Multi-Year Dynamics of Single-Step Genomic Prediction in an Applied Wheat Breeding Program
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Statistical Analysis of the Phenotypic Data
2.3. Pedigree and Genotypic Data
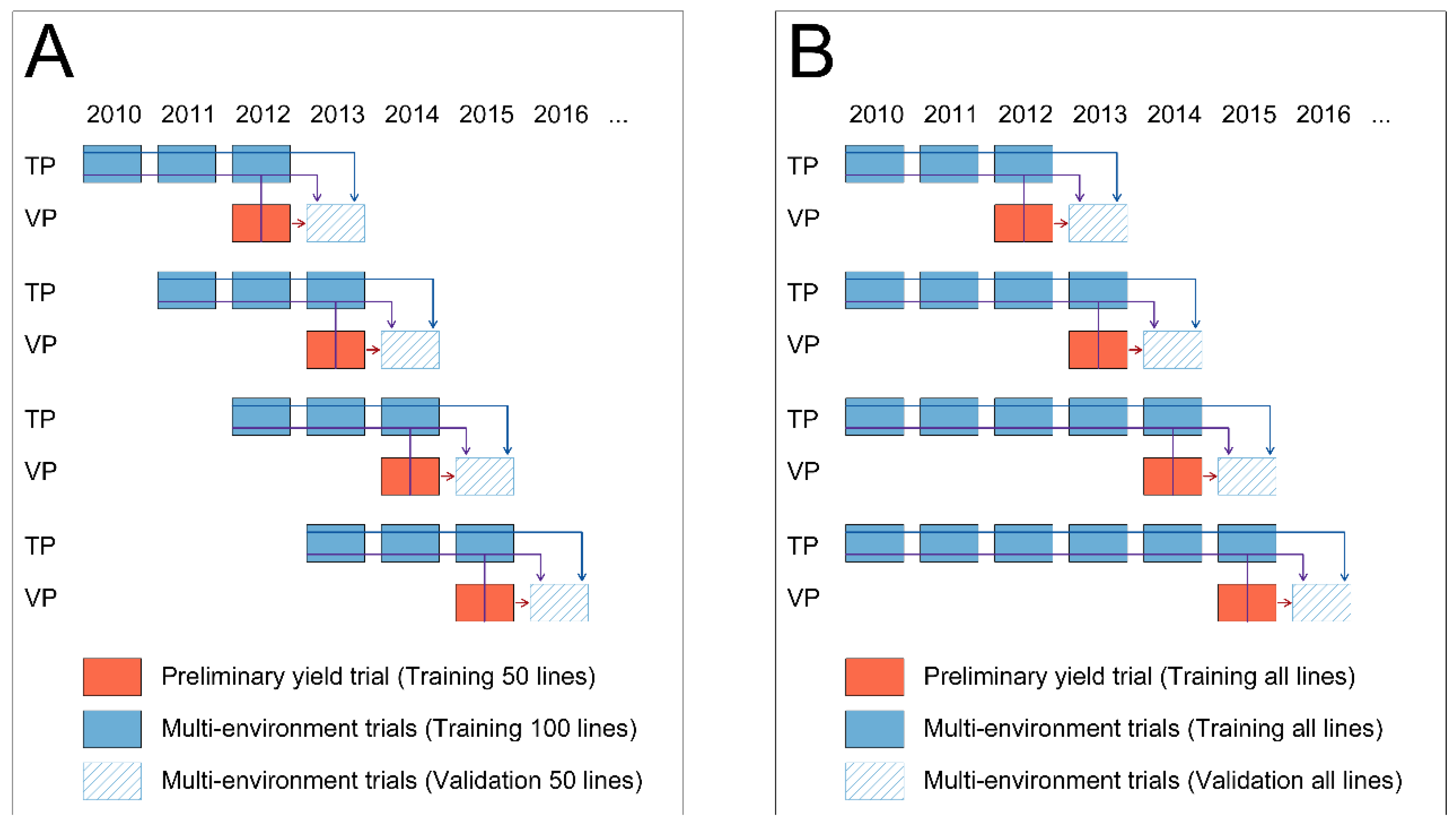
2.4. Comparison of Phenotypic with Pedigree, Genomic and Single-Step Prediction across Years
2.5. Prediction Accuracy within and across Families
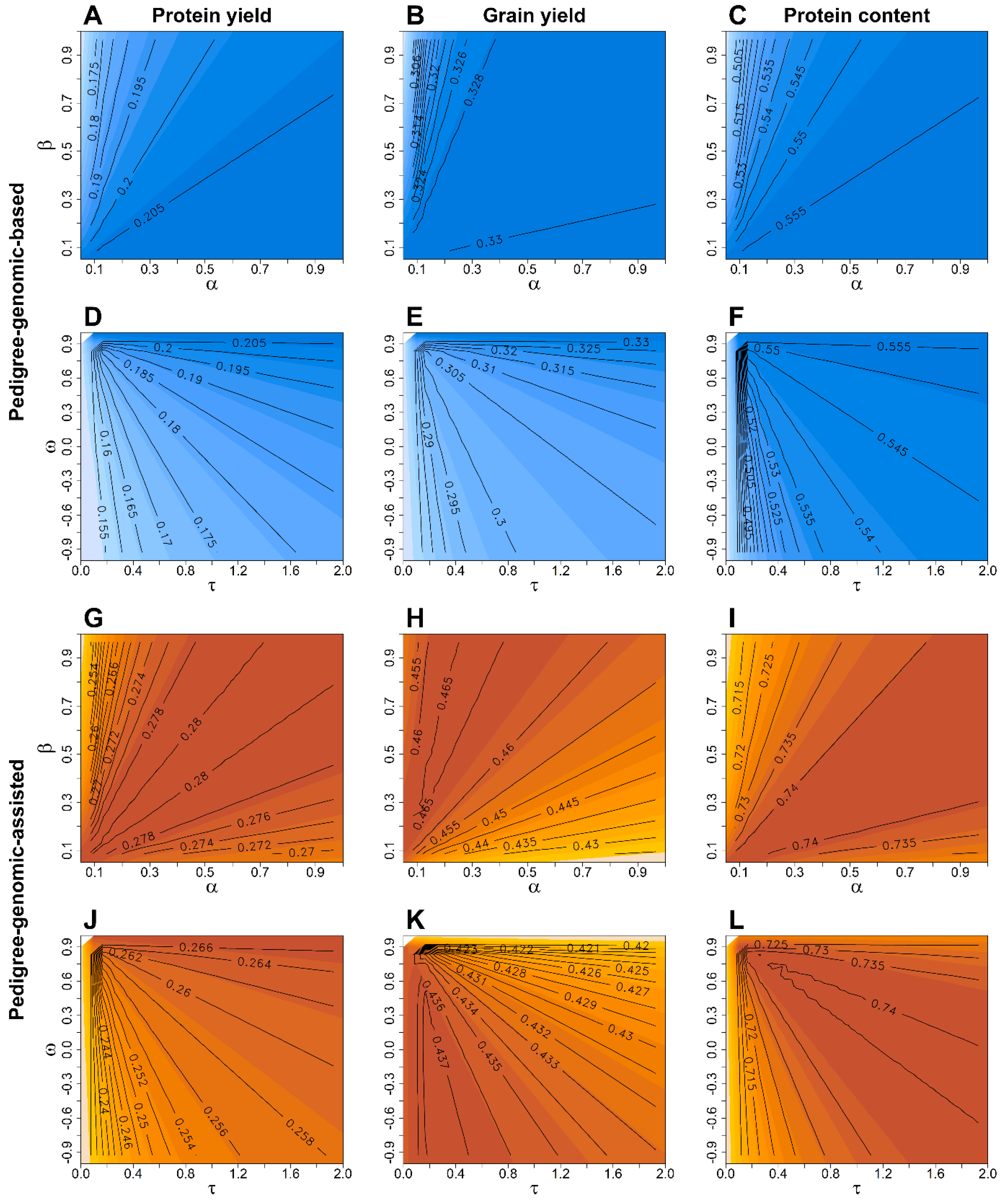
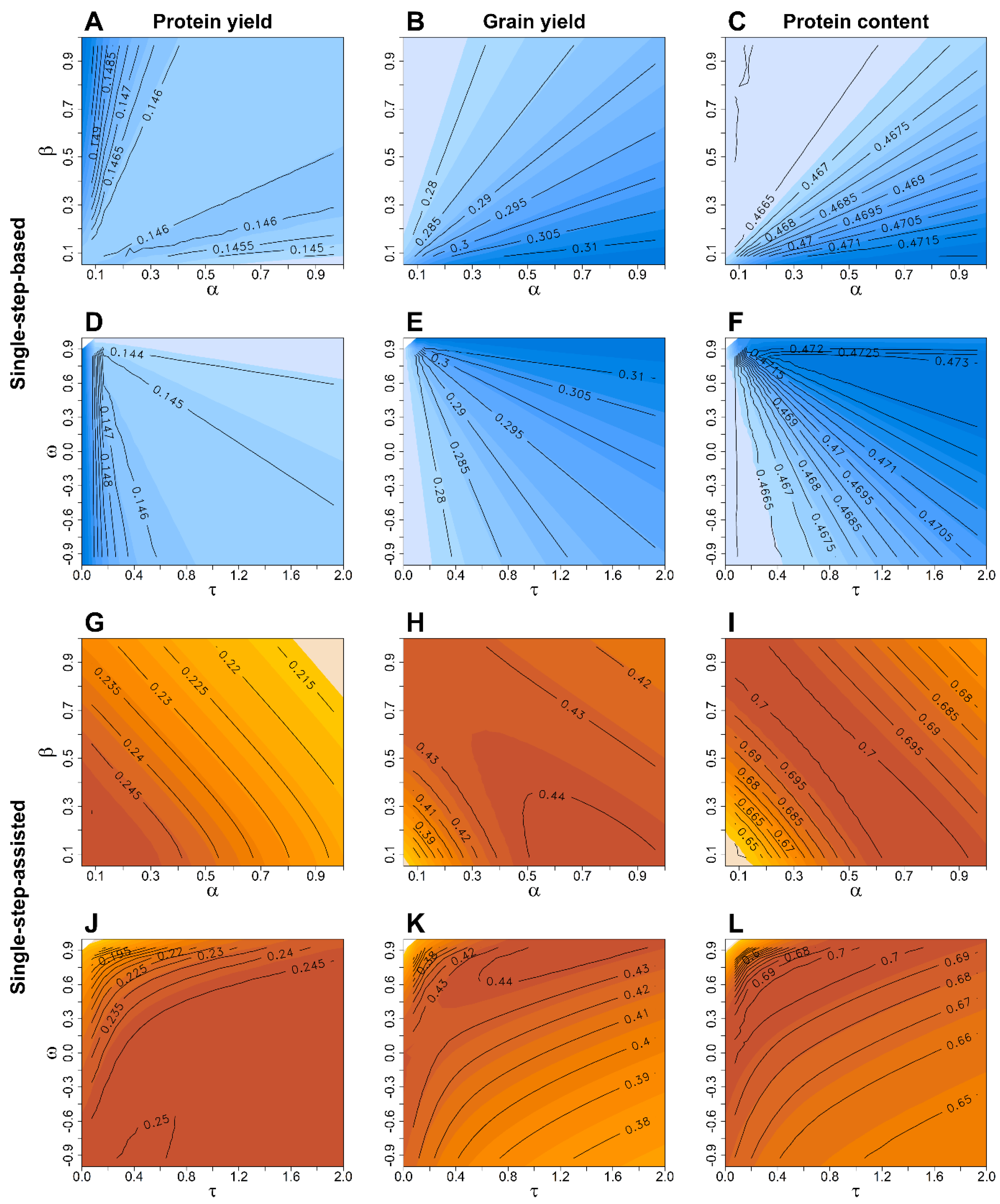
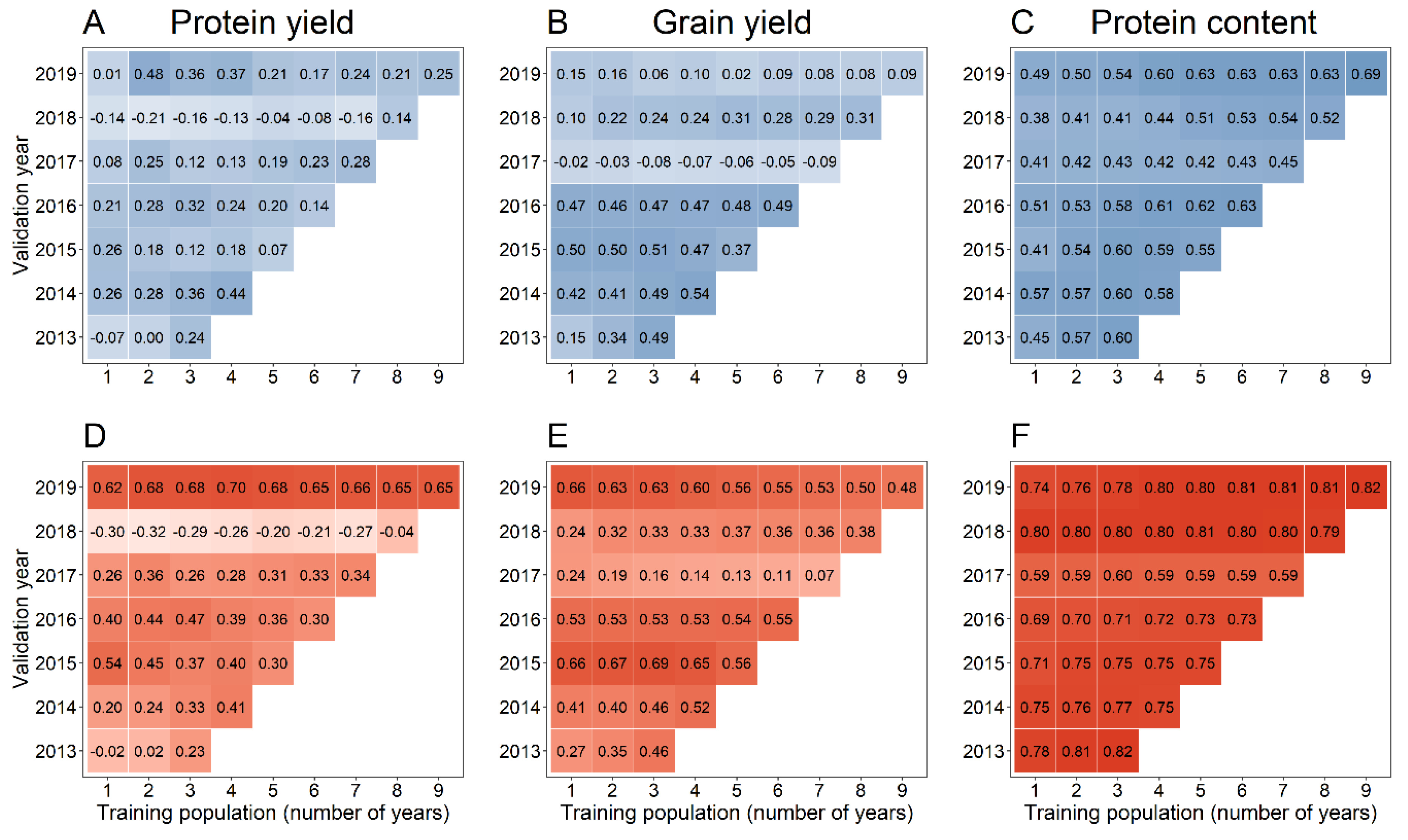
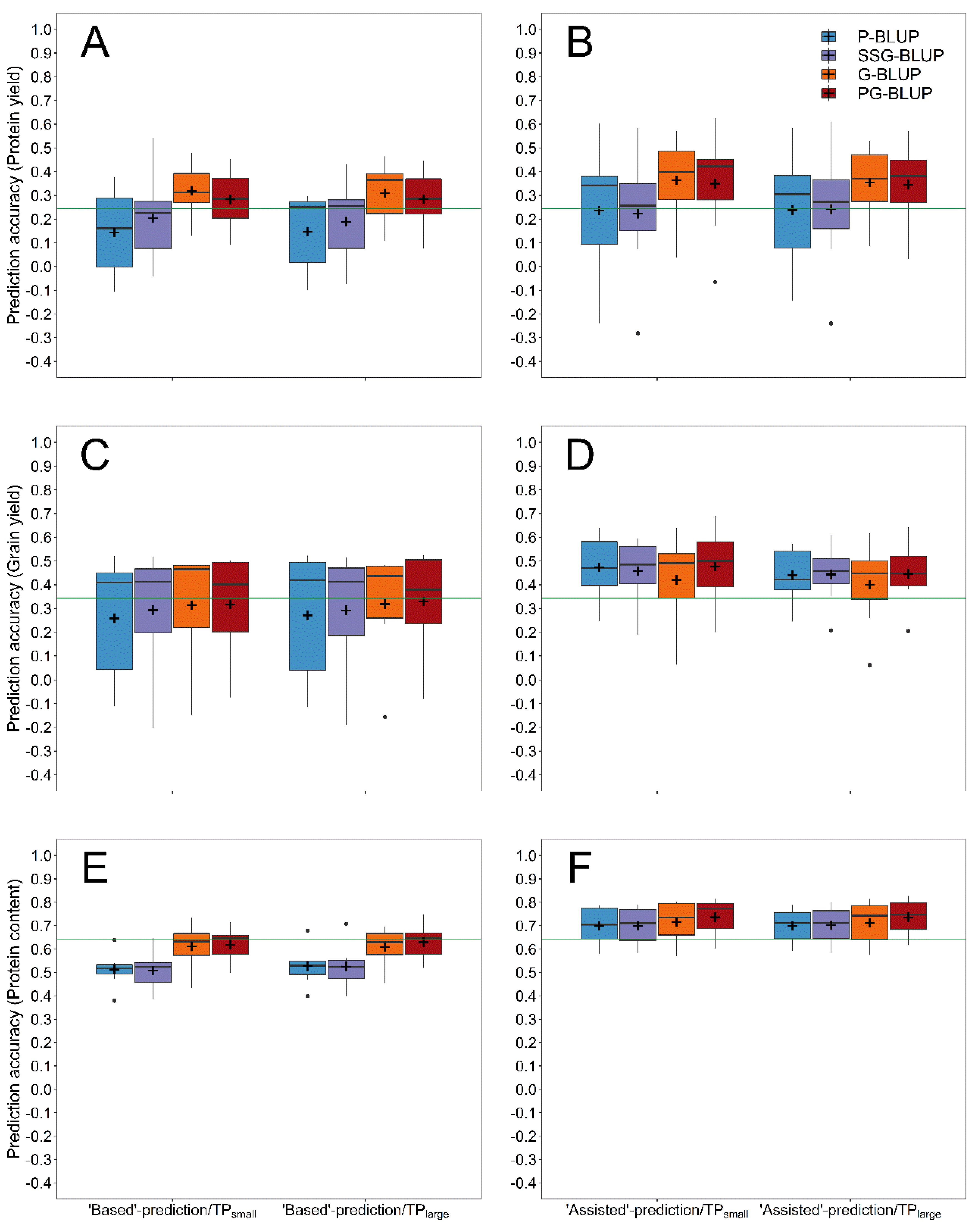
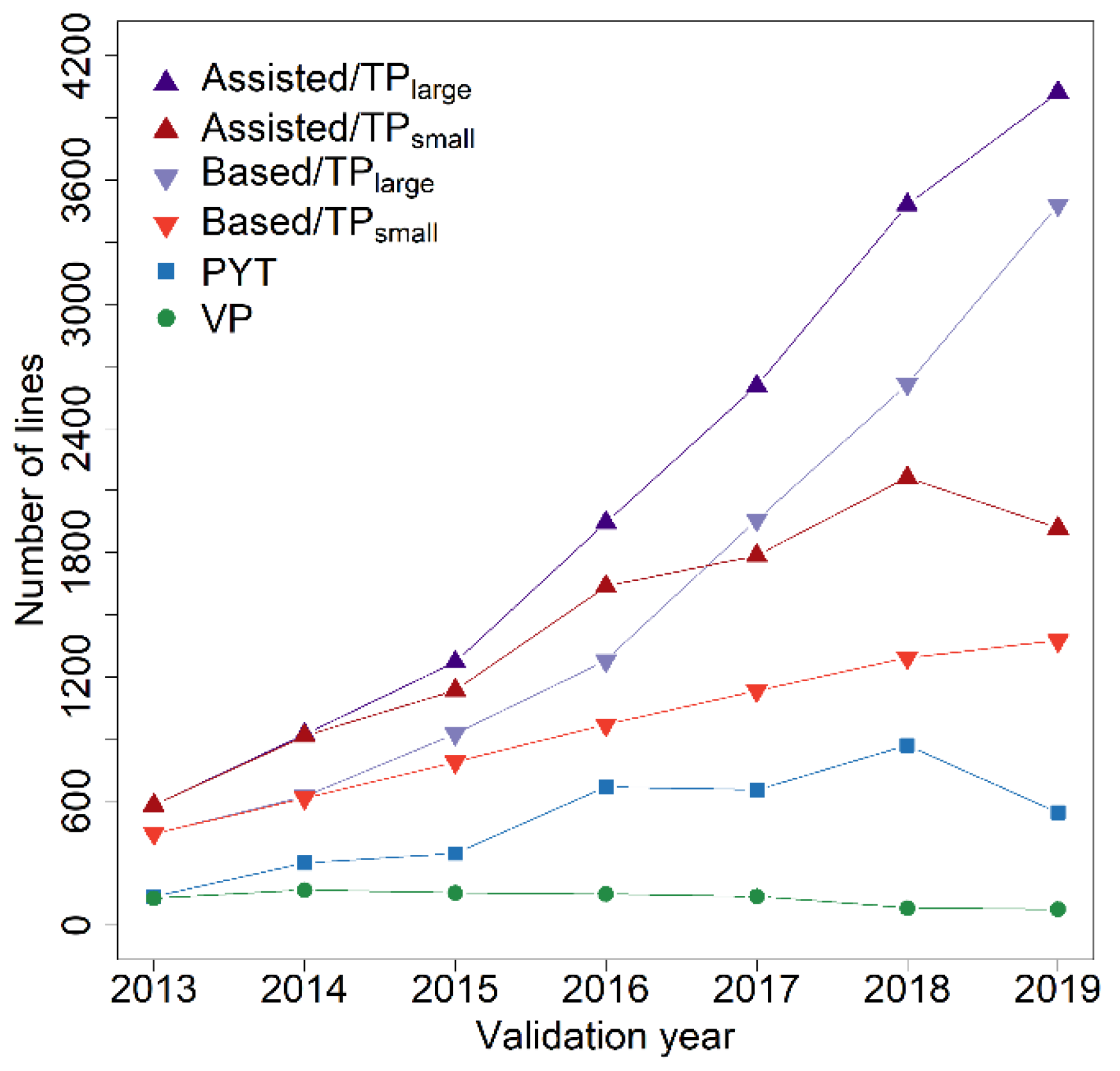
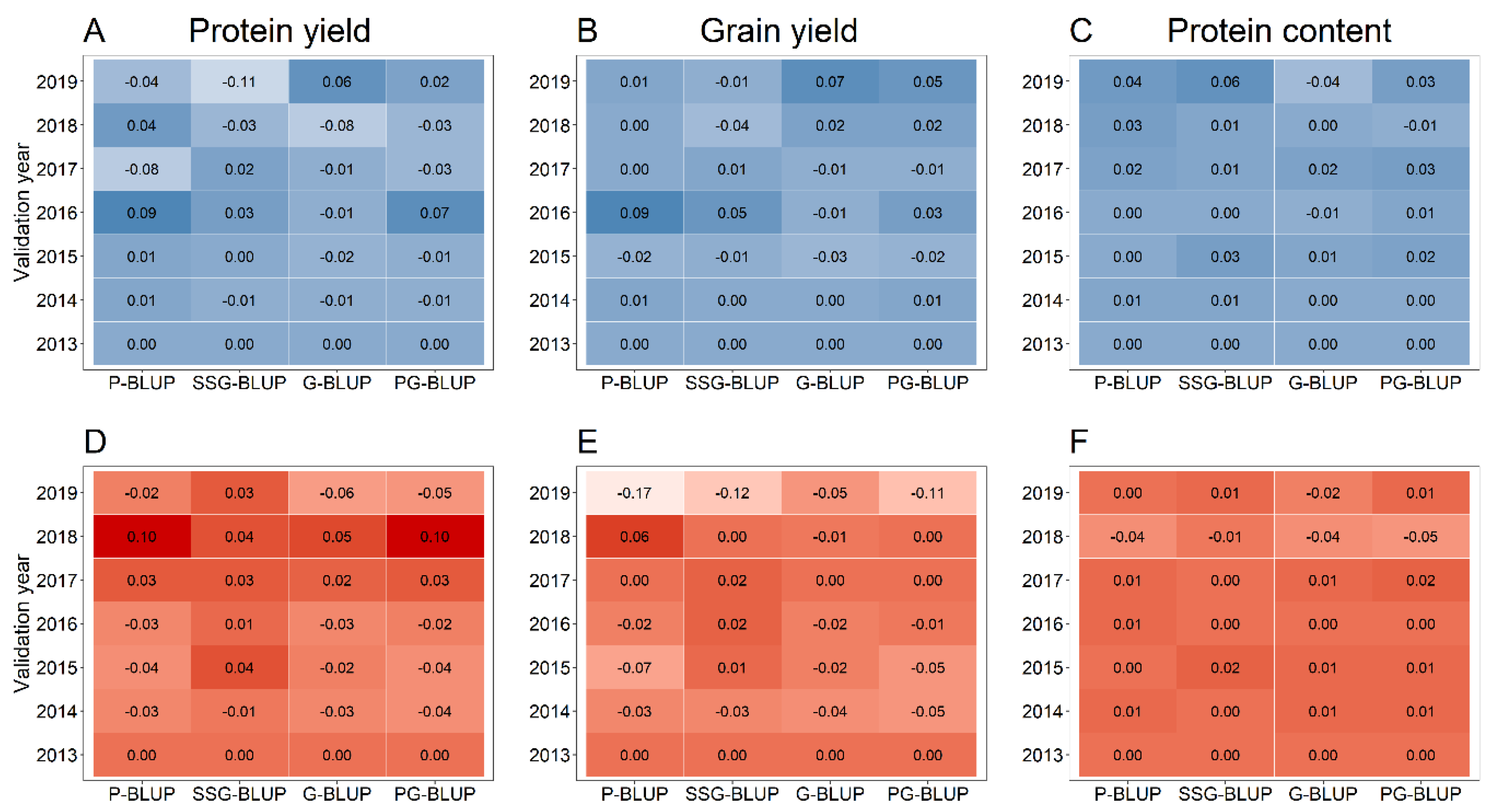
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jarquin, D.; Lemes da Silva, C.; Gaynor, R.C.; Poland, J.; Fritz, A.; Howard, R.; Battenfield, S.; Crossa, J. Increasing Genomic-Enabled Prediction Accuracy by Modeling Genotype x Environment Interactions in Kansas Wheat. Plant Genome 2017, 10. [Google Scholar] [CrossRef] [PubMed]
- Schulz-Streeck, T.; Ogutu, J.O.; Gordillo, A.; Karaman, Z.; Knaak, C.; Piepho, H.P. Genomic selection allowing for marker-by-environment interaction. Plant Breed. 2013, 132, 532–538. [Google Scholar] [CrossRef]
- Schrag, T.A.; Schipprack, W.; Melchinger, A.E. Across-years prediction of hybrid performance in maize using genomics. Theor. Appl. Genet. 2019, 132, 933–946. [Google Scholar] [CrossRef] [PubMed]
- Bernal-Vasquez, A.-M.; Gordillo, A.; Schmidt, M.; Piepho, H.-P. Genomic prediction in early selection stages using multi-year data in a hybrid rye breeding program. BMC Genet. 2017, 18, 51. [Google Scholar] [CrossRef] [PubMed]
- Dias, K.O.G.; Piepho, H.P.; Guimarães, L.J.M.; Guimarães, P.E.O.; Parentoni, S.N.; Pinto, M.O.; Noda, R.W.; Guimarães, C.T.; Garcia, A.A.F.; Pastina, M.M. Novel strategies for genomic prediction of untested single-cross maize hybrids using unbalanced historical data. Theor. Appl. Genet. 2019, 133, 443–455. [Google Scholar] [CrossRef] [PubMed]
- Haikka, H.; Knürr, T.; Manninen, O.; Pietilä, L.; Isolahti, M.; Teperi, E.; Mäntysaari, E.A.; Strandén, I. Genomic prediction of grain yield in commercial Finnish oat (Avena sativa) and barley (Hordeum vulgare) breeding programmes. Plant Breed. 2020, 139, 550–561. [Google Scholar] [CrossRef]
- Ben-Sadoun, S.; Auzanneau, R.R.J.; Rolland, F.X.O.B.; Ravel, E.H.C. Economical optimization of a breeding scheme by selective phenotyping of the calibration set in a multi-trait context: Application to bread making quality. Theor. Appl. Genet. 2020, 133, 2197–2212. [Google Scholar] [CrossRef] [PubMed]
- Bhatta, M.; Gutierrez, L.; Cammarota, L.; Cardozo, F.; Germán, S.; Gómez-Guerrero, B.; Pardo, M.F.; Lanaro, V.; Sayas, M.; Castro, A.J. Multi-trait Genomic Prediction Model Increased the Predictive Ability for Agronomic and Malting Quality Traits in Barley (Hordeum vulgare L.). G3 Genes Genomes Genet. 2020, 10, g3.400968.2019. [Google Scholar] [CrossRef]
- Tsai, H.; Cericola, F.; Edriss, V.; Andersen, J.R.; Id, J.O.; Jensen, J.D.; Jahoor, A.; Janss, L.; Jensen, J. Use of multiple traits genomic prediction, genotype by environment interactions and spatial effect to improve prediction accuracy in yield data. PLoS ONE 2020, 15. [Google Scholar] [CrossRef]
- Robertsen, C.; Hjortshøj, R.; Janss, L. Genomic Selection in Cereal Breeding. Agronomy 2019, 9, 95. [Google Scholar] [CrossRef]
- Sweeney, D.W.; Rutkoski, J.; Bergstrom, G.C.; Sorrells, M.E. A connected half-sib family training population for genomic prediction in barley. Crop Sci. 2020, 60, 262–281. [Google Scholar] [CrossRef]
- Edwards, S.M.; Buntjer, J.B.; Jackson, R.; Bentley, A.R.; Lage, J.; Byrne, E.; Burt, C.; Jack, P.; Berry, S.; Flatman, E.; et al. The effects of training population design on genomic prediction accuracy in wheat. Theor. Appl. Genet. 2019, 132, 1943–1952. [Google Scholar] [CrossRef]
- Auinger, H.-J.; Schönleben, M.; Lehermeier, C.; Schmidt, M.; Korzun, V.; Geiger, H.H.; Piepho, H.-P.; Gordillo, A.; Wilde, P.; Bauer, E.; et al. Model training across multiple breeding cycles significantly improves genomic prediction accuracy in rye (Secale cereale L.). Theor. Appl. Genet. 2016, 129, 2043–2053. [Google Scholar] [CrossRef]
- Cericola, F.; Jahoor, A.; Orabi, J.; Andersen, J.; Janss, L. Optimizing Training Population Size and Genotyping Strategy for Genomic Prediction Using Association Study Results and Pedigree Information. A Case of Study in Advanced Wheat Breeding Lines. PLoS ONE 2017, 12. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Rodríguez, P.; Crossa, J.; Rutkoski, J.; Poland, J.; Singh, R.; Legarra, A.; Autrique, E.; de los Campos, G.; Burgueño, J.; Dreisigacker, S. Single-Step Genomic and Pedigree Genotype × Environment Interaction Models for Predicting Wheat Lines in International Environments. Plant Genome 2017, 10. [Google Scholar] [CrossRef]
- Burgueño, J.; de los Campos, G.; Weigel, K.; Crossa, J. Genomic prediction of breeding values when modeling genotype × environment interaction using pedigree and dense molecular markers. Crop Sci. 2012, 52, 707–719. [Google Scholar] [CrossRef]
- Ankamah-Yeboah, T.; Janss, L.L.; Jensen, J.D.; Hjortshøj, R.; Rasmussen, S.K. Genomic Selection Using Pedigree and Marker-by-Environment Interaction for Barley Seed Quality Traits from Two Commercial Breeding Programs. Front. Plant Sci. 2020. [Google Scholar] [CrossRef] [PubMed]
- Legarra, A.; Aguilar, I.; Misztal, I. A relationship matrix including full pedigree and genomic information. J. Dairy Sci. 2009, 92, 4656–4663. [Google Scholar] [CrossRef]
- Christensen, O.; Lund, M.S. Genomic relationship matrix when some animals are not genotyped. Genet. Sel. Evol. 2010, 42, 1–8. [Google Scholar] [CrossRef]
- Misztal, I.; Aggrey, S.E.; Muir, W.M. Experiences with a single-step genome evaluation. Poult. Sci. 2013, 92, 2530–2534. [Google Scholar] [CrossRef]
- Burgueño, J.; Cadena, A.; Crossa, J. User’s Guide for Spatial Analysis of Field Variety Trials Using ASREML; CIMMYT: Mexico, Mexico, 2000; ISBN 9706480609. [Google Scholar]
- Schmidt, P.; Hartung, J.; Rath, J.; Piepho, H.P. Estimating broad-sense heritability with unbalanced data from agricultural cultivar trials. Crop Sci. 2019, 59, 525–536. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team R: A language and environment for statistical computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Saghai-Maroof, M.A.; Soliman, K.M.; Jorgensen, R.A.; Allard, R.W. Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc. Natl. Acad. Sci. USA 1984, 81, 8014–8018. [Google Scholar] [CrossRef] [PubMed]
- Diversity Arrays Technology; DArT P/L Pty Ltd.: Canberra, Australia, 2020.
- Stekhoven, D.J.; Bühlmann, P. Missforest-Non-parametric missing value imputation for mixed-type data. Bioinformatics 2012, 28, 112–118. [Google Scholar] [CrossRef]
- He, S.; Zhao, Y.; Mette, M.F.; Bothe, R.; Ebmeyer, E.; Sharbel, T.F.; Reif, J.C.; Jiang, Y. Prospects and limits of marker imputation in quantitative genetic studies in European elite wheat (Triticum aestivum L.). BMC Genom. 2015, 16, 168. [Google Scholar] [CrossRef]
- Henderson, C. A Simple Method for Computing the Inverse of a Numerator Relationship Matrix Used in Prediction of Breeding Values. Biometrics 1976, 32, 69–83. [Google Scholar] [CrossRef]
- VanRaden, P.M. Efficient methods to compute genomic predictions. J. Dairy Sci. 2008, 91, 4414–4423. [Google Scholar] [CrossRef]
- Christensen, O.; Madsen, P.; Nielsen, B.; Ostersen, T.; Su, G. Single-step methods for genomic evaluation in pigs. Animal 2012, 6, 1565–1571. [Google Scholar] [CrossRef]
- Dekkers, J.C.M. Prediction of response to marker assited and genomic selection using selection index theory. J. Anim. Breed. Genet. 2007, 124, 331–341. [Google Scholar] [CrossRef]
- Amadeu, R.R.; Cellon, C.; Olmstead, J.W.; Garcia, A.A.F.; Resende, M.F.R.; Muñoz, P.R. AGHmatrix: R Package to Construct Relationship Matrices for Autotetraploid and Diploid Species: A Blueberry Example. Plant Genome 2016, 9, 1–10. [Google Scholar] [CrossRef]
- Covarrubias-Pazaran, G. Genome-Assisted prediction of quantitative traits using the r package sommer. PLoS ONE 2016, 11, 1–15. [Google Scholar] [CrossRef]
- Broman, K.W.; Wu, H.; Sen, Ś.; Churchill, G.A. R/qtl: QTL mapping in experimental crosses. Bioinformatics 2003, 19, 889–890. [Google Scholar] [CrossRef]
- Stahl, F.W. Special Sites in Generalized Recombination. Annu. Rev. Genet. 1979, 13, 7–24. [Google Scholar] [CrossRef]
- Piepho, H.P.; Laidig, F.; Drobek, T.; Meyer, U. Dissecting genetic and non-genetic sources of long-term yield trend in German official variety trials. Theor. Appl. Genet. 2014, 127, 1009–1018. [Google Scholar] [CrossRef] [PubMed]
- Laidig, F.; Piepho, H.-P.; Rentel, D.; Drobek, T.; Meyer, U.; Huesken, A. Breeding progress, environmental variation and correlation of winter wheat yield and quality traits in German official variety trials and on-farm during 1983–2014. Theor. Appl. Genet. 2017, 130, 223–245. [Google Scholar] [CrossRef] [PubMed]
- Muellner, A.E.; Mascher, F.; Schneider, D.; Ittu, G.; Toncea, I.; Rolland, B.; Löschenberger, F. Refining breeding methods for organic and low-input agriculture: Analysis of an international winter wheat ring test. Euphytica 2014, 199, 81–95. [Google Scholar] [CrossRef]
- Legarra, A.; Christensen, O.; Aguilar, I.; Misztal, I. Single Step, a general approach for genomic selection. Livest. Sci. 2014, 166, 54–65. [Google Scholar] [CrossRef]
- Cappa, E.P.; de Lima, B.M.; da Silva-Junior, O.B.; Garcia, C.C.; Mansfield, S.D.; Grattapaglia, D. Improving genomic prediction of growth and wood traits in Eucalyptus using phenotypes from non-genotyped trees by single-step GBLUP. Plant Sci. 2019, 284, 9–15. [Google Scholar] [CrossRef]
- Imai, A.; Kuniga, T.; Yoshioka, T.; Nonaka, K.; Mitani, N.; Fukamachi, H.; Hiehata, N.; Yamamoto, M.; Hayashi, T. Single-step genomic prediction of fruit-quality traits using phenotypic records of non-genotyped relatives in citrus. PLoS ONE 2019, 14, 1–14. [Google Scholar] [CrossRef]
- Ashraf, B.; Edriss, V.; Akdemir, D.; Autrique, E.; Bonnett, D.; Crossa, J.; Janss, L.; Singh, R.; Jannink, J.L. Genomic prediction using phenotypes from pedigreed lines with no marker data. Crop Sci. 2016, 56, 957–964. [Google Scholar] [CrossRef]
- Velazco, J.G.; Malosetti, M.; Hunt, C.H.; Mace, E.S.; Jordan, D.R.; van Eeuwijk, F.A. Combining pedigree and genomic information to improve prediction quality: An example in sorghum. Theor. Appl. Genet. 2019, 132, 2055–2067. [Google Scholar] [CrossRef]
- Zapata-Valenzuela, J.; Whetten, R.W.; Neale, D.; McKeand, S.; Isik, F. Genomic estimated breeding values using genomic relationship matrices in a cloned population of loblolly pine. G3 Genes Genomes Genet. 2013, 3, 909–916. [Google Scholar] [CrossRef] [PubMed]
- Albrecht, T.; Wimmer, V.; Auinger, H.J.; Erbe, M.; Knaak, C.; Ouzunova, M.; Simianer, H.; Sch??n, C.C. Genome-based prediction of testcross values in maize. Theor. Appl. Genet. 2011, 123, 339–350. [Google Scholar] [CrossRef] [PubMed]
- Brauner, P.C.; Müller, D.; Molenaar, W.S.; Melchinger, A.E. Genomic prediction with multiple biparental families. Theor. Appl. Genet. 2020, 133, 133–147. [Google Scholar] [CrossRef]
- Schopp, P.; Müller, D.; Wientjes, Y.C.J.; Melchinger, A.E. Genomic Prediction Within and Across Biparental Families: Means and Variances of Prediction Accuracy and Usefulness of Deterministic Equations. G3 Genes Genomes Genet. 2017, 7, 3571–3586. [Google Scholar] [CrossRef]
- Molenaar, H.; Boehm, R.; Piepho, H.-P. Phenotypic Selection in Ornamental Breeding: It’s Better to Have the BLUPs Than to Have the BLUEs. Front. Plant Sci. 2018, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Cellon, C.; Amadeu, R.R.; Olmstead, J.W.; Mattia, M.R.; Ferrao, L.F.V.; Munoz, P.R. Estimation of genetic parameters and prediction of breeding values in an autotetraploid blueberry breeding population with extensive pedigree data. Euphytica 2018, 214, 1–13. [Google Scholar] [CrossRef]
- Slater, A.T.; Wilson, G.M.; Cogan, N.O.I.; Forster, J.W.; Hayes, B.J. Improving the analysis of low heritability complex traits for enhanced genetic gain in potato. Theor. Appl. Genet. 2014, 127, 809–820. [Google Scholar] [CrossRef]
- Watson, A.; Hickey, L.T.; Christopher, J.; Rutkoski, J.; Poland, J.; Hayes, B.J. Multivariate genomic selection and the potential of rapid indirect phenotypic selection with speed breeding to increase genetic gain in spring bread wheat. Crop Sci. 2019, 59, 1945–1959. [Google Scholar] [CrossRef]
- Utz, H.F.; Bohn, M.; Melchinger, A.E. Predicting progeny means and variances of winter wheat crosses from phenotypic values of their parents. Crop Sci. 2001, 41, 1470–1478. [Google Scholar] [CrossRef]
- Zhong, S.; Jannink, J.L. Using quantitative trait loci results to discriminate among crosses on the basis of their progeny mean and variance. Genetics 2007, 177, 567–576. [Google Scholar] [CrossRef]
| Year | Number of Lines † | ||
|---|---|---|---|
| Multi-Environment ‡ | Observation Trial (All) § | Observation Trial (Sel.) ¶ | |
| 2010 | 127 | ||
| 2011 | 162 | ||
| 2012 | 208 | 142 | 130 |
| 2013 | 193 | 306 | 170 |
| 2014 | 202 | 361 | 156 |
| 2015 | 206 | 676 | 151 |
| 2016 | 192 | 666 | 139 |
| 2017 | 209 | 875 | 83 |
| 2018 | 113 | 554 | 76 |
| 2019 | 114 | ||
| h² † | Model | With Pre-Existing Information | Without Pre-Existing Information | ||||
|---|---|---|---|---|---|---|---|
| Population ‡ | Across § | Within ¶ | Population ‡ | Across § | Within ¶ | ||
| 0.1 (0.3) | OBS | 0.319 | 0.835 | 0.246 | |||
| P-BLUP | 0.604 | 0.889 | 0.246 | 0.234 | 0.356 | 0.000 | |
| SSG-BLUP | 0.605 | 0.892 | 0.246 | 0.268 | 0.410 | 0.000 | |
| G-BLUP | 0.665 | 0.903 | 0.405 | 0.389 | 0.546 | 0.209 | |
| PG-BLUP | 0.670 | 0.902 | 0.405 | 0.375 | 0.512 | 0.208 | |
| 0.3 (0.5) | OBS | 0.548 | 0.949 | 0.438 | |||
| P-BLUP | 0.704 | 0.961 | 0.438 | 0.275 | 0.421 | 0.000 | |
| SSG-BLUP | 0.705 | 0.962 | 0.438 | 0.299 | 0.459 | 0.000 | |
| G-BLUP | 0.760 | 0.963 | 0.552 | 0.445 | 0.618 | 0.253 | |
| PG-BLUP | 0.772 | 0.965 | 0.576 | 0.440 | 0.598 | 0.253 | |
| 0.5 (0.7) | OBS | 0.707 | 0.977 | 0.595 | |||
| P-BLUP | 0.778 | 0.980 | 0.595 | 0.297 | 0.455 | 0.000 | |
| SSG-BLUP | 0.779 | 0.981 | 0.595 | 0.321 | 0.491 | 0.000 | |
| G-BLUP | 0.810 | 0.980 | 0.643 | 0.480 | 0.660 | 0.284 | |
| PG-BLUP | 0.832 | 0.982 | 0.687 | 0.483 | 0.660 | 0.285 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Michel, S.; Löschenberger, F.; Sparry, E.; Ametz, C.; Bürstmayr, H. Multi-Year Dynamics of Single-Step Genomic Prediction in an Applied Wheat Breeding Program. Agronomy 2020, 10, 1591. https://doi.org/10.3390/agronomy10101591
Michel S, Löschenberger F, Sparry E, Ametz C, Bürstmayr H. Multi-Year Dynamics of Single-Step Genomic Prediction in an Applied Wheat Breeding Program. Agronomy. 2020; 10(10):1591. https://doi.org/10.3390/agronomy10101591
Chicago/Turabian StyleMichel, Sebastian, Franziska Löschenberger, Ellen Sparry, Christian Ametz, and Hermann Bürstmayr. 2020. "Multi-Year Dynamics of Single-Step Genomic Prediction in an Applied Wheat Breeding Program" Agronomy 10, no. 10: 1591. https://doi.org/10.3390/agronomy10101591
APA StyleMichel, S., Löschenberger, F., Sparry, E., Ametz, C., & Bürstmayr, H. (2020). Multi-Year Dynamics of Single-Step Genomic Prediction in an Applied Wheat Breeding Program. Agronomy, 10(10), 1591. https://doi.org/10.3390/agronomy10101591





