Temporal and Cultivar-Specific Effects on Potato Root and Soil Fungal Diversity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site
2.2. Soil Chemical Analysis
2.3. Sampling and DNA Extraction
2.4. PCR Amplification and High-Throughput Sequencing
2.5. Bioinformatics
2.6. Statistical Analysis
3. Results
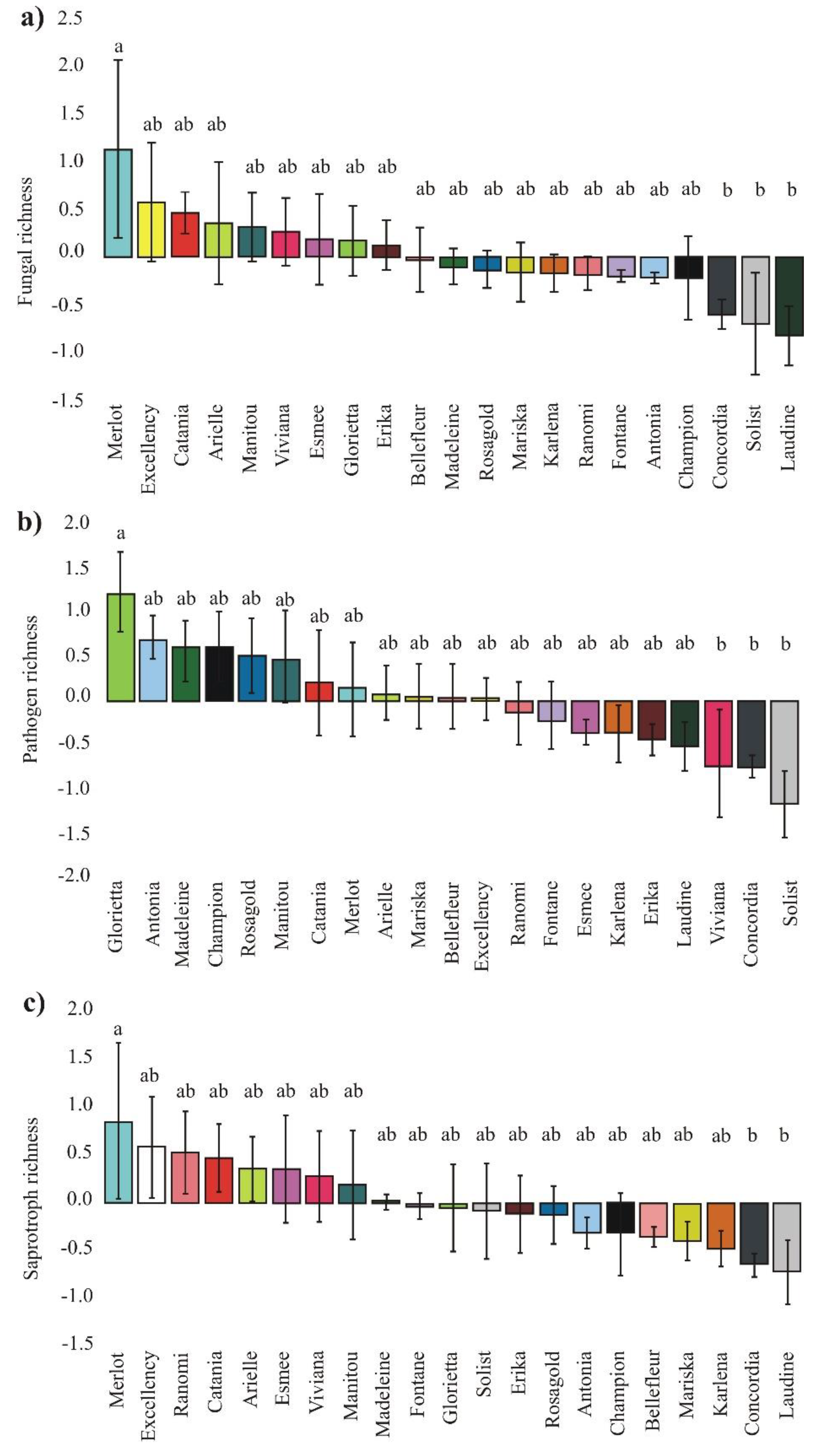
3.1. Richness of Fungal Guilds
3.2. Plant Pathogen, and Saprotroph Abundance
3.3. Factors Affecting the Abundance of Dominant Plant Pathogens
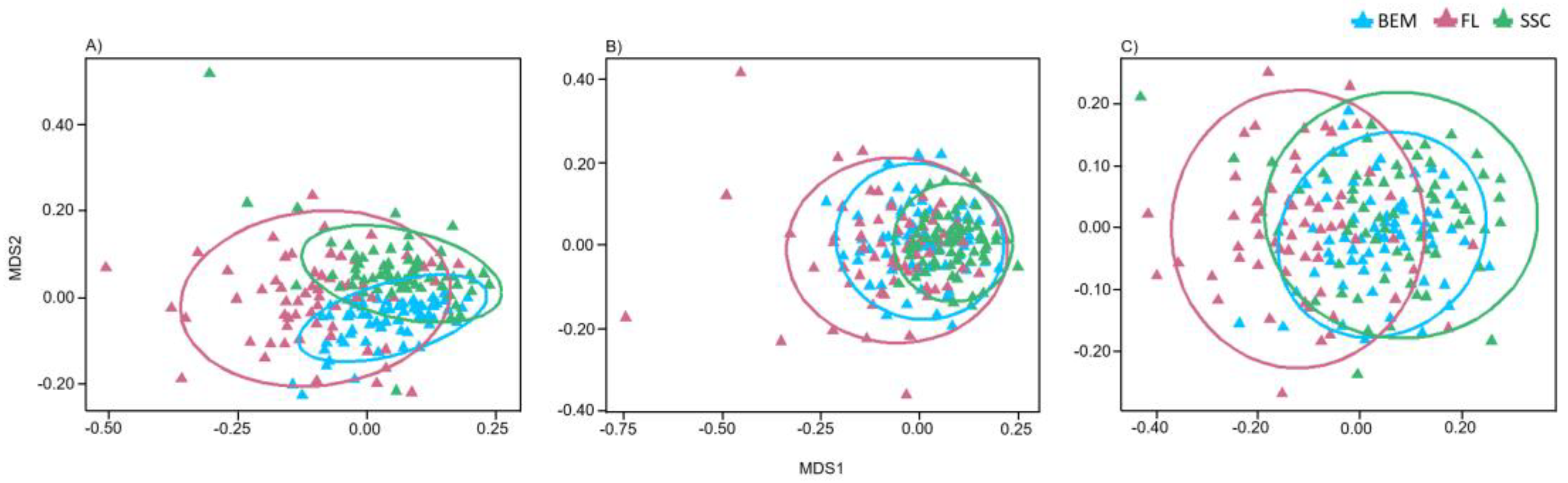
3.4. Factors Affecting Fungal Community Composition
4. Discussion
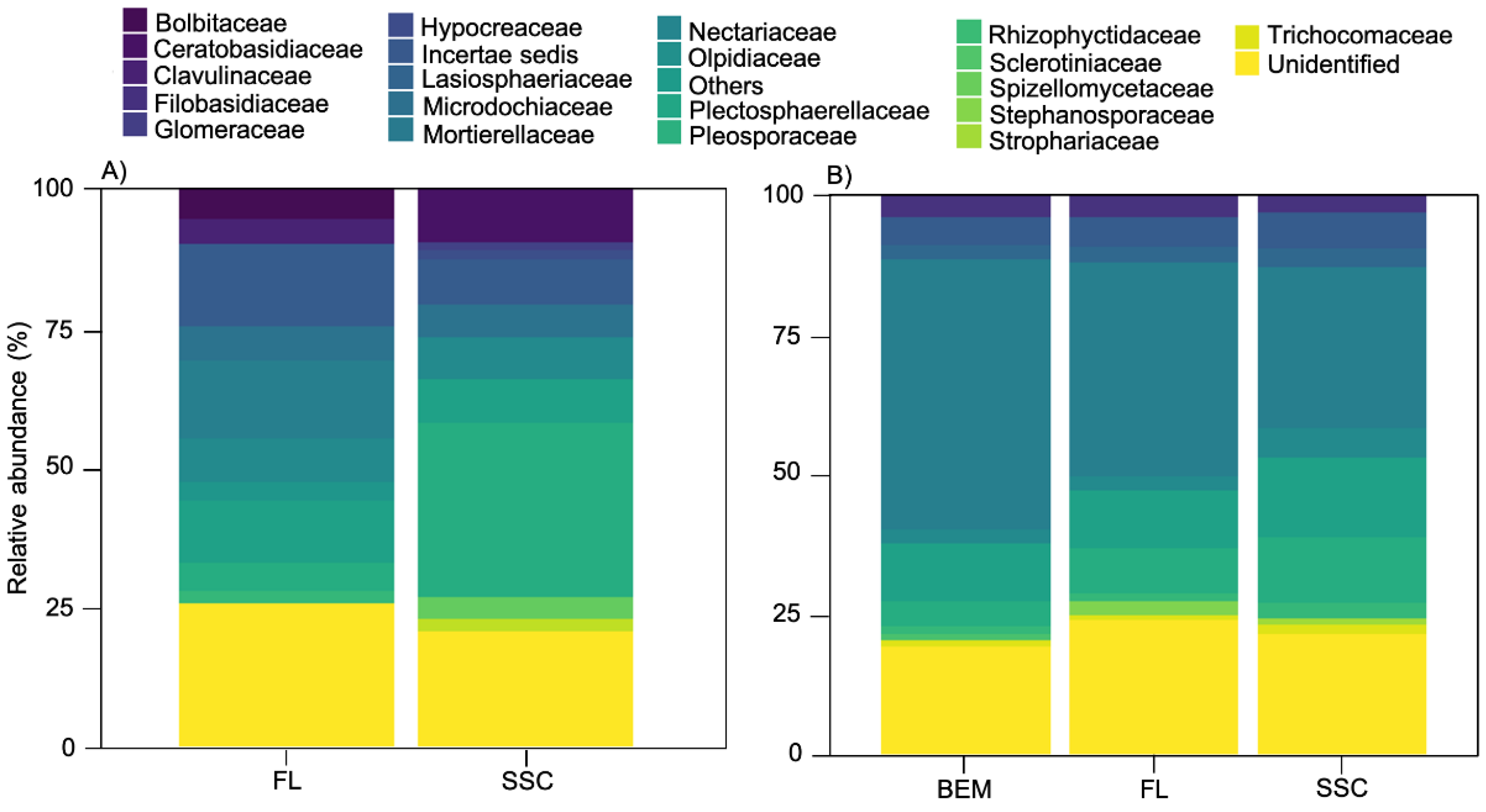
4.1. Dominant Taxa
4.2. Seasonal Variation
4.3. Effect of Cultivars
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ramankutty, N.; Mehrabi, Z.; Waha, K.; Jarvis, L.; Kremen, C.; Herrero, M.; Rieseberg, L.H. Trends in Global Agricultural Land Use: Implications for Environmental Health and Food Security. Annu. Rev. Plant Biol. 2018, 69, 789–815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tilman, D.; Balzer, C.; Hill, J.; Befort, B.L. Global food demand and the sustainable intensification of agriculture. Proc. Natl. Acad. Sci. USA 2011, 108, 20260–20264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lakshmanan, V.; Selvaraj, G.; Bais, H.P. Functional Soil Microbiome: Belowground Solutions to an Aboveground Problem. Plant Physiol. 2014, 166, 689–700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Vries, F.T.; Thébault, E.; Liiri, M.; Birkhofer, K.; Tsiafouli, M.A.; Bjørnlund, L.; Jørgensen, H.B.; Brady, M.V.; Christensen, S.; De Ruiter, P.C.; et al. Soil food web properties explain ecosystem services across European land use systems. Proc. Natl. Acad. Sci. USA 2013, 110, 14296–14301. [Google Scholar] [CrossRef] [Green Version]
- Banerjee, S.; Walder, F.; Büchi, L.; Meyer, M.; Held, A.Y.; Gattinger, A.; Keller, T.; Charles, R.; Van Der Heijden, M.G.A. Agricultural intensification reduces microbial network complexity and the abundance of keystone taxa in roots. ISME J. 2019, 13, 1722–1736. [Google Scholar] [CrossRef] [Green Version]
- Hartmann, M.; Frey, B.; Mayer, J.; Mäder, P.; Widmer, F. Distinct soil microbial diversity under long-term organic and conventional farming. ISME J. 2014, 9, 1177–1194. [Google Scholar] [CrossRef] [Green Version]
- Bardgett, R.D.; Van Der Putten, W.H. Belowground biodiversity and ecosystem functioning. Nature 2014, 515, 505–511. [Google Scholar] [CrossRef]
- Philippot, L.; Raaijmakers, J.M.; Lemanceau, P.; Van Der Putten, W.H. Going back to the roots: The microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 2013, 11, 789–799. [Google Scholar] [CrossRef]
- Van Der Heijden, M.G.A.; Bardgett, R.D.; Van Straalen, N.M. The unseen majority: Soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 2008, 11, 296–310. [Google Scholar] [CrossRef]
- Detheridge, A.P.; Brand, G.; Fychan, R.; Crotty, F.V.; Sanderson, R.; Griffith, G.W.; Marley, C.L. The legacy effect of cover crops on soil fungal populations in a cereal rotation. Agric. Ecosyst. Environ. 2016, 228, 49–61. [Google Scholar] [CrossRef] [Green Version]
- Francioli, D.; Schulz, E.; Lentendu, G.; Wubet, T.; Buscot, F.; Reitz, T. Mineral vs. Organic Amendments: Microbial Community Structure, Activity and Abundance of Agriculturally Relevant Microbes Are Driven by Long-Term Fertilization Strategies. Front. Microbiol. 2016, 7, 1446. [Google Scholar] [CrossRef] [Green Version]
- Säle, V.; Aguilera, P.; Laczko, E.; Mäder, P.; Berner, A.; Zihlmann, U.; Van Der Heijden, M.G.A.; Oehl, F. Impact of conservation tillage and organic farming on the diversity of arbuscular mycorrhizal fungi. Soil Biol. Biochem. 2015, 84, 38–52. [Google Scholar] [CrossRef]
- Schmidt, R.; Mitchell, J.; Scow, K. Cover cropping and no-till increase diversity and symbiotroph:saprotroph ratios of soil fungal communities. Soil Biol. Biochem. 2019, 129, 99–109. [Google Scholar] [CrossRef] [Green Version]
- Soonvald, L.; Loit, K.; Runno-Paurson, E.; Astover, A.; Tedersoo, L. The role of long-term mineral and organic fertilisation treatment in changing pathogen and symbiont community composition in soil. Appl. Soil Ecol. 2019, 141, 45–53. [Google Scholar] [CrossRef]
- Brader, G.; Compant, S.; Vescio, K.; Mitter, B.; Trognitz, F.; Ma, L.-J.; Sessitsch, A. Ecology and Genomic Insights into Plant-Pathogenic and Plant-Nonpathogenic Endophytes. Annu. Rev. Phytopathol. 2017, 55, 61–83. [Google Scholar] [CrossRef] [PubMed]
- Grayston, S.J.; Wang, S.; Campbell, C.D.; Edwards, A.C. Selective influence of plant species on microbial diversity in the rhizosphere. Soil Biol. Biochem. 1998, 30, 369–378. [Google Scholar] [CrossRef]
- Zimudzi, J.; Van Der Waals, J.E.; Coutinho, T.A.; Cowan, D.A.; Valverde, A. Temporal shifts of fungal communities in the rhizosphere and on tubers in potato fields. Fungal Biol. 2018, 122, 928–934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Badri, D.V.; Vivanco, J.M. Regulation and function of root exudates. Plant Cell Environ. 2009, 32, 666–681. [Google Scholar] [CrossRef]
- Canarini, A.; Kaiser, C.; Merchant, A.; Richter, A.; Wanek, W. Root Exudation of Primary Metabolites: Mechanisms and Their Roles in Plant Responses to Environmental Stimuli. Front. Plant Sci. 2019, 10, 157. [Google Scholar] [CrossRef] [Green Version]
- Wissuwa, M.; Mazzola, M.; Picard, C. Novel approaches in plant breeding for rhizosphere-related traits. Plant Soil 2009, 321, 409–430. [Google Scholar] [CrossRef]
- Food and Agriculture Organization of the United Nations. FAOSTAT Database; FAO: Rome, Italy, 2020; Available online: http://www.fao.org/faostat/en/#data/TP (accessed on 3 June 2020).
- Fry, W.E.; Goodwin, S.B. Re-emergence of Potato and Tomato Late Blight in the United States. Plant Dis. 1997, 81, 1349–1357. [Google Scholar] [CrossRef] [Green Version]
- Toth, I.K.; Van Der Wolf, J.M.; Saddler, G.; Lojkowska, E.; Hélias, V.; Pirhonen, M.; Tsoror (Lahkim), T.; Elphinstone, J.G. Dickeya species: An emerging problem for potato production in Europe. Plant Pathol. 2011, 60, 385–399. [Google Scholar] [CrossRef]
- Patil, V.U.; Girimalla, V.; Sagar, V.; Bhardwaj, V.; Chakrabarti, S.K. Draft Genome Sequencing of Rhizoctonia solani Anastomosis Group 3 (AG3- PT) Causing Stem Canker and Black Scurf of Potato. Am. J. Potato Res. 2018, 95, 87–91. [Google Scholar] [CrossRef]
- Atallah, Z.K.; Bae, J.; Jansky, S.H.; Rouse, D.I.; Stevenson, W.R. Multiplex Real-Time Quantitative PCR to Detect and Quantify Verticillium dahliae Colonization in Potato Lines that Differ in Response to Verticillium Wilt. Phytopathology 2007, 97, 865–872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stefańczyk, E.; Sobkowiak, S.; Brylińska, M.; Śliwka, J. Diversity of Fusarium spp. associated with dry rot of potato tubers in Poland. Eur. J. Plant Pathol. 2016, 145, 871–884. [Google Scholar] [CrossRef] [Green Version]
- Fry, W. Phytophthora infestans: The plant (and R gene) destroyer. Mol. Plant Pathol. 2008, 9, 385–402. [Google Scholar] [CrossRef]
- Bakker, M.G.; Manter, D.K.; Sheflin, A.M.; Weir, T.L.; Vivanco, J.M. Harnessing the rhizosphere microbiome through plant breeding and agricultural management. Plant Soil 2012, 360, 1–13. [Google Scholar] [CrossRef]
- Estonian Weather Service. Meteorological Yearbook of Estonia 2014. Available online: http://www.ilmateenistus.ee/wp-content/uploads/2013/01/aastaraamat_2014.pdf (accessed on 10 June 2020).
- Egnér, H.; Riehm, H.; Domingo, W.R. Untersuchungen über die chemische Bodenanalyse als Grundlage für die Beurteilung des Nährstoffzustandes der Böden. II. Chemische Extraktionsmethoden zur Phosphor- und Kaliumbestimmung. K. Lantbr. Ann. 1960, 26, 199–215. [Google Scholar]
- Hack, H.; Gall, H.; Klemke, T.H.; Klose, R.; Meier, U.; Stauss, R.; Witzenberger, A. The BBCH scale for phonological growth stages of potato (Solanum tuberosum L.). In Growth Stages of Mono- and Dicotyledonous Plants, BBCH Monograph; Meier, U., Ed.; Federal Biological Research Centre for Agriculture and Forestry: Berlin, Germany, 2001; pp. 7–16. [Google Scholar]
- De León, D.G.; Moora, M.; Öpik, M.; Neuenkamp, L.; Gerz, M.; Jairus, T.; Vasar, M.; Bueno, C.G.; Davison, J.; Zobel, M. Symbiont dynamics during ecosystem succession: Co-occurring plant and arbuscular mycorrhizal fungal communities. FEMS Microbiol. Ecol. 2016, 92, fiw097. [Google Scholar] [CrossRef] [Green Version]
- Tedersoo, L.; Bahram, M.; Põlme, S.; Kõljalg, U.; Yorou, N.S.; Wijesundera, R.; Ruiz, L.V.; Vasco-Palacios, A.M.; Thu, P.Q.; Suija, A.; et al. Global diversity and geography of soil fungi. Science 2014, 346, 1256688. [Google Scholar] [CrossRef] [Green Version]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [Green Version]
- Masella, A.P.; Bartram, A.K.; Truszkowski, J.M.; Brown, D.G.; Neufeld, J.D. PANDAseq: Paired-end assembler for illumina sequences. BMC Bioinform. 2012, 13, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anslan, S.; Bahram, M.; Hiiesalu, I.; Tedersoo, L.; Ashikawa, S.; Tarumoto, N.; Imai, K.; Sakai, J.; Kodana, M.; Kawamura, T.; et al. PipeCraft: Flexible open-source toolkit for bioinformatics analysis of custom high-throughput amplicon sequencing data. Mol. Ecol. Resour. 2017, 17, e234–e240. [Google Scholar] [CrossRef] [PubMed]
- Bengtsson-Palme, J.; Ryberg, M.; Hartmann, M.; Branco, S.; Wang, Z.; Godhe, A.; De Wit, P.; Sánchez-García, M.; Ebersberger, I.; De Sousa, F.; et al. Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol. Evol. 2013, 4, 914–919. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef] [PubMed]
- Kõljalg, U.; Tedersoo, L.; Nilsson, R.H.; Abarenkov, K.; Nilsson, R.H. Digital identifiers for fungal species. Science 2016, 352, 1182–1183. [Google Scholar] [CrossRef]
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Anderson, M.J.; Gorley, R.N.; Clarke, K.R. PERMANOVA+ for PRIMER: Guide to Software and Statistical Methods; PRIMER-E: Plymouth, UK, 2008. [Google Scholar]
- Guyonnet, J.P.; Cantarel, A.A.M.; Simon, L.; Haichar, F.E.Z. Root exudation rate as functional trait involved in plant nutrient-use strategy classification. Ecol. Evol. 2018, 8, 8573–8581. [Google Scholar] [CrossRef] [Green Version]
- Haichar, F.E.Z.; Santaella, C.; Heulin, T.; Achouak, W. Root exudates mediated interactions belowground. Soil Biol. Biochem. 2014, 77, 69–80. [Google Scholar] [CrossRef]
- Agrios, G.N. Plant Pathology, 5th ed.; Academic Press: Cambridge, MA, USA, 2005; ISBN 9780120445653. [Google Scholar]
- Ma, A.; Zhuang, X.; Wu, J.; Cui, M.; Lv, D.; Liu, C.; Zhuang, G. Ascomycota Members Dominate Fungal Communities during Straw Residue Decomposition in Arable Soil. PLoS ONE 2013, 8, e66146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osorio, N.W.; Habte, M. Synergistic Influence of an Arbuscular Mycorrhizal Fungus and a P Solubilizing Fungus on Growth and P Uptake ofLeucaena leucocephalain an Oxisol. Arid. Land Res. Manag. 2001, 15, 263–274. [Google Scholar] [CrossRef]
- Li, F.; Chen, L.; Redmile-Gordon, M.; Zhang, J.; Zhang, C.; Ning, Q.; Li, W. Mortierella elongata ’s roles in organic agriculture and crop growth promotion in a mineral soil. Land Degrad. Dev. 2018, 29, 1642–1651. [Google Scholar] [CrossRef]
- Carlucci, A.; Raimondo, M.; Santos, J.M.; Phillips, A.J.L. Plectosphaerella species associated with root and collar rots of horticultural crops in southern Italy. Persoonia Mol. Phylogeny Evol. Fungi 2012, 28, 34–48. [Google Scholar] [CrossRef] [Green Version]
- Raimondo, M.L.; Carlucci, A. Characterization and pathogenicity assessment ofPlectosphaerellaspecies associated with stunting disease on tomato and pepper crops in Italy. Plant Pathol. 2018, 67, 626–641. [Google Scholar] [CrossRef]
- Gao, J.; Zhang, Y.; Zhao, X.J.; Wang, K.; Zhao, J. First Report of Potato Wilt Caused by Plectosphaerella cucumerina in Inner Mongolia, China. Plant Dis. 2016, 100, 2523. [Google Scholar] [CrossRef]
- Atkins, S.D.; Clark, I.M.; Sosnowska, D.; Hirsch, P.R.; Kerry, B.R. Detection and Quantification of Plectosphaerella cucumerina, a Potential Biological Control Agent of Potato Cyst Nematodes, by Using Conventional PCR, Real-Time PCR, Selective Media, and Baiting. Appl. Environ. Microbiol. 2003, 69, 4788–4793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsror (Lahkim), L.T. Biology, Epidemiology and Management of Rhizoctonia solani on Potato. J. Phytopathol. 2010, 158, 649–658. [Google Scholar] [CrossRef]
- Leplat, J.; Friberg, H.; Abid, M.; Steinberg, C. Survival of Fusarium graminearum, the causal agent of Fusarium head blight. A review. Agron. Sustain. Dev. 2012, 33, 97–111. [Google Scholar] [CrossRef]
- Oberwinkler, F.; Riess, K.; Bauer, R.; Kirschner, R.; Garnica, S. Taxonomic re-evaluation of the Ceratobasidium-Rhizoctonia complex and Rhizoctonia butinii, a new species attacking spruce. Mycol. Prog. 2013, 12, 763–776. [Google Scholar] [CrossRef]
- Veldre, V.; Abarenkov, K.; Bahram, M.; Martos, F.; Selosse, M.-A.; Tamm, H.; Kõljalg, U.; Tedersoo, L. Evolution of nutritional modes of Ceratobasidiaceae (Cantharellales, Basidiomycota) as revealed from publicly available ITS sequences. Fungal Ecol. 2013, 6, 256–268. [Google Scholar] [CrossRef]
- Aoki, T.; O’Donnell, K.; Geiser, D.M. Systematics of key phytopathogenic Fusarium species: Current status and future challenges. J. Gen. Plant Pathol. 2014, 80, 189–201. [Google Scholar] [CrossRef]
- Walder, F.; Schlaeppi, K.; Wittwer, R.; Held, A.Y.; Vogelgsang, S.; Van Der Heijden, M.G.A. Community Profiling of Fusarium in Combination with Other Plant-Associated Fungi in Different Crop Species Using SMRT Sequencing. Front. Plant Sci. 2017, 8, 2019. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, K.; Ward, T.J.; Robert, V.A.R.G.; Crous, P.W.; Geiser, D.M.; Kang, S. DNA sequence-based identification of Fusarium: Current status and future directions. Phytoparasitica 2015, 43, 583–595. [Google Scholar] [CrossRef] [Green Version]
- Inceoğlu, O.; Salles, J.F.; Van Overbeek, L.; Van Elsas, J.D. Effects of Plant Genotype and Growth Stage on the Betaproteobacterial Communities Associated with Different Potato Cultivars in Two Fields. Appl. Environ. Microbiol. 2010, 76, 3675–3684. [Google Scholar] [CrossRef] [Green Version]
- Schlemper, T.R.; Leite, M.F.A.; Lucheta, A.R.; Shimels, M.; Bouwmeester, H.J.; Van Veen, J.A.; Kuramae, E.E. Rhizobacterial community structure differences among sorghum cultivars in different growth stages and soils. FEMS Microbiol. Ecol. 2017, 93. [Google Scholar] [CrossRef] [Green Version]
- Gao, C.; Montoya, L.; Xu, L.; Madera, M.; Hollingsworth, J.; Purdom, E.; Hutmacher, R.B.; Dahlberg, J.A.; Coleman-Derr, D.; Lemaux, P.G.; et al. Strong succession in arbuscular mycorrhizal fungal communities. ISME J. 2019, 13, 214–226. [Google Scholar] [CrossRef] [Green Version]
- Chaparro, J.M.; Badri, D.V.; Vivanco, J.M. Rhizosphere microbiome assemblage is affected by plant development. ISME J. 2014, 8, 790–803. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, C. Rhizodeposition of organic C by plants: Mechanisms and controls. Agron. EDP Sci. 2003, 23, 375–396. [Google Scholar] [CrossRef]
- Chaparro, J.M.; Badri, D.V.; Bakker, M.G.; Sugiyama, A.; Manter, D.K.; Vivanco, J.M. Root Exudation of Phytochemicals in Arabidopsis Follows Specific Patterns That Are Developmentally Programmed and Correlate with Soil Microbial Functions. PLoS ONE 2013, 8, e55731. [Google Scholar] [CrossRef]
- Kaiser, C.; Kilburn, M.R.; Clode, P.L.; Fuchslueger, L.; Koranda, M.; Cliff, J.B.; Solaiman, Z.M.; Murphy, D.V. Exploring the transfer of recent plant photosynthates to soil microbes: Mycorrhizal pathway vs direct root exudation. New Phytol. 2015, 205, 1537–1551. [Google Scholar] [CrossRef] [PubMed]
- Lemanczyk, G.; Sadowski, C.K. Fungal communities and health status of roots of winter wheat cultivated after oats and oats mixed with other crops. BioControl 2002, 47, 349–361. [Google Scholar] [CrossRef]
- Pérez-Piqueres, A.; Edel-Hermann, V.; Alabouvette, C.; Steinberg, C. Response of soil microbial communities to compost amendments. Soil Biol. Biochem. 2006, 38, 460–470. [Google Scholar] [CrossRef]
- Bains, P.S.; Bennypaul, H.S.; Lynch, D.R.; Kawchuk, L.M.; Schaupmeyer, C.A. Rhizoctonia disease of potatoes (Rhizoctonia solani): Fungicidal efficacy and cultivar susceptibility. Am. J. Potato Res. 2002, 79, 99–106. [Google Scholar] [CrossRef]
- Lundberg, D.S.; Lebeis, S.L.; Paredes, S.H.; Yourstone, S.; Gehring, J.; Malfatti, S.; Tremblay, J.; Engelbrektson, A.; Kunin, V.; Del Rio, T.G.; et al. Defining the core Arabidopsis thaliana root microbiome. Nature 2012, 488, 86–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De León, D.G.; Vahter, T.; Zobel, M.; Koppel, M.; Edesi, L.; Davison, J.; Al-Quraishy, S.; Hozzein, W.N.; Moora, M.; Oja, J.; et al. Different wheat cultivars exhibit variable responses to inoculation with arbuscular mycorrhizal fungi from organic and conventional farms. PLoS ONE 2020, 15, e0233878. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Thorngate, J.H.; Richardson, P.M.; A Mills, D. PNAS Plus: From the Cover: Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc. Natl. Acad. Sci. USA 2013, 111, E139–E148. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Sample Type | Variable | df | All Fungi | Pathogens | Saprotrophs | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| R2adj | Pseudo F | p | R2adj | Pseudo F | p | R2adj | Pseudo F | p | |||
| Soil | |||||||||||
| Cultivar | 20 | 0.030 | 1.4 | 0.157 | 0 | 0.6 | 0.908 | 0.002 | 1.1 | 0.420 | |
| Time | 2 | 0.032 | 4.2 | 0.017 * | 0.234 | 28.5 | <0.001 *** | 0.048 | 5.6 | 0.005 ** | |
| Cultivar × Time | 40 | 0 | 1.1 | 0.396 | 0 | 0.99 | 0.492 | 0 | 0.9 | 0.661 | |
| Replication block | 2 | 0 | 0.9 | 0.423 | 0 | 1.2 | 0.326 | 0 | 0.4 | 0.684 | |
| Roots | |||||||||||
| Cultivar | 20 | 0.038 | 2.2 | 0.007 ** | 0.176 | 2.3 | 0.004 ** | 0.021 | 2.3 | 0.005 ** | |
| Time | 1 | 0.184 | 43.8 | <0.001 *** | 0 | 0.6 | 0.437 | 0.305 | 79.5 | <0.001 *** | |
| Cultivar × Time | 20 | 0.106 | 2.9 | <0.001 *** | 0 | 1.0 | 0.464 | 0.024 | 2.3 | 0.004 ** | |
| Replication block | 2 | 0 | 1.2 | 0.301 | 0.002 | 1.4 | 0.259 | 0 | 1.4 | 0.260 | |
| Sample Type | Variable | df | Pathogens | Saprotrophs | ||||
|---|---|---|---|---|---|---|---|---|
| R2adj | Pseudo F | p | R2adj | Pseudo F | p | |||
| Soil | ||||||||
| Cultivar | 20 | 0.011 | 1.7 | 0.043 * | 0.012 | 1.7 | 0.039 * | |
| Time | 2 | 0.250 | 37.7 | <0.001 *** | 0.273 | 41.0 | <0.001 *** | |
| Cultivar × Time | 40 | 0 | 1.4 | 0.068 | 0 | 1.2 | 0.223 | |
| Replication block | 2 | 0 | 0.8 | 0.459 | 0 | 1.3 | 0.278 | |
| Roots | ||||||||
| Cultivar | 20 | 0.004 | 1.8 | 0.033 * | 0.012 | 1.6 | 0.072 | |
| Time | 1 | 0.296 | 66.8 | <0.001 *** | 0.242 | 46.7 | <0.001 *** | |
| Cultivar × Time | 20 | 0 | 1.1 | 0.333 | 0 | 1.0 | 0.423 | |
| Replication block | 2 | 0.048 | 7.0 | 0.002 ** | 0.019 | 3.3 | 0.042 * | |
| Sample Type | Variable | df | All Fungi | Pathogens | Saprotrophs | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| R2adj | Pseudo F | p | R2adj | Pseudo F | p | R2adj | Pseudo F | p | |||
| Soil | |||||||||||
| Cultivar | 20 | 0.008 | 1.183 | <0.001 *** | 0.004 | 1.135 | 0.144 | 0.014 | 1.260 | <0.001 *** | |
| Time | 2 | 0.062 | 4.758 | 0.002 ** | 0.102 | 9.125 | 0.004 ** | 0.058 | 4.302 | 0.004 * | |
| Replication block | 2 | 0.007 | 1.858 | <0.001 *** | 0.0109 | 2.513 | 0.001 ** | 0.004 | 1.467 | 0.012 * | |
| Cultivar × Time | 40 | 0 | 1.007 | 0.409 | 0 | 1.139 | 0.079 | 0 | 0.954 | 0.845 | |
| Cultivar × Replication block | 40 | 0 | 1.016 | 0.299 | 0 | 1.132 | 0.087 | 0 | 0.981 | 0.655 | |
| Time × Replication block | 4 | 0.009 | 1.586 | <0.001 *** | 0.003 | 1.438 | 0.053 | 0.010 | 1.621 | <0.001 *** | |
| Roots | |||||||||||
| Cultivar | 20 | 0.082 | 1.823 | <0.001 *** | 0.130 | 2.485 | <0.001 *** | 0.057 | 1.399 | 0.001 ** | |
| Time | 1 | 0.053 | 7.863 | 0.102 | 0.077 | 13.088 | 0.098 | 0.0331 | 4.994 | 0.105 | |
| Replication block | 2 | 0.012 | 2.223 | <0.001 *** | 0.004 | 1.689 | 0.040 * | 0.009 | 1.938 | 0.003 ** | |
| Cultivar × Time | 20 | 0.013 | 1.392 | <0.001 *** | 0.004 | 1.378 | 0.004 ** | 0.001 | 1.280 | 0.004 ** | |
| Cultivar × Replication block | 40 | 0 | 1.020 | 0.362 | 0 | 0.916 | 0.793 | 0 | 1.179 | 0.011 * | |
| Time × Replication block | 2 | 0 | 1.253 | 0.105 | 0 | 1.088 | 0.369 | 0 | 1.284 | 0.132 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Loit, K.; Soonvald, L.; Astover, A.; Runno-Paurson, E.; Öpik, M.; Tedersoo, L. Temporal and Cultivar-Specific Effects on Potato Root and Soil Fungal Diversity. Agronomy 2020, 10, 1535. https://doi.org/10.3390/agronomy10101535
Loit K, Soonvald L, Astover A, Runno-Paurson E, Öpik M, Tedersoo L. Temporal and Cultivar-Specific Effects on Potato Root and Soil Fungal Diversity. Agronomy. 2020; 10(10):1535. https://doi.org/10.3390/agronomy10101535
Chicago/Turabian StyleLoit, Kaire, Liina Soonvald, Alar Astover, Eve Runno-Paurson, Maarja Öpik, and Leho Tedersoo. 2020. "Temporal and Cultivar-Specific Effects on Potato Root and Soil Fungal Diversity" Agronomy 10, no. 10: 1535. https://doi.org/10.3390/agronomy10101535
APA StyleLoit, K., Soonvald, L., Astover, A., Runno-Paurson, E., Öpik, M., & Tedersoo, L. (2020). Temporal and Cultivar-Specific Effects on Potato Root and Soil Fungal Diversity. Agronomy, 10(10), 1535. https://doi.org/10.3390/agronomy10101535






