basis of the DCA5 and EMO90 (Table 3 (A)). These loci are the responsible variables in many splits (i.e., 14 and 10, respectively). However, the alleles in these loci have different splitting behaviors (Table 3 (B)).
Among the cultivars, ‘Koroneiki’ exhibits a peculiar pattern and not all samples are clustered together in the apical leaves of Figure 2B. The allele DCA5_2 is responsible for four samples of the ‘Koroneiki’, while the other two samples are clustered earlier in the tree of the same figure. Several cultivars are clustered together in the mobile of Figure 2. Such cultivars are, e.g., ‘Adramytini’, ‘Aggouromanakolia’, ‘Kalamon’, ‘Kalokairida’, ‘Valanolia’, while several others exhibit the 1–2 pattern of ‘Throuboelia’ (Figure 2D upper left). In this pattern, a sample is clustered in a different neighboring cluster with the next two samples. In the case of ‘Throuboelia’, the responsible alleles are DCA16_2 (the most frequent in splits (Table 3 (B))) and UDO043_1 (occurring in just five splits). In the other case, the cultivars are quite apart on the tree. Such a case is the ‘Aggouromanakolia’ (Figure 2C) where samples 1 and 3 are clustered together on the basis of the UDO043_1 allele, while sample 2 is separated from the other cultivars in a sequential clustering pattern (Figure 2C upper right). In this pattern, all samples belong to different cultivars and are separated sequentially.
Two cultivars that are ‘Asprolia Alexandroupolis’ and ‘Asprolia Lefkados’ are clustered quite distantly on the tree (Figure 2A,C). Moreover, they are geographically distant in very different climatic regimes in Greece. ‘Frantoio’ genotypes from Italy are located in close proximity. ‘Frantoio’ and ‘Frantoio Rhodou’are sequentially clustered in Figure 2B (lower right), discriminated by the alleles DCA18_1, Gapu71B_1, and DCA3_1. The cultivar ‘Megareitiki’ exhibits a peculiar clustering pattern since the respective samples emerge in the two main branches of the mobile immediately after the sequential splits (Figure 2B,C). ‘Pierias’ shows an extreme clustering pattern since the two samples are located in very distant sites of the mobile (Figure 2B,D)
After the root node, the largest splitting of the cultivars is done by means of the locus DCA5, which is also the responsible locus for the highest number of splits together with DCA16 (Table 3 (A)). In the left branch of the tree (Figure 2A), Italian and Spanish cultivars are sequentially split in the right part of the criterion locus. Most loci are used in this sequential split of Figure 2A and the split that forms the branch in Figure 2B is based on the DCA5 locus as a split criterion. ‘Picual’ is exceptionally located in the apical and subapical leaves of the tree in Figure 2D.
All loci participate as splitting criteria in the mobile in Figure 2. However, the alleles DCA9_1, DCA14_1, and Gapu101_1 are absent from the entire tree. Instead, the other allele, which as a rule, has more base pairs is always used as a criterion for the splits. It seems that the second allele of the locus is selected since it contains the same number of base pairs. The exception to this is the locus DCA9 which contains the same number of base pairs only for the sample DOZA1.
4. Discussion
In the present study, a combination of SSR markers with the CBT analysis was evaluated as a method for discriminating genotypes within cultivated olive as well as in relation to non-crop relative Olea europaea subsp. cuspidata, which has been used as an outgroup. The characterization of diverse olive germplasm conserved in the National Germplasm Depository of Greece was used as a case study. Findings of the current study can be used in conjunction with phenotyping of the same olive trees, a parallel task of outside the scope of the present paper, to facilitate the development of pre-breeding material with desired traits such as tolerance to abiotic and biotic stresses, high fruit yield, and nutritional value.
Ninety olive tree individuals, representing some of the major olive producing countries in the world, and maintained in the National Germplasm Depository of Greece, were scanned by 10 SSR loci, previously reported to be the most highly resolving for olive cultivars [26]. Selected loci were found to be highly polymorphic and gave reproducible amplification patterns for all 53 olive cultivars and one Olea europaea subsp. cuspidata accession which was analyzed. The average number of alleles per locus (Na), reported in this study (12.6) was higher than the equivalent reported by Mantia et al. [36] using 12 SSR on 50 olive accessions, Lopes et al. [37] using 14 SSR on 130 accessions, Belaj et al. [38] using 23 SSR on 361 accessions from 19 different countries, and by Aksehirli-Pakyurek, Koubouris, Petrakis, Hepaksoy, Metzidakis, Yalcinkaya and Doulis [25] using seven SSR on six cultivars from Greece and Turkey. Only two studies from Marra, Caruso, Costa, Di Vaio, Mafrica and Marchese [29] who investigated 68 cultivars from Southern Italy with 12 SSR loci reported N𝑎 slightly higher (Na = 13), and Sion et al. [39] who used nine SSR markers in 218 Italian accessions of olives reported a higher number of alleles (Na = 21). Compared to other published data, the average expected heterozygosity (He) 0.801 reported herein was higher than 0.76 [36], 0.68 [37], 0.62 [38], 0.79 [25], and slightly lower than the value of Marra, Caruso, Costa, Di Vaio, Mafrica and Marchese [29] (He = 0.84) and Sion et al., 2019 (He = 0.85). Correspondingly to results in the current paper, Marra et al., 2013 and Sion et al., 2019 found that DCA03, DCA09, and DCA18 yielded Ho higher than He, indicating, thus, a high genetic variability amongst the analyzed cultivars. Indeed, the mean observed heterozygosity (Ho) was lower than the mean expected heterozygosity (He), determining a positive fixation index (F) for all loci (mean = 0.203) except from, DCA3, DCA9, and DCA18 where the values were negative (Table 2). In agreement, the same results were estimated by Sion et al., 2019 where Ho was lower than He, the mean F was 0.2, and DCA3 presented a negative F value.
The average PIC value in the present study was 0.778 (Table 2), indicating that the group of 10 SSR loci employed was indeed highly informative and suitable for individual identification. Nevertheless, one marker (GAPU101) appeared relatively less informative with a PIC value of 0.489. The mean PIC value was lower than the equivalent in Marra et al., 2013 who found 0.81 but higher than the value of 0.755 determined by Aksehirli-Pakyurek et al., 2017. Furthermore, the mean value of PI was very low 1.708 × 10−13, and in fact lower than Marra et al., 2013 who found a PI value of 6.73 × 10−9 further demonstrating that the group of loci used in the present study was successful at fingerprinting olive cultivars. Furthermore, synonymies were disclosed from the LRM estimator Lynch and Ritland [32] and displayed strong relationships for: (a) ‘Chalkidiki’ and ‘Chondrolia Chalkidikis’ (0.857); which is a reasonable result considering the same geographic region and for the two cultivars and (b) cultivars ‘Frantoio’ and ‘Oblonga’ (0.601) which is in accordance with Barranco, Trujillo and Rallo [10]. Results from the Structure Harvester analysis [30] indicated two genetic pools (K = 2) for the cultivars, but must be treated with caution due to the limited number of genotypes included in the analysis. Similarly, Albertini et al. [40] found that two clusters for 22 cultivars studied from central Italy and Díez et al. [41] had the same result for ancient and cultivated cultivars in Spain; whereas Marra, Caruso, Costa, Di Vaio, Mafrica and Marchese [29] found three clusters for 68 accessions from the Southern Italian region.
From the CBT analysis, we can see that two other olive cultivars with similar names, specifically: ‘Throuba-Throuboelia’ and ‘Throuba Thassou’ purporting some kind of relationship, were found in the present study to be genetically distinct on the basis of DCA16 and UDO043 loci (Figure 2D upper left). This finding further points to the challenges of homonymy (identifying two different plant cultivars in two different geographical zones by the same name), a common obstacle in the proper characterization of plant genetic resources [38,42,43]. Indeed, this is the advantage of the employed CBT clustering method. Previously known cultivars as separate entities are found to be the same cultivar, such as ‘Frantoio’ and ‘Oblonga’ and this result is also supported by the CBT analysis and by the value of LRM estimator. Known cultivars are found to be separated, such as ‘Koroneiki’ which forms four samples closely located in the same twig apical leaves (Figure 2B bottom center) and two samples (Figure 2B top left and center right) which share the same morphological characters from ‘Koroneiki’, but they are genetically different. More importantly, we know the SSR alleles which differentiate them from the core cluster of ‘Koroneiki’ of four trees. As mentioned above, the physiological differentiation of these three ‘Koroneiki’ genotypes is our further task. Moreover, this different discrimination for various cultivars: For example, ‘Throuba-Throuboelia’ and ‘Throuba Thassou’, along with differentiation of ‘Koroneiki’ genotypes and ‘Asprolia Alexandroupolis’ and ‘Asprolia Lefkados’, further indicates the complex relationships within cultivated olive germplasm. Moreover, the same inter-cultivar variation was also reported from Omrani-Sabbaghi et al. [44] and could be due to homonyms [38,42,43], mislabeling of cultivars which was only based on morphological traits in the past, misidentification because these cultivars have been produced by vegetative propagation, or by possible and occasional outcrossing events that may have occurred spontaneously between the cultivated clones and feral forms since antiquity and the olive tree is cultivated in Greece for centuries.
In the case of CBT analysis, even though the maximum proportional reduction in error was achieved, yet the produced mobile could be further improved. Furthermore, the pattern of a sequential separation of the two trees of ‘Amfissis’ and ‘Vasilikada’ showed that the selected SSR loci functioned in a concerted action. Specifically, the two trees in ‘Vasilikada’ were separated by the function of one allele in GAPU71B and DCA18 loci while the two trees in ‘Amfissis’ were separated by the concerted action of one allele in GAPU71B and DCA16 loci. In a previous study on the Tribes Cardueae and Cichorieae (Asteraceae), steadily observed subtribe-specific features were scarce and even in conjunction, did not distinguish the subtribes [45]. It can be concluded that, classification trees are not considered suitable for hypothesis testing, however, they can be efficiently used for the identification of thresholds since tree branches are separated based on specific values [46].
In the future, characterization and utilization of plant genetic resources is expected to markedly benefit from the exploitation of new tools such as EST-SSRs [47], predictive machine learning algorithms [48], deep sequencing of gene fragments [49], and whole genome sequencing [50].
In comparison, from the UPGMA similarity dendrogram (Supplementary Figure S1), it can be seen that all genotypes originating from the same cultivar did not have the same accurate grouping as with the CBT method. For example, the six ‘Koroneiki’ genotypes clustered together in UPGMA, while in the CBT analysis performed two separate leaves (also according to their phenotypic profile). UPGMA’s greater disadvantage is that it assumes the same evolutionary speed on all lineages and this results in leaves (terminal nodes) that have an equal distance from the root. In reality, the individual branches are very unlikely to have the same mutation rate. Therefore, UPGMA frequently generates wrong tree topologies according to various studies (Belbin et al., 1992; Strobl et al., 2007). Furthermore, UPGMA starts with a matrix of pairwise distances, but in the case of SSR data where null allele frequencies are high along with scoring error distances are also calculated wrong, and this further affects the quality of the clustering. On the contrary, the CBT methodology by using the “twoing criterion” splits can separate the cultivars on the basis of the most important variable (allele in the current study) without also taking into calculation data with missing values, and provides a more accurate tree that grows according to splits that produce maximally informative and “pure” groups, according to an impurity function. From our point of view, a careful consideration of the UPGMA results should be evaluated from the scientific community and alternative methods of clustering, for example, the CBT methodology should be employed.
5. Conclusions
The present study focused on discriminating cultivars and comparing it to non-crop relative Olea europaea subsp. cuspidata, in order to characterize the diverse olive germplasm conserved in the National Germplasm Depository of Greece using the CBT-SSR analysis. All genotypes were successfully discriminated by the 10 SSR loci employed. All cultivars were efficiently assigned to different branches in the CBT and, in addition, the responsible locus and its specific allele that marks each node is written in the tree diagram. CBT was proved to be a more adequate technique over the traditional UPGMA analysis. At each node, the impurity of the corresponding sample set is written. However, the analysis should be further improved to more group individuals of the same cultivar together. The combined characterization of olive germplasm by genotyping reported here, and phenotyping reported elsewhere [6,51] would enable the utilization of existing, and breeding of new, superior cultivars for meeting specific environmental challenges in the context of climate change. Further research will focus on the usage of three-nucleotide SSR markers which have been recently discovered [9], in order to test their discriminating capacity on current Greek olive accessions and combine them with the CBT method.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4395/10/11/1662/s1. Table S1: List of samples analyzed in the present study including cultivar full name and individual genotype codes employed in the different visualization schemes; Table S2: Pairwise relatedness summary according to the LRM estimator; Table S3: The number of splits in which an allele participates and the proportional reduction in error it confers to the tree; Figure S1: Dendrogram based on the unweighted pair group method with the arithmetic mean (UPGMA) algorithm.
Author Contributions
Conceptualization, A.G.D., E.V.A. and G.C.; methodology, E.V.A. and P.V.P.; software, E.V.A. and P.V.P.; validation, G.C.K., A.G.D., I.T.M. and K.K.L.; formal analysis, E.V.A. and A.G.D.; data curation, P.V.P. and E.V.A.; writing—original draft preparation, E.V.A., P.V.P., G.C.K. and A.G.D.; writing—review and editing, all authors; visualization, P.V.P, G.C.K. and E.V.A.; supervision, E.V.A. and A.G.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Acknowledgments
The authors want to acknowledge the assistance they received from Dr. Ermioni Malliarou, who supported the experiments in the laboratory.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Aybar, V.E.; de Melo, E.A.; Mourão, J.P.; Searles, P.S.; Matias, A.C.; del Río, C.; Reig, J.M.C.; Rousseaux, M.C. Evaluation of olive flowering at low latitude sites in Argentina using a chilling requirement model. Span. J. Agric. Res. 2015, 13, e09-001. [Google Scholar] [CrossRef]
- Ponti, L.; Gutierrez, A.P.; Ruti, P.M.; Dell’Aquila, A. Fine-scale ecological and economic assessment of climate change on olive in the Mediterranean basin reveals winners and losers. Proc. Natl. Acad. Sci. USA 2014, 111, 5598–5603. [Google Scholar] [CrossRef] [PubMed]
- Koubouris, G.; Kavroulakis, N.; Metzidakis, I.; Vasilakakis, M.; Sofo, A. Ultraviolet-B radiation or heat cause changes in photosynthesis, antioxidant enzyme activities and pollen performance in olive tree. Photosynthetica 2015, 53, 279–287. [Google Scholar] [CrossRef]
- Brito, C.; Dinis, L.-T.; Moutinho-Pereira, J.; Correia, C.M. Drought stress effects and olive tree acclimation under a changing climate. Plants 2019, 8, 232. [Google Scholar] [CrossRef] [PubMed]
- Avramidou, E.V.; Doulis, A.G.; Petrakis, P.V. Chemometrical and molecular methods in olive oil analysis: A review. J. Food Process. Preserv. 2018, 42, e13770. [Google Scholar] [CrossRef]
- Koubouris, G.; Avramidou, E.; Metzidakis, I.; Petrakis, P.; Sergentani, C.; Doulis, A. Phylogenetic and evolutionary applications of analyzing endocarp morphological characters by classification binary tree and leaves by SSR markers for the characterization of olive germplasm. Tree Genet. Genomes 2019, 15, 26. [Google Scholar] [CrossRef]
- Sebastiani, L.; Busconi, M. Recent developments in olive (Olea europaea L.) genetics and genomics: Applications in taxonomy, varietal identification, traceability and breeding. Plant Cell Rep. 2017, 36, 1345–1360. [Google Scholar] [CrossRef] [PubMed]
- Belaj, A.; De La Rosa, R.; Lorite, I.J.; Mariotti, R.; Cultrera, N.G.; Beuzón, C.R.; González-Plaza, J.J.; Muñoz-Mérida, A.; Trelles, O.; Baldoni, L. Usefulness of a new large set of high throughput EST-SNP markers as a tool for olive germplasm collection management. Front. Plant Sci. 2018, 9, 1320. [Google Scholar] [CrossRef]
- Li, D.; Long, C.; Pang, X.; Ning, D.; Wu, T.; Dong, M.; Han, X.; Guo, H. The newly developed genomic-SSR markers uncover the genetic characteristics and relationships of olive accessions. PeerJ 2020, 8, e8573. [Google Scholar] [CrossRef]
- Barranco, D.; Trujillo, I.; Rallo, P. Are Oblonga’ and Frantoio’ Olives the Same Cultivar? HortScience 2000, 35, 1323–1325. [Google Scholar] [CrossRef]
- Breiman, L.; Friedman, J.; Stone, C.J.; Olshen, R.A. Classification and Regression Trees; CRC Press: Boca Raton, FL, USA, 1984. [Google Scholar]
- Petrakis, P.V.; Agiomyrgianaki, A.; Christophoridou, S.; Spyros, A.; Dais, P. Geographical characterization of Greek virgin olive oils (Cv. Koroneiki) using 1H and 31P NMR fingerprinting with canonical discriminant analysis and classification binary trees. J. Agric. Food Chem. 2008, 56, 3200–3207. [Google Scholar] [CrossRef]
- Agiomyrgianaki, A.; Petrakis, P.V.; Dais, P. Detection of refined olive oil adulteration with refined hazelnut oil by employing NMR spectroscopy and multivariate statistical analysis. Talanta 2010, 80, 2165–2171. [Google Scholar] [CrossRef] [PubMed]
- Agiomyrgianaki, A.; Petrakis, P.V.; Dais, P. Influence of harvest year, cultivar and geographical origin on Greek extra virgin olive oils composition: A study by NMR spectroscopy and biometric analysis. Food Chem. 2012, 135, 2561–2568. [Google Scholar] [CrossRef] [PubMed]
- Belbin, L.; Faith, D.P.; Milligan, G.W. A comparison of two approaches to beta-flexible clustering. Multivar. Behav. Res. 1992, 27, 417–433. [Google Scholar] [CrossRef] [PubMed]
- Strobl, C.; Boulesteix, A.-L.; Augustin, T. Unbiased split selection for classification trees based on the Gini index. Comput. Stat. Data Anal. 2007, 52, 483–501. [Google Scholar] [CrossRef]
- Rokach, L.; Maimon, O. Classification trees. In Data Mining and Knowledge Discovery Handbook; Springer: Berlin/Heidelberg, Germany, 2009; pp. 149–174. [Google Scholar]
- Steinberg, D.; Colla, P. CART: Classification and regression trees. Top Ten Algorithms Data Min. 2009, 9, 179. [Google Scholar]
- Wilkinson, L. Systat. Wiley Interdiscip. Rev. Comput. Stat. 2010, 2, 256–257. [Google Scholar] [CrossRef]
- Nisbet, R.; Elder, J.; Miner, G. Handbook of Statistical Analysis and Data Mining Applications; Academic Press: Cambridge, MA, USA, 2009. [Google Scholar]
- Paradis, E. Analysis of Phylogenetics and Evolution with R.; Springer: Berlin/Heidelberg, Germany, 2011. [Google Scholar]
- Sneath, P.; Sokal, R. Unweighted pair group method with arithmetic mean. In Numerical Taxonomy; Springer: Berlin/Heidelberg, Germany, 1973; pp. 230–234. [Google Scholar]
- Hart, G. The occurrence of multiple UPGMA phenograms. In Numerical Taxonomy; Springer: Berlin/Heidelberg, Germany, 1983; pp. 254–258. [Google Scholar]
- Loewenstein, Y.; Portugaly, E.; Fromer, M.; Linial, M. Efficient algorithms for accurate hierarchical clustering of huge datasets: Tackling the entire protein space. Bioinformatics 2008, 24, i41–i49. [Google Scholar] [CrossRef]
- Aksehirli-Pakyurek, M.; Koubouris, G.; Petrakis, P.; Hepaksoy, S.; Metzidakis, I.; Yalcinkaya, E.; Doulis, A. Cultivated and Wild Olives in Crete, Greece—Genetic Diversity and Relationships with Major Turkish Cultivars Revealed by SSR Markers. Plant Mol. Biol. Report. 2017, 35, 575–585. [Google Scholar] [CrossRef]
- Baldoni, L.; Cultrera, N.G.; Mariotti, R.; Ricciolini, C.; Arcioni, S.; Vendramin, G.G.; Buonamici, A.; Porceddu, A.; Sarri, V.; Ojeda, M.A. A consensus list of microsatellite markers for olive genotyping. Mol. Breed. 2009, 24, 213–231. [Google Scholar] [CrossRef]
- Marshall, T.C.; Slate, J.; Kruuk, L.E.B.; Pemberton, J.M. Statistical confidence for likelihood-based paternity inference in natural populations. Mol. Ecol. 1998, 7, 639–655. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [PubMed]
- Marra, F.; Caruso, T.; Costa, F.; Di Vaio, C.; Mafrica, R.; Marchese, A. Genetic relationships, structure and parentage simulation among the olive tree (Olea europaea L. Subsp. Europaea) cultivated in Southern Italy revealed by SSR markers. Tree Genet. Genomes 2013, 9, 961–973. [Google Scholar] [CrossRef]
- Earl, D.A. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Lynch, M.; Ritland, K. Estimation of pairwise relatedness with molecular markers. Genetics 1999, 152, 1753–1766. [Google Scholar] [PubMed]
- Atkinson, E.J.; Therneau, T.M. An Introduction to Recursive Partitioning Using the RPART Routines; Mayo Foundation: Rochester, NY, USA, 2000. [Google Scholar]
- Therneau, T.M.; Atkinson, E.J. An Introduction to Recursive Partitioning Using the RPART Routines; Technical Report; Mayo Foundation: Rochester, NY, USA, 1997. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Mantia, L.; Lain, T.; Caruso, T.; Testolin, R. SSR-based DNA fingerprints reveal the genetic diversity of Sicilian olive (Olea europaea L.) germplasm. J. Hortic. Sci. Biotechnol. 2005, 80, 628–632. [Google Scholar] [CrossRef]
- Lopes, M.S.; Mendonça, D.; Sefc, K.M.; Gil, F.S.; da Câmara Machado, A. Genetic evidence of intra-cultivar variability within Iberian olive cultivars. HortScience 2004, 39, 1562–1565. [Google Scholar] [CrossRef]
- Belaj, A.; del Carmen Dominguez-García, M.; Atienza, S.G.; Urdíroz, N.M.; De la Rosa, R.; Satovic, Z.; Martín, A.; Kilian, A.; Trujillo, I.; Valpuesta, V. Developing a core collection of olive (Olea europaea L.) based on molecular markers (DArTs, SSRs, SNPs) and agronomic traits. Tree Genet. Genomes 2012, 8, 365–378. [Google Scholar] [CrossRef]
- Sion, S.; Taranto, F.; Montemurro, C.; Mangini, G.; Camposeo, S.; Falco, V.; Gallo, A.; Mita, G.; Saddoud Debbabi, O.; Ben Amar, F. Genetic Characterization of Apulian Olive Germplasm as Potential Source in New Breeding Programs. Plants 2019, 8, 268. [Google Scholar] [CrossRef]
- Albertini, E.; Torricelli, R.; Bitocchi, E.; Raggi, L.; Marconi, G.; Pollastri, L.; Di Minco, G.; Battistini, A.; Papa, R.; Veronesi, F. Structure of genetic diversity in Olea europaea L. cultivars from central Italy. Mol. Breed. 2011, 27, 533–547. [Google Scholar] [CrossRef]
- Díez, C.M.; Trujillo, I.; Barrio, E.; Belaj, A.; Barranco, D.; Rallo, L. Centennial olive trees as a reservoir of genetic diversity. Ann. Bot. 2011, 108, 797–807. [Google Scholar] [CrossRef]
- Khadari, B.; Breton, C.; Moutier, N.; Roger, J.; Besnard, G.; Bervillé, A.; Dosba, F. The use of molecular markers for germplasm management in a French olive collection. Theor. Appl. Genet. 2003, 106, 521–529. [Google Scholar] [CrossRef] [PubMed]
- Abdessemed, S.; Muzzalupo, I.; Benbouza, H. Assessment of genetic diversity among Algerian olive (Olea europaea L.) cultivars using SSR marker. Sci. Hortic. 2015, 192, 10–20. [Google Scholar] [CrossRef]
- Omrani-Sabbaghi, A.; Shahriari, M.; Falahati-Anbaran, M.; Mohammadi, S.; Nankali, A.; Mardi, M.; Ghareyazie, B. Microsatellite markers based assessment of genetic diversity in Iranian olive (Olea europaea L.) collections. Sci. Hortic. 2007, 112, 439–447. [Google Scholar] [CrossRef]
- Ginko, E.; Dobeš, C.; Saukel, J. Suitability of root and rhizome anatomy for taxonomic classification and reconstruction of phylogenetic relationships in the tribes cardueae and cichorieae (asteraceae). Sci. Pharm. 2016, 84, 585. [Google Scholar] [CrossRef]
- Germino, M.J.; Barnard, D.M.; Davidson, B.E.; Arkle, R.S.; Pilliod, D.S.; Fisk, M.R.; Applestein, C. Thresholds and hotspots for shrub restoration following a heterogeneous megafire. Landsc. Ecol. 2018, 33, 1177–1194. [Google Scholar] [CrossRef]
- Mousavi, S.; Mariotti, R.; Regni, L.; Nasini, L.; Bufacchi, M.; Pandolfi, S.; Baldoni, L.; Proietti, P. The first molecular identification of an olive collection applying standard simple sequence repeats and novel expressed sequence tag markers. Front. Plant Sci. 2017, 8, 1283. [Google Scholar] [CrossRef]
- Beiki, A.H.; Saboor, S.; Ebrahimi, M. A new avenue for classification and prediction of olive cultivars using supervised and unsupervised algorithms. PLoS ONE 2012, 7, e44164. [Google Scholar] [CrossRef]
- Cultrera, N.G.; Sarri, V.; Lucentini, L.; Ceccarelli, M.; Alagna, F.; Mariotti, R.; Mousavi, S.; Ruiz, C.G.; Baldoni, L. High levels of variation within gene sequences of Olea europaea L. Front. Plant Sci. 2019, 9, 1932. [Google Scholar] [CrossRef] [PubMed]
- Cruz, F.; Julca, I.; Gómez-Garrido, J.; Loska, D.; Marcet-Houben, M.; Cano, E.; Galán, B.; Frias, L.; Ribeca, P.; Derdak, S. Genome sequence of the olive tree, Olea europaea. Gigascience 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Garantonakis, N.; Varikou, K.; Birouraki, A. Parasitism of psytallia concolor (hymenoptera: Braconidae) on bactrocera oleae (diptera: Tephritidae) infesting different olive varieties. Phytoparasitica 2017, 45, 461–469. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).