Multi-Year Dynamics of Single-Step Genomic Prediction in an Applied Wheat Breeding Program
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Statistical Analysis of the Phenotypic Data
2.3. Pedigree and Genotypic Data
2.4. Comparison of Phenotypic with Pedigree, Genomic and Single-Step Prediction across Years
2.5. Prediction Accuracy within and across Families
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jarquin, D.; Lemes da Silva, C.; Gaynor, R.C.; Poland, J.; Fritz, A.; Howard, R.; Battenfield, S.; Crossa, J. Increasing Genomic-Enabled Prediction Accuracy by Modeling Genotype x Environment Interactions in Kansas Wheat. Plant Genome 2017, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schulz-Streeck, T.; Ogutu, J.O.; Gordillo, A.; Karaman, Z.; Knaak, C.; Piepho, H.P. Genomic selection allowing for marker-by-environment interaction. Plant Breed. 2013, 132, 532–538. [Google Scholar] [CrossRef]
- Schrag, T.A.; Schipprack, W.; Melchinger, A.E. Across-years prediction of hybrid performance in maize using genomics. Theor. Appl. Genet. 2019, 132, 933–946. [Google Scholar] [CrossRef] [PubMed]
- Bernal-Vasquez, A.-M.; Gordillo, A.; Schmidt, M.; Piepho, H.-P. Genomic prediction in early selection stages using multi-year data in a hybrid rye breeding program. BMC Genet. 2017, 18, 51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dias, K.O.G.; Piepho, H.P.; Guimarães, L.J.M.; Guimarães, P.E.O.; Parentoni, S.N.; Pinto, M.O.; Noda, R.W.; Guimarães, C.T.; Garcia, A.A.F.; Pastina, M.M. Novel strategies for genomic prediction of untested single-cross maize hybrids using unbalanced historical data. Theor. Appl. Genet. 2019, 133, 443–455. [Google Scholar] [CrossRef] [PubMed]
- Haikka, H.; Knürr, T.; Manninen, O.; Pietilä, L.; Isolahti, M.; Teperi, E.; Mäntysaari, E.A.; Strandén, I. Genomic prediction of grain yield in commercial Finnish oat (Avena sativa) and barley (Hordeum vulgare) breeding programmes. Plant Breed. 2020, 139, 550–561. [Google Scholar] [CrossRef]
- Ben-Sadoun, S.; Auzanneau, R.R.J.; Rolland, F.X.O.B.; Ravel, E.H.C. Economical optimization of a breeding scheme by selective phenotyping of the calibration set in a multi-trait context: Application to bread making quality. Theor. Appl. Genet. 2020, 133, 2197–2212. [Google Scholar] [CrossRef] [PubMed]
- Bhatta, M.; Gutierrez, L.; Cammarota, L.; Cardozo, F.; Germán, S.; Gómez-Guerrero, B.; Pardo, M.F.; Lanaro, V.; Sayas, M.; Castro, A.J. Multi-trait Genomic Prediction Model Increased the Predictive Ability for Agronomic and Malting Quality Traits in Barley (Hordeum vulgare L.). G3 Genes Genomes Genet. 2020, 10, g3.400968.2019. [Google Scholar] [CrossRef] [Green Version]
- Tsai, H.; Cericola, F.; Edriss, V.; Andersen, J.R.; Id, J.O.; Jensen, J.D.; Jahoor, A.; Janss, L.; Jensen, J. Use of multiple traits genomic prediction, genotype by environment interactions and spatial effect to improve prediction accuracy in yield data. PLoS ONE 2020, 15. [Google Scholar] [CrossRef]
- Robertsen, C.; Hjortshøj, R.; Janss, L. Genomic Selection in Cereal Breeding. Agronomy 2019, 9, 95. [Google Scholar] [CrossRef] [Green Version]
- Sweeney, D.W.; Rutkoski, J.; Bergstrom, G.C.; Sorrells, M.E. A connected half-sib family training population for genomic prediction in barley. Crop Sci. 2020, 60, 262–281. [Google Scholar] [CrossRef] [Green Version]
- Edwards, S.M.; Buntjer, J.B.; Jackson, R.; Bentley, A.R.; Lage, J.; Byrne, E.; Burt, C.; Jack, P.; Berry, S.; Flatman, E.; et al. The effects of training population design on genomic prediction accuracy in wheat. Theor. Appl. Genet. 2019, 132, 1943–1952. [Google Scholar] [CrossRef] [Green Version]
- Auinger, H.-J.; Schönleben, M.; Lehermeier, C.; Schmidt, M.; Korzun, V.; Geiger, H.H.; Piepho, H.-P.; Gordillo, A.; Wilde, P.; Bauer, E.; et al. Model training across multiple breeding cycles significantly improves genomic prediction accuracy in rye (Secale cereale L.). Theor. Appl. Genet. 2016, 129, 2043–2053. [Google Scholar] [CrossRef] [Green Version]
- Cericola, F.; Jahoor, A.; Orabi, J.; Andersen, J.; Janss, L. Optimizing Training Population Size and Genotyping Strategy for Genomic Prediction Using Association Study Results and Pedigree Information. A Case of Study in Advanced Wheat Breeding Lines. PLoS ONE 2017, 12. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Rodríguez, P.; Crossa, J.; Rutkoski, J.; Poland, J.; Singh, R.; Legarra, A.; Autrique, E.; de los Campos, G.; Burgueño, J.; Dreisigacker, S. Single-Step Genomic and Pedigree Genotype × Environment Interaction Models for Predicting Wheat Lines in International Environments. Plant Genome 2017, 10. [Google Scholar] [CrossRef] [Green Version]
- Burgueño, J.; de los Campos, G.; Weigel, K.; Crossa, J. Genomic prediction of breeding values when modeling genotype × environment interaction using pedigree and dense molecular markers. Crop Sci. 2012, 52, 707–719. [Google Scholar] [CrossRef] [Green Version]
- Ankamah-Yeboah, T.; Janss, L.L.; Jensen, J.D.; Hjortshøj, R.; Rasmussen, S.K. Genomic Selection Using Pedigree and Marker-by-Environment Interaction for Barley Seed Quality Traits from Two Commercial Breeding Programs. Front. Plant Sci. 2020. [Google Scholar] [CrossRef] [PubMed]
- Legarra, A.; Aguilar, I.; Misztal, I. A relationship matrix including full pedigree and genomic information. J. Dairy Sci. 2009, 92, 4656–4663. [Google Scholar] [CrossRef] [Green Version]
- Christensen, O.; Lund, M.S. Genomic relationship matrix when some animals are not genotyped. Genet. Sel. Evol. 2010, 42, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Misztal, I.; Aggrey, S.E.; Muir, W.M. Experiences with a single-step genome evaluation. Poult. Sci. 2013, 92, 2530–2534. [Google Scholar] [CrossRef]
- Burgueño, J.; Cadena, A.; Crossa, J. User’s Guide for Spatial Analysis of Field Variety Trials Using ASREML; CIMMYT: Mexico, Mexico, 2000; ISBN 9706480609. [Google Scholar]
- Schmidt, P.; Hartung, J.; Rath, J.; Piepho, H.P. Estimating broad-sense heritability with unbalanced data from agricultural cultivar trials. Crop Sci. 2019, 59, 525–536. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team R: A language and environment for statistical computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Saghai-Maroof, M.A.; Soliman, K.M.; Jorgensen, R.A.; Allard, R.W. Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc. Natl. Acad. Sci. USA 1984, 81, 8014–8018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diversity Arrays Technology; DArT P/L Pty Ltd.: Canberra, Australia, 2020.
- Stekhoven, D.J.; Bühlmann, P. Missforest-Non-parametric missing value imputation for mixed-type data. Bioinformatics 2012, 28, 112–118. [Google Scholar] [CrossRef] [Green Version]
- He, S.; Zhao, Y.; Mette, M.F.; Bothe, R.; Ebmeyer, E.; Sharbel, T.F.; Reif, J.C.; Jiang, Y. Prospects and limits of marker imputation in quantitative genetic studies in European elite wheat (Triticum aestivum L.). BMC Genom. 2015, 16, 168. [Google Scholar] [CrossRef] [Green Version]
- Henderson, C. A Simple Method for Computing the Inverse of a Numerator Relationship Matrix Used in Prediction of Breeding Values. Biometrics 1976, 32, 69–83. [Google Scholar] [CrossRef]
- VanRaden, P.M. Efficient methods to compute genomic predictions. J. Dairy Sci. 2008, 91, 4414–4423. [Google Scholar] [CrossRef] [Green Version]
- Christensen, O.; Madsen, P.; Nielsen, B.; Ostersen, T.; Su, G. Single-step methods for genomic evaluation in pigs. Animal 2012, 6, 1565–1571. [Google Scholar] [CrossRef] [Green Version]
- Dekkers, J.C.M. Prediction of response to marker assited and genomic selection using selection index theory. J. Anim. Breed. Genet. 2007, 124, 331–341. [Google Scholar] [CrossRef]
- Amadeu, R.R.; Cellon, C.; Olmstead, J.W.; Garcia, A.A.F.; Resende, M.F.R.; Muñoz, P.R. AGHmatrix: R Package to Construct Relationship Matrices for Autotetraploid and Diploid Species: A Blueberry Example. Plant Genome 2016, 9, 1–10. [Google Scholar] [CrossRef]
- Covarrubias-Pazaran, G. Genome-Assisted prediction of quantitative traits using the r package sommer. PLoS ONE 2016, 11, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Broman, K.W.; Wu, H.; Sen, Ś.; Churchill, G.A. R/qtl: QTL mapping in experimental crosses. Bioinformatics 2003, 19, 889–890. [Google Scholar] [CrossRef] [Green Version]
- Stahl, F.W. Special Sites in Generalized Recombination. Annu. Rev. Genet. 1979, 13, 7–24. [Google Scholar] [CrossRef]
- Piepho, H.P.; Laidig, F.; Drobek, T.; Meyer, U. Dissecting genetic and non-genetic sources of long-term yield trend in German official variety trials. Theor. Appl. Genet. 2014, 127, 1009–1018. [Google Scholar] [CrossRef] [PubMed]
- Laidig, F.; Piepho, H.-P.; Rentel, D.; Drobek, T.; Meyer, U.; Huesken, A. Breeding progress, environmental variation and correlation of winter wheat yield and quality traits in German official variety trials and on-farm during 1983–2014. Theor. Appl. Genet. 2017, 130, 223–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muellner, A.E.; Mascher, F.; Schneider, D.; Ittu, G.; Toncea, I.; Rolland, B.; Löschenberger, F. Refining breeding methods for organic and low-input agriculture: Analysis of an international winter wheat ring test. Euphytica 2014, 199, 81–95. [Google Scholar] [CrossRef]
- Legarra, A.; Christensen, O.; Aguilar, I.; Misztal, I. Single Step, a general approach for genomic selection. Livest. Sci. 2014, 166, 54–65. [Google Scholar] [CrossRef]
- Cappa, E.P.; de Lima, B.M.; da Silva-Junior, O.B.; Garcia, C.C.; Mansfield, S.D.; Grattapaglia, D. Improving genomic prediction of growth and wood traits in Eucalyptus using phenotypes from non-genotyped trees by single-step GBLUP. Plant Sci. 2019, 284, 9–15. [Google Scholar] [CrossRef]
- Imai, A.; Kuniga, T.; Yoshioka, T.; Nonaka, K.; Mitani, N.; Fukamachi, H.; Hiehata, N.; Yamamoto, M.; Hayashi, T. Single-step genomic prediction of fruit-quality traits using phenotypic records of non-genotyped relatives in citrus. PLoS ONE 2019, 14, 1–14. [Google Scholar] [CrossRef]
- Ashraf, B.; Edriss, V.; Akdemir, D.; Autrique, E.; Bonnett, D.; Crossa, J.; Janss, L.; Singh, R.; Jannink, J.L. Genomic prediction using phenotypes from pedigreed lines with no marker data. Crop Sci. 2016, 56, 957–964. [Google Scholar] [CrossRef] [Green Version]
- Velazco, J.G.; Malosetti, M.; Hunt, C.H.; Mace, E.S.; Jordan, D.R.; van Eeuwijk, F.A. Combining pedigree and genomic information to improve prediction quality: An example in sorghum. Theor. Appl. Genet. 2019, 132, 2055–2067. [Google Scholar] [CrossRef] [Green Version]
- Zapata-Valenzuela, J.; Whetten, R.W.; Neale, D.; McKeand, S.; Isik, F. Genomic estimated breeding values using genomic relationship matrices in a cloned population of loblolly pine. G3 Genes Genomes Genet. 2013, 3, 909–916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albrecht, T.; Wimmer, V.; Auinger, H.J.; Erbe, M.; Knaak, C.; Ouzunova, M.; Simianer, H.; Sch??n, C.C. Genome-based prediction of testcross values in maize. Theor. Appl. Genet. 2011, 123, 339–350. [Google Scholar] [CrossRef] [PubMed]
- Brauner, P.C.; Müller, D.; Molenaar, W.S.; Melchinger, A.E. Genomic prediction with multiple biparental families. Theor. Appl. Genet. 2020, 133, 133–147. [Google Scholar] [CrossRef]
- Schopp, P.; Müller, D.; Wientjes, Y.C.J.; Melchinger, A.E. Genomic Prediction Within and Across Biparental Families: Means and Variances of Prediction Accuracy and Usefulness of Deterministic Equations. G3 Genes Genomes Genet. 2017, 7, 3571–3586. [Google Scholar] [CrossRef] [Green Version]
- Molenaar, H.; Boehm, R.; Piepho, H.-P. Phenotypic Selection in Ornamental Breeding: It’s Better to Have the BLUPs Than to Have the BLUEs. Front. Plant Sci. 2018, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Cellon, C.; Amadeu, R.R.; Olmstead, J.W.; Mattia, M.R.; Ferrao, L.F.V.; Munoz, P.R. Estimation of genetic parameters and prediction of breeding values in an autotetraploid blueberry breeding population with extensive pedigree data. Euphytica 2018, 214, 1–13. [Google Scholar] [CrossRef]
- Slater, A.T.; Wilson, G.M.; Cogan, N.O.I.; Forster, J.W.; Hayes, B.J. Improving the analysis of low heritability complex traits for enhanced genetic gain in potato. Theor. Appl. Genet. 2014, 127, 809–820. [Google Scholar] [CrossRef]
- Watson, A.; Hickey, L.T.; Christopher, J.; Rutkoski, J.; Poland, J.; Hayes, B.J. Multivariate genomic selection and the potential of rapid indirect phenotypic selection with speed breeding to increase genetic gain in spring bread wheat. Crop Sci. 2019, 59, 1945–1959. [Google Scholar] [CrossRef] [Green Version]
- Utz, H.F.; Bohn, M.; Melchinger, A.E. Predicting progeny means and variances of winter wheat crosses from phenotypic values of their parents. Crop Sci. 2001, 41, 1470–1478. [Google Scholar] [CrossRef]
- Zhong, S.; Jannink, J.L. Using quantitative trait loci results to discriminate among crosses on the basis of their progeny mean and variance. Genetics 2007, 177, 567–576. [Google Scholar] [CrossRef] [Green Version]
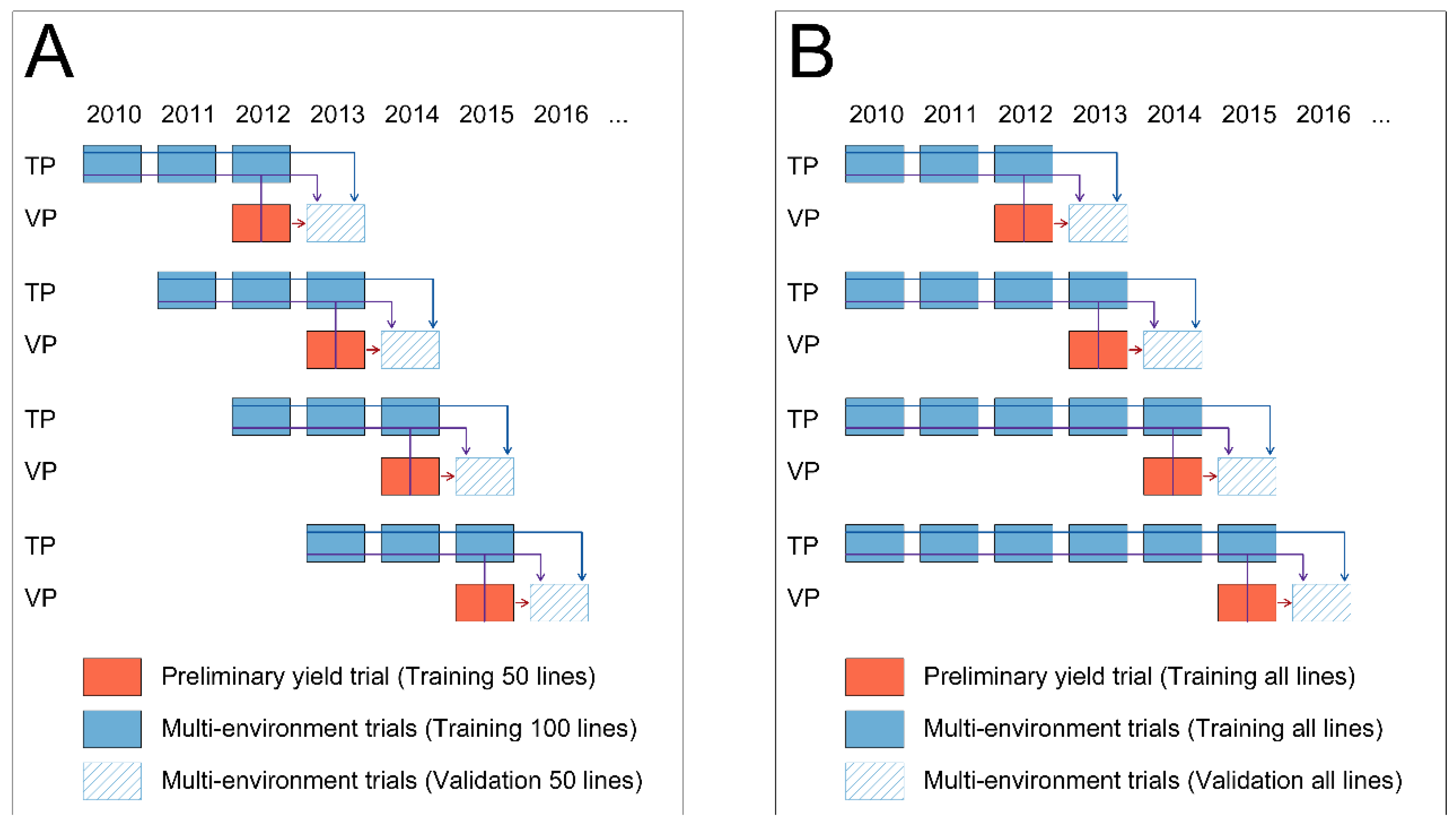
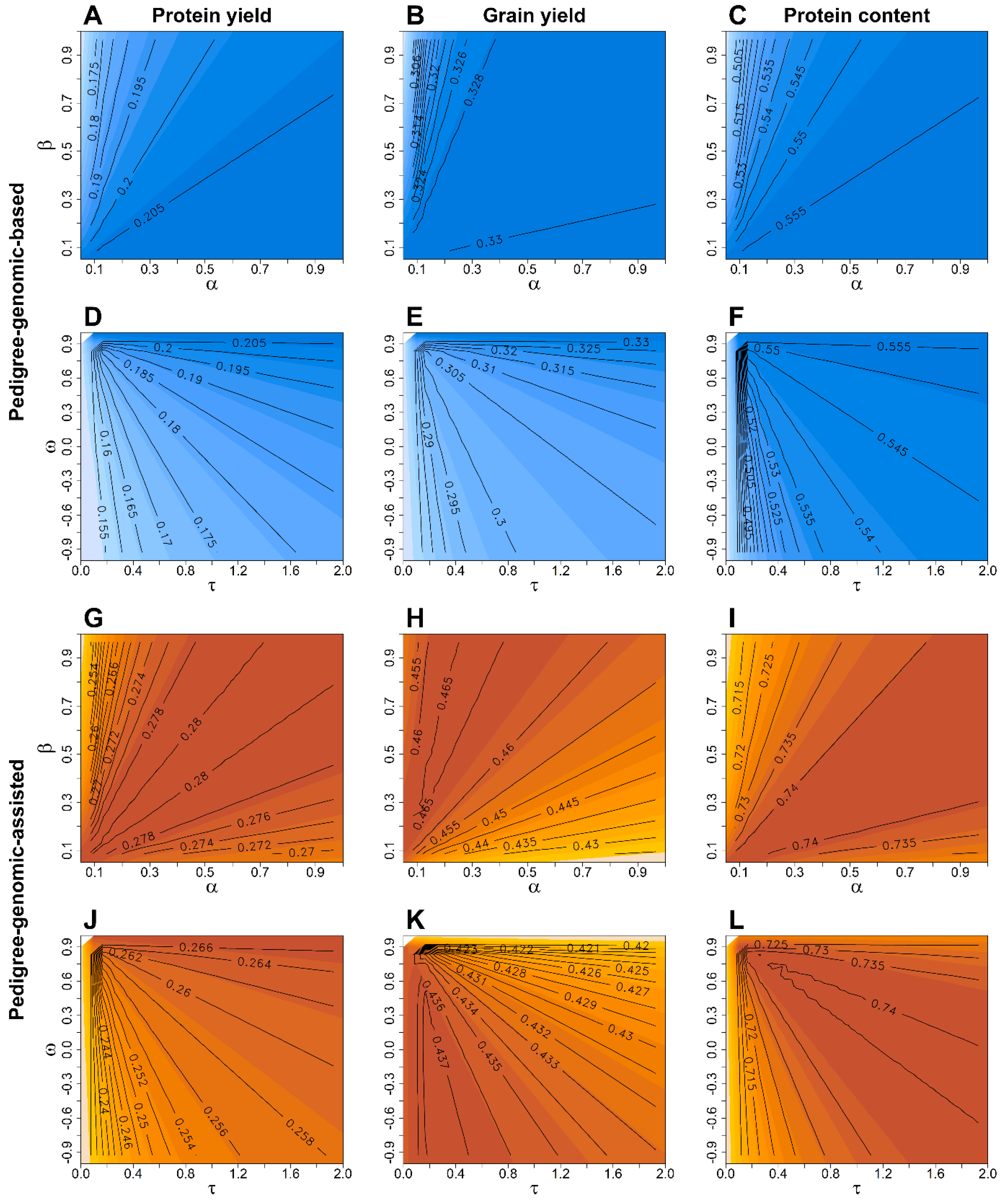
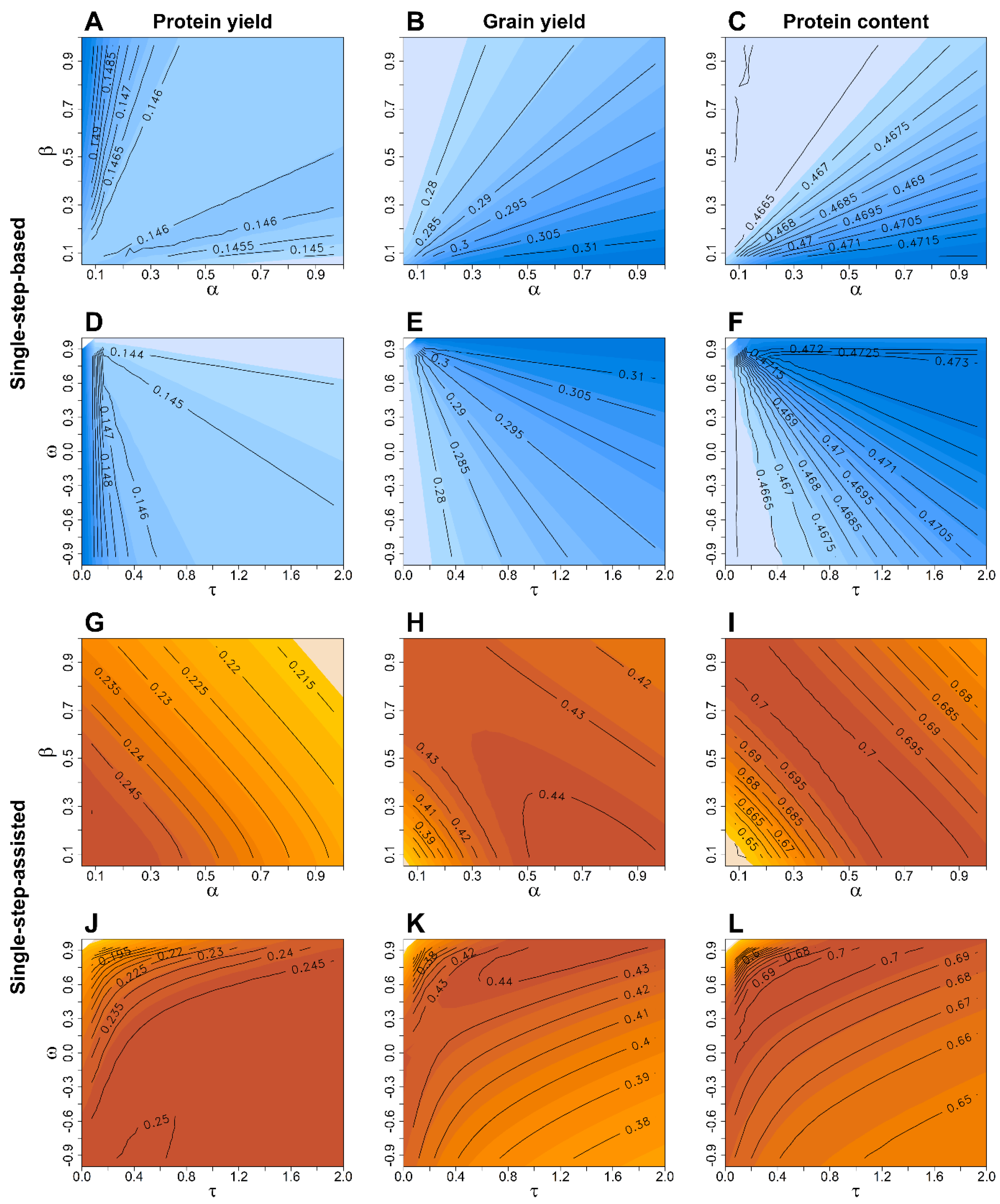
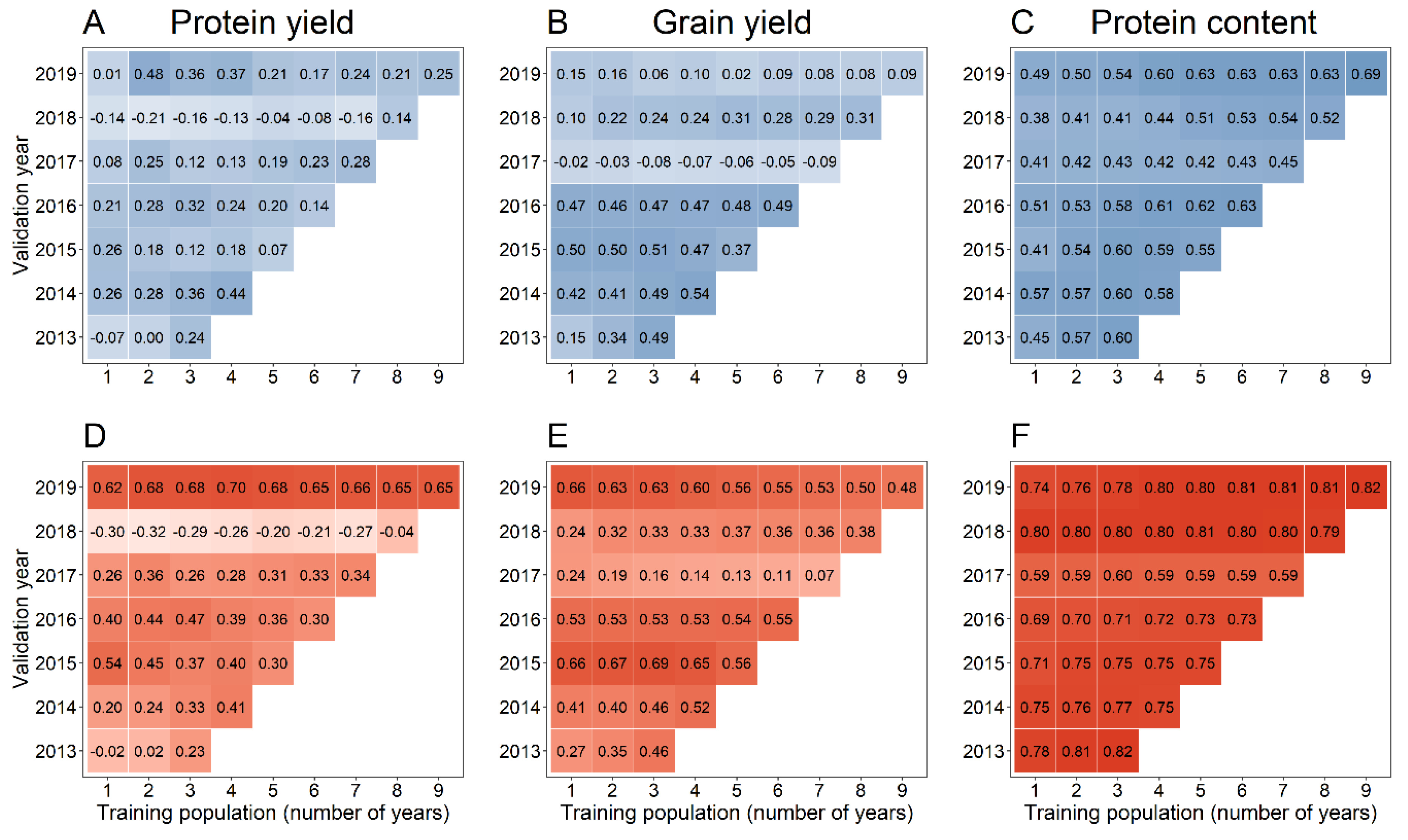
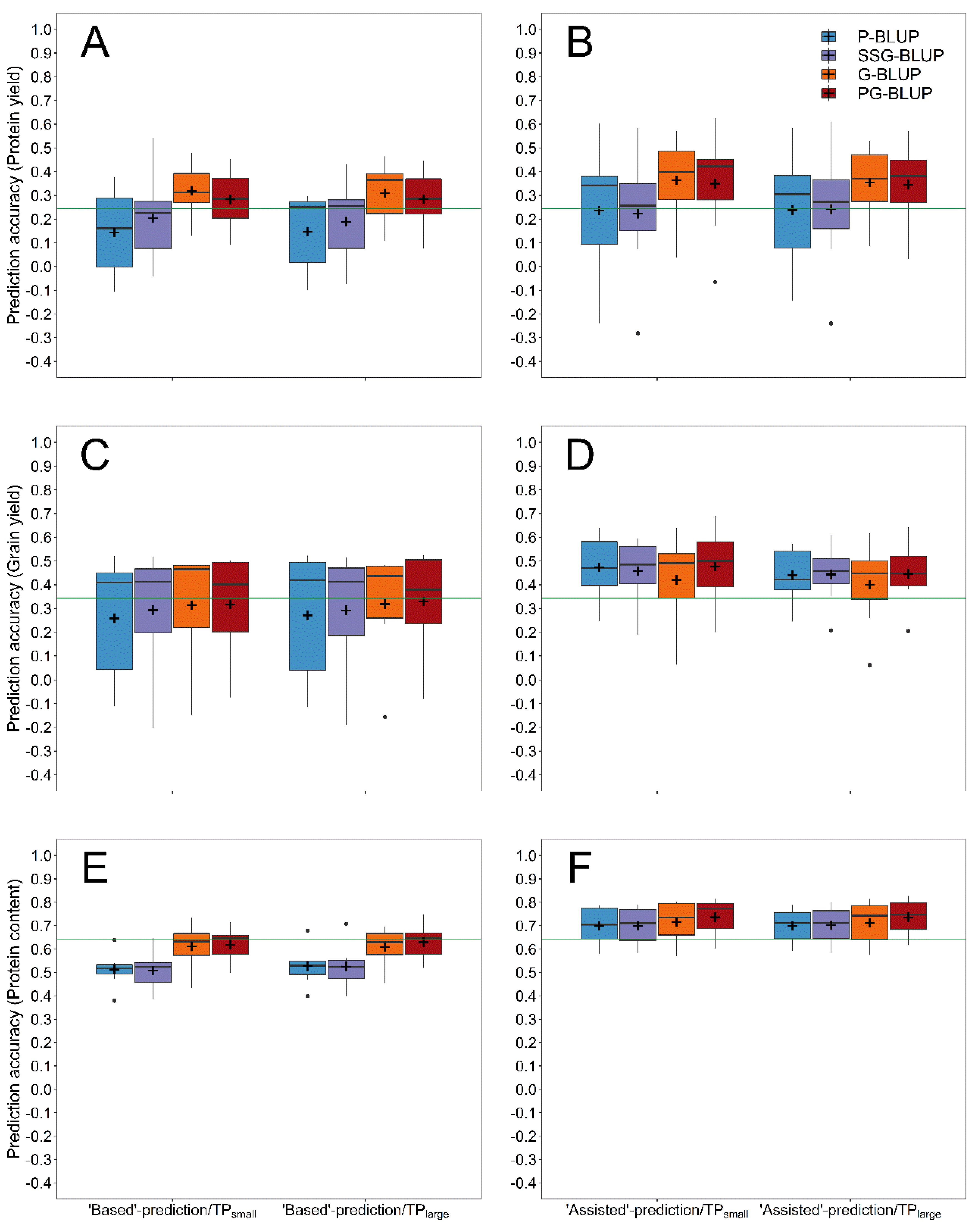
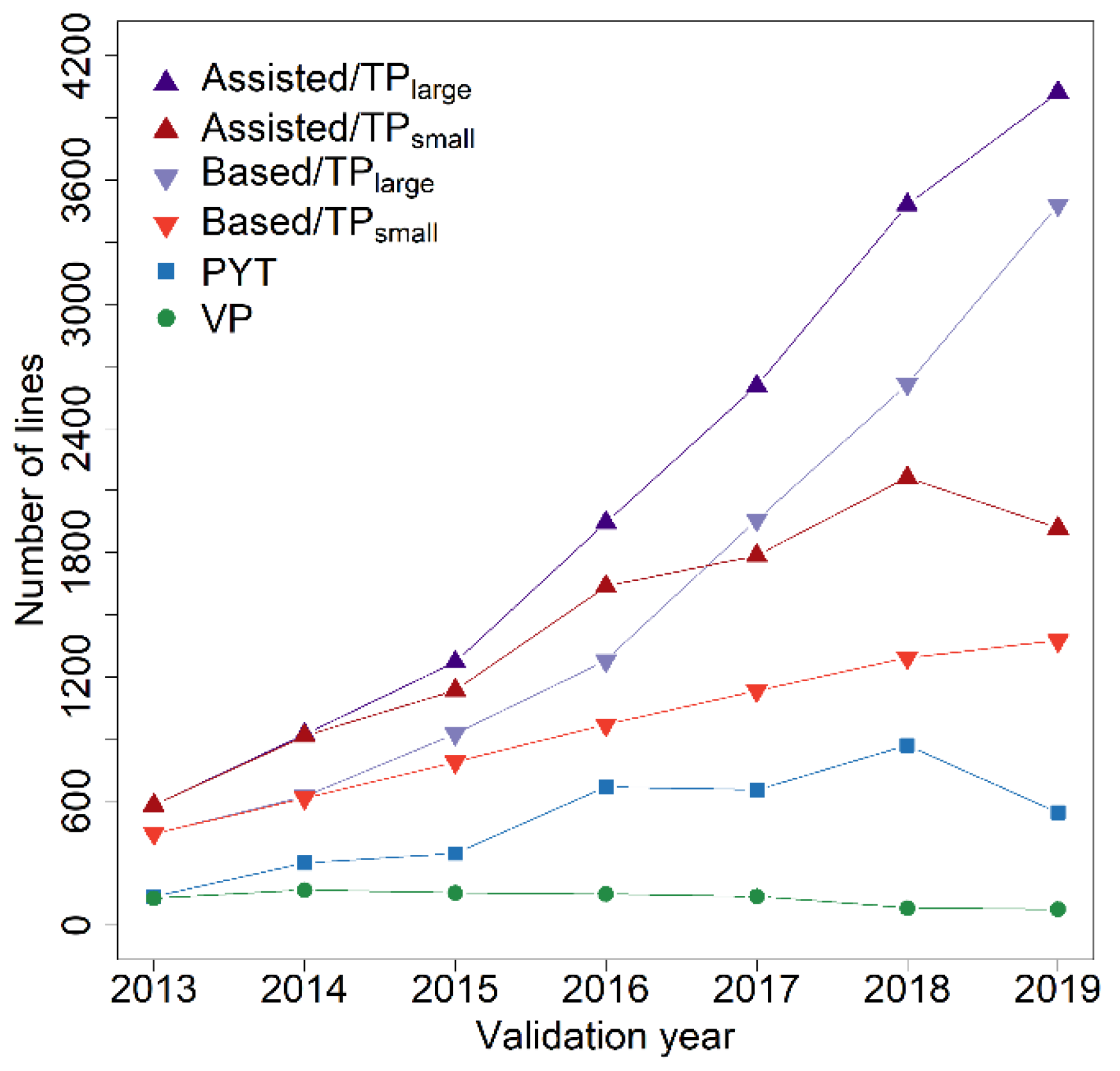
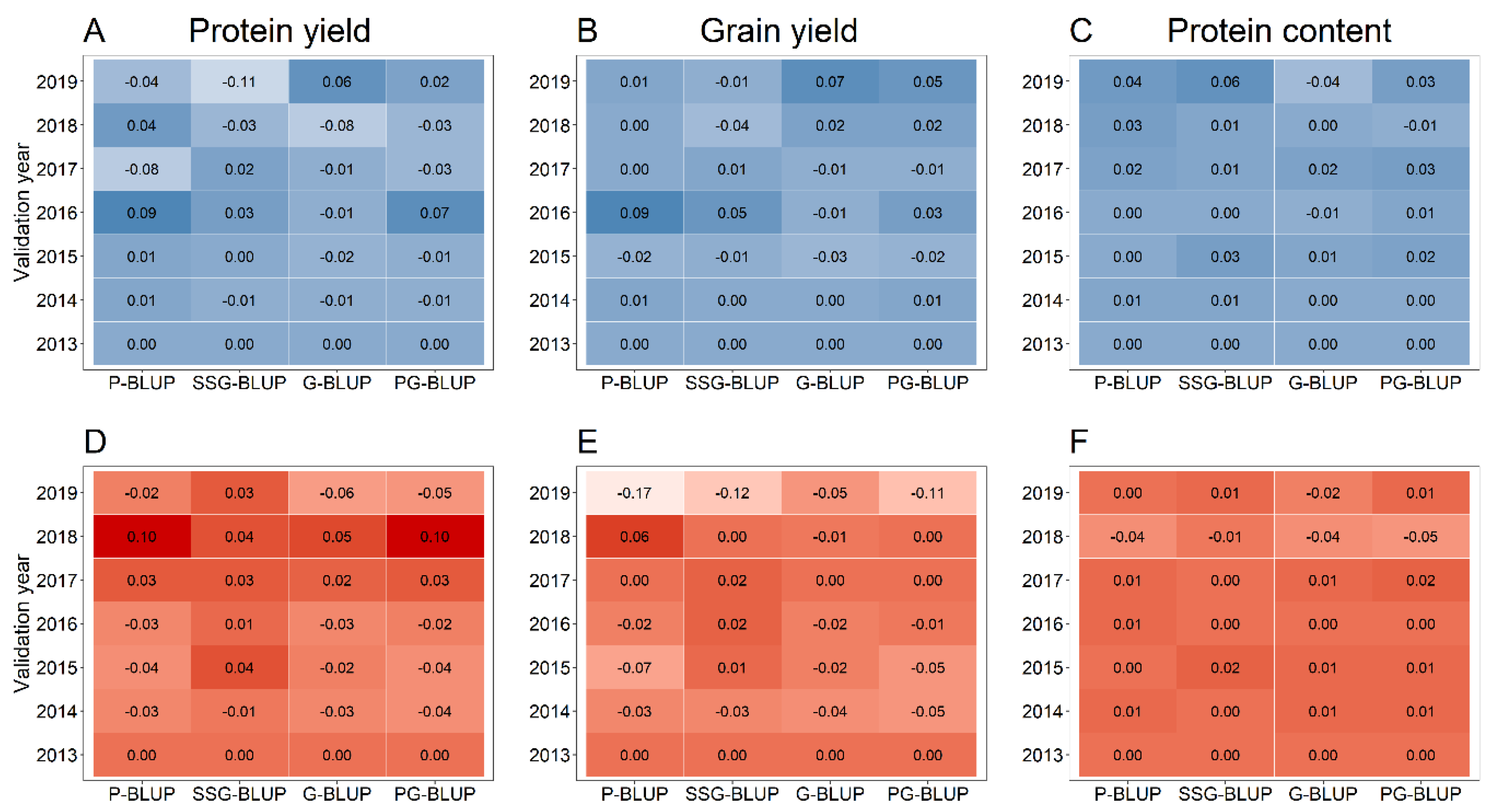
| Year | Number of Lines † | ||
|---|---|---|---|
| Multi-Environment ‡ | Observation Trial (All) § | Observation Trial (Sel.) ¶ | |
| 2010 | 127 | ||
| 2011 | 162 | ||
| 2012 | 208 | 142 | 130 |
| 2013 | 193 | 306 | 170 |
| 2014 | 202 | 361 | 156 |
| 2015 | 206 | 676 | 151 |
| 2016 | 192 | 666 | 139 |
| 2017 | 209 | 875 | 83 |
| 2018 | 113 | 554 | 76 |
| 2019 | 114 | ||
| h² † | Model | With Pre-Existing Information | Without Pre-Existing Information | ||||
|---|---|---|---|---|---|---|---|
| Population ‡ | Across § | Within ¶ | Population ‡ | Across § | Within ¶ | ||
| 0.1 (0.3) | OBS | 0.319 | 0.835 | 0.246 | |||
| P-BLUP | 0.604 | 0.889 | 0.246 | 0.234 | 0.356 | 0.000 | |
| SSG-BLUP | 0.605 | 0.892 | 0.246 | 0.268 | 0.410 | 0.000 | |
| G-BLUP | 0.665 | 0.903 | 0.405 | 0.389 | 0.546 | 0.209 | |
| PG-BLUP | 0.670 | 0.902 | 0.405 | 0.375 | 0.512 | 0.208 | |
| 0.3 (0.5) | OBS | 0.548 | 0.949 | 0.438 | |||
| P-BLUP | 0.704 | 0.961 | 0.438 | 0.275 | 0.421 | 0.000 | |
| SSG-BLUP | 0.705 | 0.962 | 0.438 | 0.299 | 0.459 | 0.000 | |
| G-BLUP | 0.760 | 0.963 | 0.552 | 0.445 | 0.618 | 0.253 | |
| PG-BLUP | 0.772 | 0.965 | 0.576 | 0.440 | 0.598 | 0.253 | |
| 0.5 (0.7) | OBS | 0.707 | 0.977 | 0.595 | |||
| P-BLUP | 0.778 | 0.980 | 0.595 | 0.297 | 0.455 | 0.000 | |
| SSG-BLUP | 0.779 | 0.981 | 0.595 | 0.321 | 0.491 | 0.000 | |
| G-BLUP | 0.810 | 0.980 | 0.643 | 0.480 | 0.660 | 0.284 | |
| PG-BLUP | 0.832 | 0.982 | 0.687 | 0.483 | 0.660 | 0.285 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Michel, S.; Löschenberger, F.; Sparry, E.; Ametz, C.; Bürstmayr, H. Multi-Year Dynamics of Single-Step Genomic Prediction in an Applied Wheat Breeding Program. Agronomy 2020, 10, 1591. https://doi.org/10.3390/agronomy10101591
Michel S, Löschenberger F, Sparry E, Ametz C, Bürstmayr H. Multi-Year Dynamics of Single-Step Genomic Prediction in an Applied Wheat Breeding Program. Agronomy. 2020; 10(10):1591. https://doi.org/10.3390/agronomy10101591
Chicago/Turabian StyleMichel, Sebastian, Franziska Löschenberger, Ellen Sparry, Christian Ametz, and Hermann Bürstmayr. 2020. "Multi-Year Dynamics of Single-Step Genomic Prediction in an Applied Wheat Breeding Program" Agronomy 10, no. 10: 1591. https://doi.org/10.3390/agronomy10101591





