Abstract
Final grain production and quality in durum wheat are affected by biotic and abiotic stresses. The association mapping (AM) approach is useful for dissecting the genetic control of quantitative traits, with the aim of increasing final wheat production under stress conditions. In this study, we used AM analyses to detect quantitative trait loci (QTL) underlying agronomic and quality traits in a collection of 294 elite durum wheat lines from CIMMYT (International Maize and Wheat Improvement Center), grown under different water regimes over four growing seasons. Thirty-seven significant marker-trait associations (MTAs) were detected for sedimentation volume (SV) and thousand kernel weight (TKW), located on chromosomes 1B and 2A, respectively. The QTL loci found were then confirmed with several AM analyses, which revealed 12 sedimentation index (SDS) MTAs and two additional loci for SV (4A) and yellow rust (1B). A candidate gene analysis of the identified genomic regions detected a cluster of 25 genes encoding blue copper proteins in chromosome 1B, with homoeologs in the two durum wheat subgenomes, and an ubiquinone biosynthesis O-methyltransferase gene. On chromosome 2A, several genes related to photosynthetic processes and metabolic pathways were found in proximity to the markers associated with TKW. These results are of potential use for subsequent application in marker-assisted durum wheat-breeding programs.
1. Introduction
Wheat is one of the most widely grown crops worldwide (FAO, 2015), and is essential for the human diet []. Its importance and worldwide dominance are due, in part, to its agronomic adaptability. Durum wheat (Triticum durum) is a tetraploid wheat species (AABB genomes) mainly grown in the Mediterranean basin, in the Northern Plains (between the USA and Canada), in the arid areas of South Western USA and in Northern Mexico []. Durum wheat is well-adapted to a broad range of climatic conditions (including dry environments) and marginal soils, and has low water requirements [,]. Climatic conditions, as temperature and water availability, together with biotic stresses, can strongly affect durum wheat development and production [,,,]. Crop adaptation is a central objective for breeding progress, driving improvement in final production, quantity and quality [,]. For over two decades, CIMMYT (International Maize and Wheat Improvement Center) has had an intensive breeding and improvement programme focused on the acceleration of durum productivity in developing countries.
Grain quality is an important breeding aim determining product end-use linked to financial returns. It is influenced by both genetic and environmental conditions [], and biotic and abiotic stresses during growth and at key development stages []. Temperature, water availability and soil properties, especially nitrogen content, influence the final quality and protein content of wheat and its end-products [,,].
There is a growing need to increase wheat yield without losing grain quality [,]. Key end-use grain quality traits include grain protein content (GPC), gluten strength, kernel size and vitreousness [,] and are all influenced by climatic conditions []. A number of agronomic components influence final productivity, including phenology (maturity) and plant architecture (plant height and lodging resistance). The majority of important agronomic traits, including yield, are controlled or influenced by multiple genes and are quantitatively inherited []. In addition, most are influenced by the environment and interactions between environmental and genetic (GxE) effects [,,,,]. One of the most common methods currently used for dissection of quantitative agronomic and quality traits is the association mapping (AM) approach [].
AM, originating in human genetics, was initially combined with linkage disequilibrium (LD) to identify the role of genes and linked markers for the determination of disease loci []. It is now widely used in plant and crop genetics. Some of the first studies based on LD mapping applied in plants were done in maize [], rice [] and oat []. AM has the main objective of determining, based on LD, correlations between genotypes and phenotypes in a panel of selected individuals []. It can support the development of new genetic markers for use in marker-assisted plant breeding []. It also facilitates the analysis of genetic variation underlying traits for further characterisation of the loci of interest [].
Single nucleotide polymorphism (SNP) markers are commonly used in quantitative trait loci (QTL) mapping experiments [,] and genome-wide association studies (GWAS) for the detection of marker-traits associations (MTAs) in wheat [,,,,]. DArTseq, a variant of the microarray-based DArT technology, has also been widely used in QTL mapping [,,]. It reduces the complexity of the genome, using combinations of restriction enzymes [] and next-generation sequencing. Several studies have assessed MTAs in durum wheat. The analyzed traits include grain yield, yield and yield components [,,,], heading date [], and grain quality traits (thousand kernel weight, vitreousness, protein content, sedimentation index [,,,], yellow colour [,]).
In this study, three panels of elite durum wheat lines from CIMMYT were assessed in field trials conducted over multiple seasons and with differing water regimes. The AM approach was used to detect SNP and DArT markers associated with heterogeneous agronomic and quality trait data in order to test the approach as a tool for marker discovery within a live and ongoing breeding programme.
2. Material and Methods
2.1. Plant Material, Phenotyping and Genotyping
Panels of elite durum lines from CIMMYT wheat preliminary yield trials (PYT), comprising a total of 294 accessions (Supplementary Materials Table S1) were used for agronomic and quality assessment. PYT trials consisted of the best advanced breeding lines which were promoted to unreplicated trials, including one or two repeated checks. The trials were sown, assessed and analysed according to their specific statistical designs [] and consisted of two blocks with different water treatments, one with full irrigation (FI) and the other with reduced irrigation (RI). In the FI treatment four to five irrigations were applied during the field season to maintain the optimal soil moisture conditions, whilst in the RI block a single irrigation was applied at planting, in order to ensure establishment. In both water treatments the irrigation was applied by gravity in furrows. The rainfall data (https://www.meteoblue.com/en/weather/historyclimate/weatherarchive/ciudad-obreg%c3%b3n_mexico_4013704) for the four field seasons is included in Supplementary Materials Figure S1. The agronomic and quality assessment of the panels over seasons is summarised in Table 1.

Table 1.
Agronomic and quality assessment of wheat field trials. The number of lines, year, location and water regime applied is shown.
Field experiments were conducted at CIMMYT’s experimental station in the Yaqui Valley, Mexico (27.282848° N; 109.923878° W) over four field seasons (2012 to 2015 harvest years, inclusive). Wheat panels 2 and 3 were phenotypically assessed across years, while panel 1 was only grown in 2012. The experimental plots (1.6 × 3 m) were sown in November/December of each year and harvested in May of the following year. Data for yellow rust were assessed in semi-controlled conditions at CIMMYT’s experimental station in Toluca (Mexico).
Plant material for genetic analysis was harvested for each line at the 4th leaf stage (growth stage 14 on the Zadoks scale []) and immediately frozen in dry-ice. Samples were stored at −80 °C until DNA extraction. Approximately 100 mg of frozen tissue was used for DNA isolation with a DNeasy Plant Mini Kit from Qiagen, following the manufacturer’s protocol. DNA sample quality and concentration were assessed using electrophoresis on a 0.8% agarose gel and the restriction enzyme Tru1I (ThermoFisher) was used to check for the absence of nucleases in DNA prior to genotyping.
Samples were genotyped by Diversity Arrays Technology Pty Ltd. (Montana St, University of Camberra, Bruce ACT 2617, Australia) (DArT) using DartSeqTM. A total of 35,509 polymorphic dominant DArT markers and 9142 biallelic SNP markers were generated. Both datasets were thinned by removing one marker from each pair with a correlation coefficient of >0.95. The final dataset consisted on 14,588 DArT markers (of which 8411 were mapped) and 5716 SNP markers (4142 mapped markers). DartSeqTM genotyping and mapping of the corresponding markers to the wheat reference genome sequence RefSeq v1 from the International Wheat Genome Sequencing Consortium (IWGSC, http://www.wheatgenome.org/) was performed by DArT (diversityarrays.com), as described by Sukumaran et al. []. The distribution of markers across the A and B subgenomes is given in Table 2.

Table 2.
Molecular markers distribution across the wheat genome. The distribution of DArT (Diversity Arrays Technology, left) and single nucleotide polymorphism (SNP, right) markers across the durum wheat A and B subgenomes and the number of unmapped markers are shown.
2.2. Phenotypic Data
Ten agronomic and quality traits were assessed for three durum wheat elite line panels: days to heading (days, DTH); plant height (cm, PH); lodging (%, LOD); yellow rust (%, YR); yellow colour (YC); sedimentation index (cm3, SDS); sedimentation volume (cm3, SV); grain protein content (%, GPC); thousand kernel weight (g, TKW); and grain yield (Kg/ha, GY). Agronomic traits (DTH, PH, LOD, YR, TKW and GY) were assessed under both water treatments (FI and RI), and quality traits (YC, SV, SDS and GPC) were only evaluated under FI conditions. Visual disease evaluation and phenology assessments were made in the field, while quality parameters were evaluated on grain samples post-harvest. DTH, PH and LOD and YR were visually assessed at the field trials in Yaqui, while YR was assessed at Toluca. To assess DTH, heading date was recorded as the time when 50% of the spikes have emerged from the flag leaf sheath (stage 59 in Zadoks scale []); PH was recorded by measuring the distance between the stem’s base and the top of the spike (awns not included); LOD was assessed as the percentage of lodging plot; and YR was assessed as the percentage of leaves with rust pustules. YC was assessed by a rapid colorimetric method with a Minolta color meter following CEN/TS 15,465:2008 [,,]; SDS was evaluated following UNE 34,903:2014 [,]; SV and GPC were assessed by Near-infrared spectroscopy (NIRs) []; TKW was measured by weighing 2 samples of 100 entire kernels randomly chosen previously dried at 70 °C for 48 h.
The correlation between the assessed traits was analysed using the ‘cor’ function in R [,,]. Then, an analysis of variance (ANOVA) was undertaken, using the ‘aov’ function in R [], to obtain the descriptive statistics for each trait.
Traits were analysed using a Q + K linear mixed-model [,] which follows the model equation:
where y is a vector of observed phenotypes; X, S and Z are matrices related to β, α and µ, respectively; β is a vector of fixed effects; α is a vector of marker effects; Qv is a vector of population effect; µ is a vector of polygenic effects (with covariance proportional to a kindship or relationship matrix); and ε is a vector of residuals.
y = Xβ + Sα + Qv + Zµ + ε
These analyses were carried out using GenStat (14th Edition) to generate the best linear unbiased estimates (BLUEs) of variety performance in different ways: (i) across years and blocks; (ii) across years for each block (FI and RI); (iii) across a reduced dataset (years 2013 and 2014) and blocks; and (iv) across the reduced dataset for each block. The resulting datasets (available in Supplementary Material Table S2) were then used in different association mapping analyses.
2.3. Population Structure and Linkage Disequilibrium
Population structure was assessed using principal component analysis (PCA) based on the combined DArT and SNP genotyping datasets. Euclidean distances were calculated using the R package ‘ggfortify’ [] and the PCA was visualised with ‘ggplot2’ [].
The pattern of linkage disequilibrium (LD) was assessed between each pair of SNP markers on the same chromosome across the two constitutive genomes with the allele frequency correlation (r2) using the ‘popgen’ package in R []. A heatmap was obtained with the D’ and r2 values for each chromosome and a scatterplot to determine LD decay (genetic distance in cM).
2.4. Association Mapping (AM)
The AM analyses were performed on the BLUEs obtained above using an additive model with ‘rrBLUP’ [] and ‘GWASpoly’ [] packages in R in different ways. Two marker-based kinship matrices (k-matrix), created from a subset of 14,588 DArT and 5716 SNP markers, respectively, were used for the adjustment based on relatedness of individuals (Supplementary Materials Tables S3 and S4). A minor allele frequency (maf) threshold of 0.05 was used. To establish a p-value detection threshold for statistical significance of associations, the Bonferroni correction, which employs a threshold of α/m to ensure the genome-wide type error I of 0.05, was applied with a total of 1000 permutations.
Associated DartSeqTM and SNP markers were blasted against the wheat reference assembly RefSeqv1 [] with no indels or mismatches allowed, using an ad hoc Java program, to confirm their physical mapping location on the A or B genomes. The molecular markers were also mapped against the durum wheat genome (https://www.interomics.eu/durum-wheat-genome) to confirm their physical positions. In addition, to identify candidate genes, results were filtered, selecting the hits with best e-value for each molecular marker, and candidate genes were manually selected based on their annotations.
3. Results
3.1. Phenotypic Assessment
Results from the ANOVA for all the traits across years and water treatments are shown in Table 3. The mean phenotypic values across years were calculated for each block and panel to evaluate the influence of water conditions on the assessed traits (Table 3). Days to heading during the field seasons assessed ranged from 63 to 94 days. In plots with lower water availability (RI block), the spike emergence from the flag leaf took place approximately 11 days earlier than in FI plots. Plant height ranged from 39 to 110 cm showing differences between water regimes, with a decrease of 25–30 cm under RI conditions. Likewise, and as result of the RI treatment, GY (ranging from 1.35 to 10.63 ton/ha) and TKW (from 29.6 to 63.2 g) also varied, being reduced by 4–5 tons/ha and 7–10 g, respectively, in the low water availability RI treatment. This strong RI treatment resulted in very low heritability values for DTH, PH and LOD.

Table 3.
Summary of overall phenotypic data including analysis of variance (ANOVA) results and broad sense heritability (h2) for agronomic and quality traits.
Several significant phenotypic correlations were observed between the analysed traits (Figure 1 and Supplementary Materials Table S5). The most correlated traits were PH and GY (r = 0.90, p-value = <2.2 × 10−16), followed by DTH and GY (r = 0.87, p-value = <2.2 × 10−16), SDS and SV (r = 0.85, p-value = <2.2 × 10−16) and also DTH and PH (r = 0.82, p-value = <2.2 × 10−16). Other traits showed important positive correlations too, including YC and DTH (r = 0.69, p-value = 2.82 × 10−09), GY and TKW (r = 0.66, p-value = <2.2 × 10−16), PH and TKW (r = 0.62, p-value = <2.2 × 10−16), GY and YC (r = 0.53, p-value = 0.039) and DTH and TKW (r = 0.49, p-value = <2.2 × 10−16). Negative correlations were also observed for SDS and GPC (r = −0.38, p-value = <2.2 × 10−16), and for GPC and YC (r = −0.08, p-value = <2.2 × 10−16) (Figure 1).
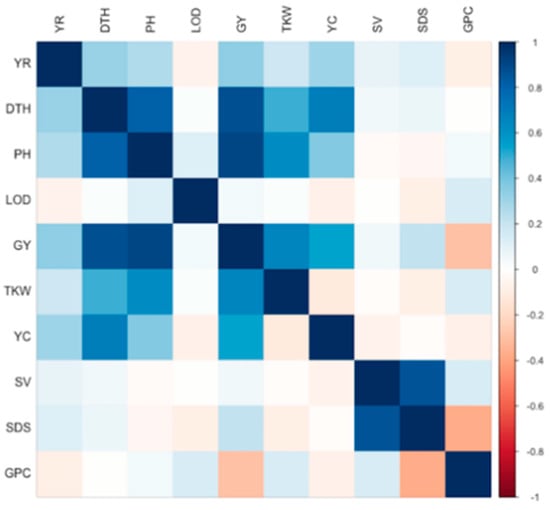
Figure 1.
Phenotypic correlations. YR: yellow rust; DTH: days to heading; PH: plant height; GY: grain yield; TKW: thousand kernel weight; YC: yellow colour; SV: sedimentation volume; SDS: sedimentation index; and GPC: grain protein content.
3.2. Population Structure and Linkage Disequilibrium
The PCA used a total of 14,588 DArT and 5716 SNP markers. The first and second principal components explained 3.91% of the genetic variation (Figure 2). No underlying genetic structure was detected within or between the panels assessed. LD was estimated using the mapped SNP markers dataset. LD decay was determined within 20–30 cM for all the chromosomes (Figure 3). Using the classification defined by Maccaferri et al. [], the markers presented loose linkage (Class 2), showing a distance value between 21 to 50 cM.
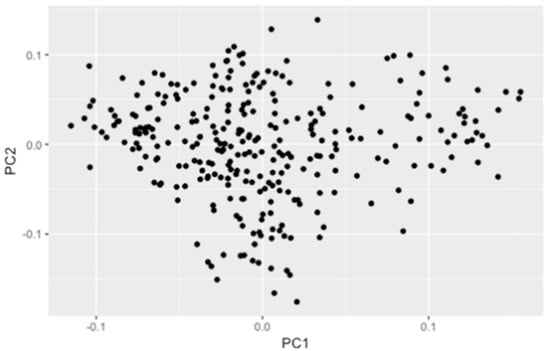
Figure 2.
Principal components analysis (PCA) of the genotypic data.
Figure 3.
Linkage disequilibrium (LD) decay analysis using SNP data. Estimates f r2 versus linkage distance on chromosome in centimorgan (cM) was represented. The LD decay was established between 20–30 cM.
3.3. AM Analysis
Thirty-seven significant marker-trait associations (MTAs) were detected for TKW and SV across all years and water treatments with most of the significant markers located on chromosome 2A (Table 4). Twenty DArT and seven SNP markers were found in association with TKW on chromosome 2A (with additive effects ranging from −3.41 to 3.46). In addition, eight unmapped DArT and one SNP marker were also associated with TKW (additive effects ranged from −3.39 to 3.46 g). Most of these MTAs showed a negative additive effect, reducing the final weight value (ranging from −2.84 to −3.19 g), and only two MTAs were found to increase TKW (values of 2.97 and 3.09 g). Finally, a single SNP associated with SV was located on chromosome 1B (showing a positive effect increasing the final value by 1.26 mL). The resulting Manhattan and QQ-plots from this AM analysis are included in Supplementary Materials Figures S2–S5.

Table 4.
Significant marker-trait associations found in durum wheat elite lines across years and water treatments.
The AM analyses on partitioned subsets of the data consistently detected the QTLs for TKW and SV. Nevertheless, the individual assessment of the water treatments significantly reduced the number of MTAs found, due in part to less available data for the RI block (Supplementary Material Table S6). The initial dataset of 294 durum wheat elite lines was reduced to 200 lines (assessed during the 2013 and 2014 seasons) to give a dataset balanced across assessment years. Using this reduced dataset for AM analysis, the results confirmed the QTLs previously found for the full dataset (Supplementary Materials Table S6). The analysis also detected an additional locus for SV on chromosome 4A, and a locus for YR on chromosome 1B (with additive effects of −0.84 and 2.79, respectively).
3.4. Candidate Genes Analysis
The marker SNP620, located on chromosome 1B and detected in association with SV, was found included into a cluster of 12 genes encoding blue copper proteins (BCP), with homoeologs in the two durum wheat subgenomes (Figure 4, Table 5 and Supplementary Material Figure S6). In the hexaploid wheat genome, this set of genes form a cluster of homeolog triads [] with a total of 31 genes (Supplementary Materials Table S7 and Figure S7). Additionally, another interesting gene (TraesCS1B01G568400LC.1) was found closer this marker, coding for the ubiquinone biosynthesis O-methyltransferase.
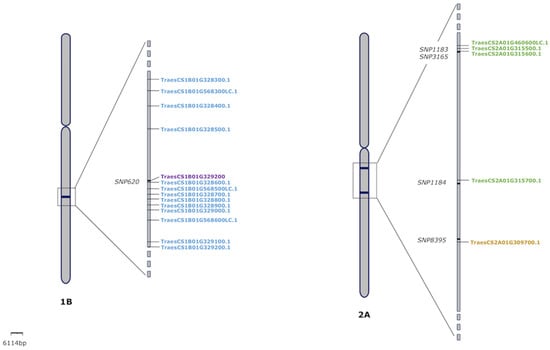
Figure 4.
Location of candidate genes on chromosomes 1B and 2A. Markers are indicated with the symbol ‘-’; blue copper proteins are shown in blue colour; ubiquinone biosynthesis O-methyltransferase in purple colour; the regulator response gene in brown colour; and for reductase 1 genes in green colour.

Table 5.
Candidate genes for markers with significant marker-traits associations. Genes located in proximity of markers found in association with TKW and SV across years and water treatments (within a ±50 kb window). Values for physical position and distance are indicated in base pairs (bp). Chr: chromosome position. Blue copper proteins are shown in blue colour; ubiquinone biosynthesis O-methyltransferase in purple colour; the regulator response gene in brown colour; and for reductase 1 genes in green colour. Physical positions and gene annotations are based on the wheat reference assembly RefSeqv1 [].
There were several markers located in chromosome 2A, in close proximity to some interesting genes. Markers SNP1183, SNP1184 and DArT3165 were found next to several genes encoding reductase-1 (Figure 4 and Table 5). In addition, the marker SNP8395, also located on the same chromosome, was found in proximity to the gene TraesCS2A01G309700.1, which encodes a type A response regulator 1 (Figure 4 and Table 5).
Significant MTAs from the partitioned analysis also allowed the identification of potentially interesting genes. On chromosome 1B, marker DArT1744, previously described by Mérida-García et al. [] related to high molecular-weight glutenin subunits, was found in proximity to genes encoding isocitrate dehydrogenase kinase/phosphatase G and leucine-rich repeat receptor-like protein kinase family protein. These genes participate in the carbohydrate metabolism during the Krebs cycle and play a crucial role in plant development and stress responses, respectively [,]. On this chromosome, another marker (SNP809) was found in proximity to some interesting genes encoding sugar transporter proteins. Additionally, some markers located on chromosome 2A were found in proximity to Acyl-CoA N-acyltransferase genes (SNP1206, SNP8395 and DArT3180) and chloroplastic zeaxanthin epoxidase (SNP1189) (Supplementary Materials Table S8).
4. Discussion
The maintenance of crop production is a current and pressing need given growing populations and reduced availability of arable land []. There is an increasing need for breeding programs to improve yield potential and the adaptation of new varieties to different biotic and abiotic stresses []. Abiotic stresses, including drought and heat, are impacting productivity on the million hectares of wheat grown worldwide each year []. Detailed molecular and phenotypic characterization are valuable tools in the dissection of complex traits [], and especially those that are influenced by water availability [].
The improvement of key traits is essential for better end-use production quantity and quality in wheat []. In this study, we analysed a set of 10 agronomic and quality traits under full irrigation conditions (FI), with an additional six traits also assessed under low water availability (RI) in order to understand trait variation under contrasting water regimes in the CIMMYT durum wheat breeding programme. Irrigation conditions influenced some important yield and yield-related traits such as GY and TKW, as well as adaptive traits including DTH and PH (Table 3). The RI treatments had decreased final yields in line with previous observations [,]. Previous reports have also shown TKW to be reduced by high temperatures [], most likely related to water availability. Groos et al. [] assessed the genetic basis of grain yield and protein content in a segregating population of wheat RILs grown at six locations and also identified effects from GxE interactions involving protein content and yield. Our mean trait values corroborated this, with the highest values for GY recorded for FI blocks across panels (see Table 3). A similar trend was shown for DTH, PH and TKW, which decreased under low-water regimes.
Correlations between the assessed traits showed that GY was positively correlated with two different phenology traits (PH and DTH). This is in agreement with Maccaferri et al. [], who showed important positive and negative correlations for GY and DTH, and also positive correlations for GY and PH in several environments with different water regimes. DTH and PH were also positively correlated (Supplementary Materials Table S5), with taller plants having a longer time period to the emergence of the tip of the spike stage.
Wheat TKW is an important yield component with a direct effect on grain yield [,]. In line with this, our results showed a significant and positive correlation between TKW and GY. However, the previously reported negative correlation between TKW and DTH [] was not observed, potentially as result of temperatures and water availability from emergence to heading, and also from heading to harvest. Rharrabti et al. [] previously highlighted a positive correlation between protein content and TKW, which is in agreement with the results obtained in the present study. They highlighted that warm temperatures during grain filling could negatively affect this correlation.
Significant associations between endosperm proteins (gliadin and glutenin subunits) and SV have been previously highlighted [,]. Here we found a positive correlation (r = 0.15) between SV and GPC. This correlation is thought to be the result of grain protein content influencing the sedimentation volume value [], which depends on the degree of protein hydration and oxidation []. Finally, for sedimentation index (SDS) analysis, we observed a negative correlation with protein content, in agreement with results presented by Rharrabti et al. [,]. This is also in agreement with Oelofse et al. [] who highlighted the significant influence of protein content on the SDS sedimentation test [,,].
The SNP and DArT markers used to analyze patterns of genetic structure (Figure 2) and LD (Figure 3) for the durum wheat lines revealed no detectable genetic structure and consistent patterns of LD across chromosomes (LD was estimated to extend ~20 cM). These results support the suitability of durum elite line sets currently in use in breeding programmes for the practical application of GWAs analysis. The rate of unmapped markers was lower for SNP than for DArT markers (27.5% vs. 42.0%, Table 2), indicating higher precision in genetically mapping SNP markers, probably as a result of co-dominance.
In the assessment of MTAs for quality and yield-related traits, different AM analyses were performed on subsets of the dataset. Several MTAs for SV and TKW were detected across years and water regimes, located on chromosomes 1B and 2A, respectively. All GWAS analyses corroborated the major QTLs previously detected, and reported two new QTLs, one for YR in chromosome 1B, and another for SV in chromosome 4A.
Associations on chromosome 1B were significant for wheat quality. There are known loci including Gli-B1/Glu-B3 on this chromosome, which are of great importance for some gliadin and glutenin subunits []. In fact, the candidate gene analysis reported the presence of a high molecular-weight glutenin subunit (HMW-GS) gene in the proximity of marker DArT1744 (found to be significantly associated with SV and SDS), which was previously described by Mérida-García et al. [] in association with specific weight. In line with this, Pogna et al. [] highlighted the importance of Glu-B3 for determining protein quality with these endosperm proteins showing significant effects on SV, which also showed a high and positive correlation with SDS in our study (Figure 1 and Supplementary Materials Table S5). Likewise, Blanco et al. [] reported three QTLs on chromosomes 1B, 6A and 7B (based on the analysis of 259 polymorphic markers) associated with SDS and SV in a recombinant inbred line population. In the present study, we found a SNP marker (SNP620) associated with SV, showing a positive additive effect of 1.26 (see Table 4) and also with SDS (marker effect of 0.11) (Supplementary Materials Table S6). This marker was previously placed on chromosome 1B, in the same location as MTAs for gluten index and sedimentation index []. Other previous studies, such as Reif et al. [] and Fiedler et al. [], also reported markers associated with SV on chromosome 1B, but with differing genetic positions. The additional locus for SV found on chromosome 4A (marker DArT9459) has not been previously reported.
Marker SNP620, significantly associated with SV, is located within a cluster of homoeolog gene triads coding for blue copper proteins (Table 5 and Supplementary Materials Figure S6). These proteins have been described containing a copper atom, and participate in redox processes [], with a crucial role in the electron shuttle in plants []. In addition, Yao et al. [] described the blue copper protein genes as the targets of miR408 in wheat, which is involved in the regulation of gene transcription required for heading time []. In our study SNP620 was also found in proximity to a gene coding for an ubiquinone biosynthesis O-methyltransferase. Liu et al. [] highlighted its crucial role as an electron transporter in the electron transport chain of the aerobic respiratory chain. This ubiquinone gene is involved in plant growth and development, is implied in chemical compounds biosynthesis and metabolism which are involved in plant responses to stress, and also participates in gene expression regulation and cell signal transduction [].
On chromosome 1B we also found a significant MTA for yellow rust, in agreement with previous studies in durum and bread wheat, which placed different markers significantly associated with this trait, but in differing genetic positions [,,,]. The candidate gene analysis revealed the proximity of this marker (SNP809) to sugar transporter protein genes. Sugars are formed during the photosynthetic process and are essential for plant nutrition. The sucrose transport has been considered of great importance for plant productivity []. In line with this, the sucrose is involved in the gene expression regulation of the supposedly sugar-sensing pathway [,].
The majority of MTAs for TKW were located on chromosome 2A, showing both positive and negative effects. Previous studies have reported different markers in association with this quality trait, including a number mapped on chromosome 2A [,,,]. One of the markers found by Yao et al. [] (SSR marker gwm445 on chromosome 2A (68.2 cM), belongs to the same QTL found in this study for the marker SNP1153 (chromosome 2A, 68.6 cM), and also found by Juliana et al. [] in bread wheat lines from CIMMYT’s first year-yield trials. Sukumaran et al. [] analysed a durum wheat panel of 208 lines under yield potential, heat and drought stress conditions, and identified markers on chromosome 2A with a similar position to those detected in this study (4 markers at 70 cM and 6 markers at 69 cM) under heat stress conditions. They highlighted that several SNP markers were related to transmembranes or were uncharacterized proteins. We found several candidate genes for this important TKW QTL (Table 5) among which the most striking feature is the presence of four reductase 1 genes (NADPH-dependent 6′-deoxychalcone synthase) and a type A response regulator 1 (Figure 4). These genes are both related with photosynthesis. Hu et al. [] highlighted that NADPH plays a crucial role in biological processes in plants, such as the regulation of the production of reactive oxygen species (ROS) for the stress tolerance [,]. Additional GWAS analyses using reduced datasets revealed other interesting genes for this QTL (chromosome 2A, Supplementary Materials Table S8), encoding for the Acyl-CoA N-acyltransferase and the chloroplastic zeaxanthin epoxidase. The first gene has several functions in signaling and metabolic pathways []. The zeaxanthin epoxidase plays an important role in the xanthophyll cycle and abscisic acid (ABA) biosynthesis. The xanthophyll cycle has a main function in the dissipation of light energy excess and also increasing the photosynthetic system stability [].
The proposed approach has successfully detected genetic markers with significant associations with TKW, SV, SDS and YR. These are of potential use in durum wheat breeding programs, and can be further interrogated to the candidate gene level using the RefSeqv1 bread wheat genome reference [] and the durum wheat genome reference [].
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4395/10/1/144/s1: Figure S1: Rainfall data for Ciudad Obregon (Mexico) for the growing seasons 2012–2015; Figure S2: Manhattan plots for durum wheat mapped DArT markers. DTH: days to heading; PH: plant height; GY: grain yield; TKW: thousand kernel weight; YC: yellow color; SV: sedimentation volume; SDS: sedimentation index; and GPC: grain protein content; Figure S3: Manhattan plots for durum wheat mapped SNP markers. DTH: days to heading; PH: plant height; GY: grain yield; TKW: thousand kernel weight; YC: yellow color; SV: sedimentation volume; SDS: sedimentation index; and GPC: grain protein content; Figure S4: Quantile quantile-plots from genome-wide association studies (GWAS) analysis for durum wheat DArT markers (mapped and unmapped). DTH: days to heading; PH: plant height; LOD: lodging; GY: grain yield; TKW: thousand kernel weight; YC: yellow color; SV: sedimentation volume; SDS: sedimentation index; and GPC: grain protein content; Figure S5: Quantile quantile-plots from GWAS analysis for durum wheat SNP markers (mapped and unmapped). DTH: days to heading; PH: plant height; LOD: lodging; GY: grain yield; TKW: thousand kernel weight; YC: yellow color; SV: sedimentation volume; SDS: sedimentation index; and GPC: grain protein content; Figure S6: Blue copper protein gene cluster on durum wheat chromosome 1B. High confidence genes are shown in green colour, low confidence genes are shown in yellow; Figure S7: Cluster tree of blue copper protein gene homoeologs in bread wheat (RefSeqv1 []). For chromosome 1A, high confidence (HC) and low confidence (LC) genes are shown in brown and orange colour, respectively; for chromosome 1B, HC and LC genes are shown in dark and light green colour, respectively; for chromosome 1D, HC and LC genes are shown in dark and light blue colour, respectively; Table S1: Durum wheat elite lines assessed; Table S2: Best Linear Unbiased Estimates (BLUEs) outputs for all assessed traits and the association mapping analyses performed in durum wheat: [i] across years and blocks; [ii] across years for each block (FI and RI); [iii] across years and blocks for a reduced dataset (years 2013 and 2014); and [iv] across the reduced dataset for each block. DTH: days to heading; GPC: grain protein content; GY: grain yield; PH: plant height; SDS: sedimentation index; SV: sedimentation volume; TKW: thousand kernel weight; and YC: yellow colour; YR: yellow rust; LOD: lodging; Table S3: Kinship matrix for durum wheat DArT markers; Table S4: Kinship matrix for durum wheat SNP markers; Table S5: Phenotypic correlations between the assessed traits in durum wheat and their corresponding p values. YR: yellow rust; DTH: days to heading; PH: plant height; LOD: lodging; GY: grain yield; TKW: thousand kernel weight; YC: yellow color; SV: sedimentation volume; SDS: sedimentation index; and GPC: grain protein content; Table S6: Marker-trait associations found for the association mapping analyses performed in durum wheat: [i] across years and blocks; [ii] across years for each block (FI and RI); [iii] across years and blocks for a reduced dataset (years 2013 and 2014); and [iv] across the reduced dataset for each block. SV: sedimentation volume; TKW: thousand kernel weight; SDS: sedimentation index; YR: yellow rust; “-”: unmapped marker; Table S7: Homoeolog triads for blue copper protein genes mapped in the wheat reference assembly RefSeqv1 []; Table S8: Candidate genes for GWAS analyses performed in durum wheat: [i] across years for each block (FI and RI); [ii] across years and blocks for a reduced dataset (years 2013 and 2014); and [iii] across the reduced dataset for each block. Molecular markers mapping positions are shown both in the durum wheat genome [] and the wheat reference assembly RefSeqv1 []; Supplementary Material S1. R script used to perform the GWAS analysis; Supplementary Material S2. R script used to perform the LD analysis.
Author Contributions
P.H., I.S. and K.A. conceived the experiment. K.A. and I.S. selected the plant materials and agronomic traits. K.A. managed the field trials and contributed the phenotypic data. I.S., P.H., G.D., S.G., R.M.-G. and A.R.B. analysed the genotypic and phenotypic data. R.M.-G. isolated the DNA. R.M.-G., A.R.B. and P.H. carried out the AM analyses. R.M.-G., A.R.B. and P.H. drafted the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by project P12-AGR-0482 from Junta de Andalucía (Andalusian Regional Government), Spain (Co-funded by FEDER). ARB is supported by the UK Biotechnology and Biological Sciences Research Council (BBSRC) ‘Designing Future Wheat’ cross-institute strategic programme (BB/P016855/1). PH is supported by the Spanish Ministry of Economy, Industry and Competitiveness (MINECO) project AGL2016-77149-C2-1-P.
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Curtis, B.C. Wheat in the world. In Bread Wheat Improvement and Production; Curtis, B.C., Rajaram, S., Gomez Macpherson, H., Eds.; Plant Production and Protection Series; Food and Agriculture Organization of the United Nations: Rome, Italy, 2002; Volume 30, pp. 1–17. [Google Scholar]
- Ranieri, R. Geography of Durum Wheat Crop; Pastaria Open Fields: Collecchio, Italy, 2015. [Google Scholar]
- Bozzini, A.; Fabriani, G.; Lintas, C. Origin, Distribution, and Production of Durum Wheat in the World. In American Associate of Cereal Chemists International, Durum Wheat, 2nd ed.; AACC International Press: Saint Paul, MN, USA, 2012. [Google Scholar] [CrossRef]
- Nachit, M.M. Durum wheat breeding for Mediterranean drylands of north Africa and west Asia. In Durum Wheats: Challenges and Opportunities; Rajaram, S., Saari, E.E., Hettel, G.P., Eds.; Wheat Special Report; CIMMYT: Ciudad Obregón, Mexico, 1992. [Google Scholar]
- Singh, R.P.; Huerta-Espino, J.; Fuentes, G.; Duveiller, E.; Gilchrist, L.; Henry, M.; Nicol, M.J. Resistance to diseases. In Durum Wheat Breeding: Current Approaches and Future Strategies; Royo, C., Nachit, M.M., Fonzo, D., Araus, J.L., Er, W.H.P., Slafer, G.A., Eds.; Food Prod Press: Binghamton, NY, USA, 2005; pp. 291–315. [Google Scholar]
- Maccaferri, M.; Sanguineti, M.C.; Demontis, A.; El-Ahmed, A.; Garcia del Moral, L.; Maalouf, F.; Nachit, M.; Nserallah, N.; Ouabbou, H.; Rhouma, S.; et al. Association mapping in durum wheat grown across a broad range of water regimes. J. Exp. Bot. 2011, 62, 409–438. [Google Scholar] [CrossRef]
- Abdalla, O.; Dieseth, J.A.; Singh, R.P. Breeding durum wheat at CIMMYT. In Durum Wheats: Challenges and Opportunities; Rajaram, S., Saari, E.E., Hettel, G.P., Eds.; Wheat Special Report; CIMMYT: Ciudad Obregón, Mexico, 1992. [Google Scholar]
- Wrigley, C.W. Wheat: An overview of the grain that provides our daily bread. In Encyclopedia of Food Grains; Colin, W., Wrigley, H.C., Koushik, S., Jonathan, F., Eds.; Elsevier: Oxford, UK, 2016; pp. 105–116. [Google Scholar] [CrossRef]
- Triboi, E.; Abad, A.; Michelena, A.; Lloveras, J.; Ollier, J.L.; Daniel, C. Environmental effects on the quality of two wheat genotypes: 1. quantitative and qualitative variation of storage proteins. Eur. J. Agron. 2000, 13, 47–64. [Google Scholar] [CrossRef]
- Halford, N.G.; Curtis, T.Y.; Chen, Z.; Huang, J. Effects of abiotic stress and crop management on cereal grain composition: Implications for food quality and safety. J. Exp. Bot. 2015, 66, 1145–1156. [Google Scholar] [CrossRef]
- Blumenthal, F.; Batey, I.L.; Wrigley, C.W.; Moss, H.J.; Mares, D.J.; Barlow, E.W.C. Interpretation of grain quality results from wheat variety trials with reference to high temperatures stress. Aust. J. Agric. Res. 1991, 42, 325–334. [Google Scholar] [CrossRef]
- Campbell, C.A.; Davidson, H.R.; Winkleman, G.E. Effect of nitrogen, temperature, growth stage and duration of moisture stress on yield components and protein content of manitou spring wheat. Can. J. Plant Sci. 1981, 61, 549–563. [Google Scholar] [CrossRef]
- Uhlen, A.K.; Hafskjold, R.; Kalhovd, A.H.; Sahlström, S.; Longva, Å.; Magnus, E.M. Effects of cultivar and temperature during grain filling on wheat protein content, composition, and dough mixing properties. Cereal Chem. 1998, 75, 460–465. [Google Scholar] [CrossRef]
- Araus, J.L.; Slafer, G.A.; Royo, C.; Serret, M.D. Breeding for Yield Potential and Stress Adaptation in Cereals. Crit. Rev. Plant Sci. 2008, 27, 377–412. [Google Scholar] [CrossRef]
- Curtis, T.; Halford, N.G. Food security: The challenge of increasing wheat yield and the importance of not compromising food safety. Ann. Appl. Biol. 2014, 164, 354–372. [Google Scholar] [CrossRef]
- Steiger, D.K.; Elias, E.M.; Joppa, L.R.; Cantrell, R.G. Quality Evaluation of Lines Derived from Crosses of Langdon (Triticum dicoccoides) Substitution Lines to a Common Durum Wheat. In Durum Wheats: Challenges and Opportunities; Rajaram, S., Saari, E.E., Hettel, G.P., Eds.; Wheat Special Report; CIMMYT: Ciudad Obregón, Mexico, 1992. [Google Scholar]
- Rharrabti, Y.; Villegas, D.; Royo, C.; Martos-Nuñez, V.; Garcia del Moral, L.F. Durum wheat quality in Mediterranean environments II. Influence of climatic variables and relationships between quality parameters. Field Crops Res. 2003, 80, 133–140. [Google Scholar] [CrossRef]
- Falconer, D.S.; Mackay, T.F.C. Introduction to Quantitative Genetics, 4th ed.; Pearson Education Limited: Harlow, UK, 1997; p. 464. [Google Scholar]
- Borghi, B.; Corbellino, M.; Ciuffi, M.; La fiandra, D.; Destefanis, E.; Sgrulletta, D.; Boggini, G.; Di Fonzo, N. Effect of heat-shock during grain filling on grain quality of bread and durum wheats. Aust. J. Agric. Res. 1995, 46, 1365–1380. [Google Scholar] [CrossRef]
- Garrido-Lestache, E.; Lopez-Bellido, R.J.; Lopez-Bellido, L. Durum wheat quality under Mediterranean conditions as affected by N rate, timing and splitting, N form and S fertilization. Eur. J. Agron. 2005, 23, 265–278. [Google Scholar] [CrossRef]
- Holland, J.B. Genetic architecture of complex traits in plants. Plant Biol. 2007, 10, 156–161. [Google Scholar] [CrossRef]
- Jackson, P.; Robertson, M.; Cooper, M.; Hammer, G. The role of physiological understanding in plant breeding; from a breeding perspective. Field Crops Res. 1996, 49, 11–39. [Google Scholar] [CrossRef]
- Taghouti, M.; Gaboun, F.; Nsarellah, N.; Rhrib, R.; El-Haila, M.; Kamar, M.; Abbad-Andaloussi, F.; Udapa, S.M. Genotype x Envitonment interactin for quality traits in durum wheat cultivars adapted to different environments. Afr. J. Biotechnol. 2010, 9, 3054–3062. [Google Scholar]
- Flint-Garcia, S.A.; Thuillet, A.C.; Yu, J.; Pressoir, G.; Romero, S.M.; Mitchell, S.E.; Doebley, J.; Kresovich, S.; Goodman, M.M.; Buckler, E.S. Maize association population: A high-resolution platform for quantitative trait locus dissection. Plant J. 2005, 44, 1054–1064. [Google Scholar] [CrossRef]
- Bodmer, W.F. Human genetics: The molecular challenge. BioEssays: News and Reviews in Molecular. Cell. Dev. Biol. 1987, 7, 41–45. [Google Scholar] [CrossRef]
- Bar-Hen, A.; Charcosset, A.; Bourgoin, M.; Guiard, J. Relationship between genetic markers and morphological traits in a Maize inbred lines collection. Euphytica 1995, 84, 145–154. [Google Scholar] [CrossRef]
- Virk, P.S.; Ford-Lloyd, B.V.; Jackson, M.T.; Pooni, H.S.; Clemeno, T.P. Predicting quantitative variation within rice germplasm using molecular markers. Heredity 1996, 76, 296–304. [Google Scholar] [CrossRef]
- Beer, M.U.; Wood, P.J.; Weisz, J.; Fillion, N. Effect of cooking and storage on the amount and molecular weight of (1→3) (1→4)-d-glucan extracted from oat products by an in vitro digestion system. Cereal Chem. 1997, 74, 705–709. [Google Scholar] [CrossRef]
- Zondervan, K.T.; Cardon, L.R. The complex interplay among factors that influence allelic association. Nat. Rev. Genet. 2004, 5, 89–100. [Google Scholar] [CrossRef]
- Breseghello, F.; Sorrells, M.E. Association mapping of kernel size and milling quality in wheat (Triticum aestivum L.) cultivars. Genetics 2006, 172, 1165–1177. [Google Scholar] [CrossRef]
- Maccaferri, M.; Sanguineti, M.C.; Mantovani, P.; Demontis, A.; Massi, A.; Ammar, K.; Kolmer, J.A.; Czembor, J.H.; Ezrati, S.; Tuberosa, R. Association mapping of leaf rust response in durum wheat. Mol. Breed. 2010, 26, 189–228. [Google Scholar] [CrossRef]
- Marcotuli, I.; Gadaleta, A.; Mangini, G.; Signorile, A.M.; Zacheo, S.A.; Blanco, A.; Simeone, R.; Colasuonno, P. Development of a High-Density SNP-Based Linkage Map and Detection of QTL for beta-Glucans, Protein Content, Grain Yield per Spike and Heading Time in Durum Wheat. Int. J. Mol. Sci. 2017, 18, 1329. [Google Scholar] [CrossRef]
- Liu, J.; Luo, W.; Qin, N.; Ding, P.; Zhang, H.; Yang, C.; Mu, Y.; Tang, H.; Liu, Y.; Li, W.; et al. A 55 K SNP array-based genetic map and its utilization in QTL mapping for productive tiller number in common wheat. Theor. Appl. Genet. 2018, 131, 2439–2450. [Google Scholar] [CrossRef]
- Marcotuli, I.; Houston, K.; Schwerdt, J.G.; Waugh, R.; Fincher, G.B.; Burton, R.A.; Blanco, A.; Gadalena, A. Genetic Diversity and Genome Wide Association Study of beta-Glucan Content in Tetraploid Wheat Grains. PLoS ONE 2016, 11, e0152590. [Google Scholar] [CrossRef]
- Poland, J.A.; Brown, P.J.; Sorrells, M.E.; Jannink, J.L. Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLoS ONE 2012, 7, e32253. [Google Scholar] [CrossRef]
- Somers, D.J.; Banks, T.; Depauw, R.; Fox, S.; Clarke, J.; Pozniak, C.; McCartney, C. Genome-wide linkage disequilibrium analysis in bread wheat and durum wheat. Genome 2007, 50, 557–567. [Google Scholar] [CrossRef]
- Wang, S.X.; Zhu, Y.L.; Zhang, D.X.; Shao, H.; Liu, P.; Hu, J.B.; Zhang, H.; Zhang, H.P.; Chang, C.; Lu, J.; et al. Genome-wide association study for grain yield and related traits in elite wheat varieties and advanced lines using SNP markers. PLoS ONE 2017, 12, e0188662. [Google Scholar] [CrossRef]
- Yao, J.; Wang, L.; Liu, L.; Zhao, C.; Zheng, Y. Association mapping of agronomic traits on chromosome 2A of wheat. Genetica 2009, 137, 67–75. [Google Scholar] [CrossRef]
- Li, H.; Vikram, P.; Singh, R.P.; Kilian, A.; Carling, J.; Song, J.; Burgueno-Ferreira, J.A.; Bhavani, S.; Huerta-Espino, J.; Payne, T.; et al. A high density GBS map of bread wheat and its application for dissecting complex disease resistance traits. BMC Genom. 2015, 16, 216. [Google Scholar] [CrossRef]
- Ren, R.; Ray, R.; Li, P.; Xu, J.; Zhang, M.; Liu, G.; Yao, X.; Kilian, A.; Yang, X. Construction of a high-density DArTseq SNP-based genetic map and identification of genomic regions with segregation distortion in a genetic population derived from a cross between feral and cultivated-type watermelon. Mol. Genet. Genom. 2015, 290, 1457–1470. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Sevilla, J.F.; Horvath, A.; Botella, M.A.; Gaston, A.; Folta, K.; Kilian, A.; Denoyes, B.; Amaya, I. Diversity Arrays Technology (DArT) Marker Platforms for Diversity Analysis and Linkage Mapping in a Complex Crop, the Octoploid Cultivated Strawberry (Fragaria x ananassa). PLoS ONE 2015, 10, e0144960. [Google Scholar] [CrossRef] [PubMed]
- Kilian, A.; Huttner, E.; Wenzl, P.; Jaccoud, D.; Carling, J.; Caig, V.; Evers, M.; Heller-Uszynska, K.; Cayla, C.; Patarapuwadol, S.; et al. The fast and the cheap, SNP and DArT-based whole genome profiling for crop improvement. In The Wake of the Double Helix: From the Green Revolution to the Gene Revolution, Proceedings of the International Congress Avenue Media, Bologna, Italy, 27–31 May 2005; Tuberosa, R., Phillips, R.L., Gale, M., Eds.; Avenue Media: Bologna, Italy, 2005; pp. 443–461. [Google Scholar]
- Sukumaran, S.; Reynolds, M.P.; Sansaloni, C. Genome-Wide Association Analyses Identify QTL Hotspots for Yield and Component Traits in Durum Wheat Grown under Yield Potential, Drought, and Heat Stress Environments. Front. Plant Sci. 2018, 9, 81. [Google Scholar] [CrossRef]
- Turuspekov, Y.; Baibulatova, A.; Yermekbayev, K.; Tokhetova, L.; Chudinov, V.; Sereda, G.; Ganal, M.; Griffiths, S.; Abugalieva, S. GWAS for plant growth stages and yield components in spring wheat (Triticum aestivum L.) harvested in three regions of Kazakhstan. BMC Plant Biol. 2017, 17 (Suppl. 1), 190. [Google Scholar] [CrossRef]
- Johnson, M.; Kumar, A.; Oladzad-Abbasabadi, A.; Salsman, E.; Aoun, M.; Manthey, F.A.; Elias, E.M. Association Mapping for 24 Traits Related to Protein Content, Gluten Strength, Color, Cooking, and Milling Quality Using Balanced and Unbalanced Data in Durum Wheat [Triticum turgidum L. var. durum (Desf)]. Front. Genet. 2019, 10. [Google Scholar] [CrossRef]
- Mangini, G.; Gadaleta, A.; Colasuonno, P.; Marcotuli, I.; Signorile, A.M.; Simeone, R.; De Vita, P.; Mastrangelo, A.M.; Laido, G.; Pecchioni, N.; et al. Genetic dissection of the relationships between grain yield components by genome-wide association mapping in a collection of tetraploid wheats. PLoS ONE 2018, 13, e0190162. [Google Scholar] [CrossRef]
- Mengistu, D.K.; Kidane, Y.G.; Catellani, M.; Frascaroli, E.; Fadda, C.; Pe, M.E.; Dell’Acua, M. High-density molecular characterization and association mapping in Ethiopian durum wheat landraces reveals high diversity and potential for wheat breeding. Plant Biotechnol. J. 2016, 14, 1800–1812. [Google Scholar] [CrossRef]
- Pozniak, C.J.; Knox, R.E.; Clarke, F.R.; Clarke, J.M. Identification of QTL and association of a phytoene synthase gene with endosperm colour in durum wheat. Theor. Appl. Genet. 2007, 114, 525–537. [Google Scholar] [CrossRef]
- Reimer, S.; Pozniak, C.J.; Clarke, F.R.; Clarke, J.M.; Somers, D.J.; Knox, R.E.; Singh, A.K. Association mapping of yellow pigment in an elite collection of durum wheat cultivars and breeding lines. Genome 2008, 51, 1016–1025. [Google Scholar] [CrossRef]
- Van Ginkel, M.R.; Trethowan, R.; Cukadar, B. A Guide to the CIMMY Bread Wheat Program; Wheat Special Report No 5; CIMMYT: Ciudad Obregón, Mexico, 1998. [Google Scholar]
- Zadoks, J.C.; Chang, T.T.; Konzak, C.F. A decimal code for the growth stages of cereals. Weed Res. 1974, 14, 415–421. [Google Scholar] [CrossRef]
- Henstchel, V.; Kranl, K.; Hollmann, J.; Lindhauer, M.G.; Bohm, V.; Bitsch, R. Spectrophotometric determination of yellow pigment content and evaluation of carotenoids by high-performance liquid chromatography in durum wheat grain. J. Agric. Food Chem. 2002, 50, 6663–6668. [Google Scholar]
- Martinez, C.S.; Ribotta, P.D.; León, A.E.; Añon, M.C. Colour assessment on bread wheat and triticale fresh pasta. Int. J. Food Prop. 2010, 15. [Google Scholar] [CrossRef]
- Beleggia, R.; Platani, C.; Nigro, F.; Papa, R. Yellow pigment determination for single kernels of durum wheat (Triticum durum Desf.). Cereal Chem. 2011, 88, 504–508. [Google Scholar] [CrossRef]
- Axford, D.W.E.; McDermott, E.E.; Redman, D.G. Small-scale test for breadmaking quality of wheat. Cereal Foods World 1978, 23, 477–478. [Google Scholar]
- Seabourn, B.W.; Xiao, Z.S.; Tilley, T.; Herald, T.J.; Park, S.H. A rapid, small-scale sedimentation method to predict bread-making quality of hard winter wheat. Crop Sci. 2012, 52, 1306–1315. [Google Scholar] [CrossRef]
- Williams, P.C.; Norris, K. Near Infrared Technology in the Agricultural and Food Industries, 2nd ed.; American Association of Cereal Chemistry, Inc.: St Paul, MN, USA, 2001. [Google Scholar]
- Becker, R.A.; Chambers, J.M.; Wilks, A.R. The New S Language: A Programming Environment for Data Analysis and Graphics; Wadsworth & Brooks/Cole: Pacific Grove, CA, USA, 1988. [Google Scholar]
- Kendall, M.G. A new measure of rank correlation. Biometrika 1938, 30, 81–93. [Google Scholar] [CrossRef]
- Kendall, M.G. The treatment of ties in rank problems. Biometrika 1945, 33, 239–251. [Google Scholar] [CrossRef]
- Chambers, J.M.; Freeny, A.; Heiberger, R.M. Chapter 5: Analysis of Variance; Designed Experiment. In Statistical Models in S; Chambers, J.M., Hastie, T.J., Eds.; Wadsworth & Brooks/Cole: Pacific Grove, CA, USA, 1992; pp. 145–193. [Google Scholar]
- Yu, J.; Pressoir, G.; Briggs, W.H.; Vroh Bi, I.; Yamasaki, M.; Doebley, J.F.; McMullen, M.D.; Gaut, B.S.; Nielsen, D.M.; Holland, J.B.; et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 2006, 38, 203–208. [Google Scholar] [CrossRef]
- Kang, H.M.; Zaitlen, N.A.; Wade, C.M.; Kirby, A.; Heckerman, D.; Daly, M.J.; Eskin, E. Efficient control of population structure in model organism association mapping. Genetics 2008, 178, 1709–1723. [Google Scholar] [CrossRef]
- Horikoshi, M.; Tang, Y. ggfortify: Data Visualization Tools for Statistical Analysis Results. R J. 2016, 8, 474–489. [Google Scholar]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2009. [Google Scholar]
- Marchini, J.L. Popgen: Statistical and Population Genetics. R Package Version 1.0-3. 2013. Available online: http://CRANR-projectorg/package=popgen/ (accessed on 14 January 2020).
- Endelman, J.B. Ridge regression and other kernels for genomic selection with R package rrBLUP. Plant Genome 2011, 4, 250–255. [Google Scholar] [CrossRef]
- Rosyara, U.R.; De Jong, W.S.; Douches, D.S.; Endelman, J.B. Software for Genome-Wide Association Studies in Autopolyploids and Its Application to Potato. Plant Genome 2016, 9. [Google Scholar] [CrossRef] [PubMed]
- Appels, R.; Eversole, K.; Feuillet, C.; Keller, B.; Rogers, J.; Stein, N.; Pozniak, C.J.; Choulet, F.; Distelfeld, A.; Poland, J.; et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar]
- Maccaferri, M.; Sanguineti, M.C.; Noli, E.; Tuberosa, R. Population structure and long-range linkage disequilibrium in a durum wheat elite collection. Mol. Breed. 2005, 15, 271–290. [Google Scholar] [CrossRef]
- Maccaferri, M.; Harris, N.S.; Twardziok, S.O.; Pasam, R.K.; Gundlach, H.; Spannagl, M.; Ormanbekova, D.; Lux, T.; Prade, V.M.; Milner, S.G.; et al. Durum wheat genome highlights past domestication signatures and future improvement targets. Nat. Genet. 2019, 51, 885–895. [Google Scholar] [CrossRef]
- Galvez, S.; Merida-Garcia, R.; Camino, C.; Borrill, P.; Abrouk, M.; Ramirez-Gonzalez, R.H.; Biyiklioglu, S.; Amil-Ruiz, F.; Dorado, G.; Budak, H.; et al. Hotspots in the genomic architecture of field drought responses in wheat as breeding targets. Funct. Integr. Genom. 2019. [Google Scholar] [CrossRef]
- Mérida-García, R.; Guozheng, L.; He, S.; González-Dugo, V.; Dorado, G.; Gálvez, S.; Solís, I.; Zarco-Tejada, P.; Reif, J.C.; Hernández, P. Genetic dissection of agronomic and quality traits based on association mapping and genomic selection approaches in durum wheat grown in Southern Spain. PLoS ONE 2019. [Google Scholar] [CrossRef]
- Ikeda, T.; Laporte, D.C. Isocitrate Dehydrogenase Kinase/Phospatase: AceK alleles that express kinase but not phosphatase activity. J. Bacteriol. 1991, 1801–1806. [Google Scholar] [CrossRef][Green Version]
- Liu, P.L.; Du, L.; Huang, Y.; Gao, S.M.; Yu, M. Origin and diversification of leucine-rich repeat receptor-like protein kinase (LRR-RLK) genes in plants. BMC Evol. Biol. 2017, 17, 47. [Google Scholar] [CrossRef]
- Cassman, K.G.; Dobermann, A.; Walters, D.T.; Yang, H. Meeting cereal demand while protecting natural resources and improving environmental quality. Annu. Rev. Environ. Resour. 2003, 28, 315–358. [Google Scholar] [CrossRef]
- Araus, J.L.; Slafer, G.A.; Reynolds, M.P.; Royo, C. Plant Breeding and Drought in C3 Cereals: What Should We Breed For? Ann. Bot. 2002, 89, 925–940. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, R.; Sayre, K.D.; Govaerts, B.; Gupta, R.K.; Subbarao, G.V.; Ban, T.; Hodson, D.; Dixon, J.; Ortiz-Monasterio, I.; Reynolds, M. Climate change: Can wheat beat the heat? Agric. Ecosyst. Environ. 2008, 126, 46–58. [Google Scholar] [CrossRef]
- Alonso-Blanco, C.; Aarts, M.G.; Bentsink, L.; Keurentjes, J.J.; Reymond, M.; Vreugdenhil, D.; Koornneef, M. What has natural variation taught us about plant development, physiology, and adaptation? Plant Cell 2009, 21, 1877–1896. [Google Scholar] [CrossRef] [PubMed]
- Amaya, A.; Peña, R.J. Utilization and quality of durum wheat. In Durum Wheats: Challenges and Opportunities; Rajaram, S., Saari, E.E., Hettel, G.P., Eds.; Wheat Special Report; CIMMYT: Ciudad Obregón, Mexico, 1992. [Google Scholar]
- Maccaferri, M.; Sanguineti, M.C.; Corneti, S.; Ortega, J.L.A.; Salem, M.B.; Bort, J.; DeAmbrogio, E.; García del Moral, L.F.; Demontis, A.; El-Ahmed, A.; et al. Quantitative trait loci for grain yield and adaptation of durum wheat (Triticum durum Desf.) across a wide range of water availability. Genetics 2008, 178, 489–511. [Google Scholar] [CrossRef]
- Groos, C.; Robert, N.; Bervas, E.; Charmet, G. Genetic analysis of grain protein-content, grain yield and thousand-kernel weight in bread wheat. Theor. Appl. Genet. 2003, 106, 1032–1040. [Google Scholar] [CrossRef]
- Sen, C.; Toms, B. Character association and component analysis in wheat (Triticum aestivum L.). Crop. Res. 2007, 34, 166–170. [Google Scholar]
- Shamsi, K.; Petrosyan, M.; Noor-mohammadi, G.; Haghparast, A.; Kobrace, S.; Rasekhi, B. Differential agronomic responses of bread wheat cultivars to drought stress in the west of Iran. Afr. J. Biotechnol. 2011, 10, 2708–2715. [Google Scholar]
- Blanco, A.; de Giovanni, C.; Laddomada, B.; Sciancalepore, A.; Simeone, R.; Devos, K.M.; Gale, M.D. Quantitative trait loci influencing grain protein content in tetraploid wheats. Plant Breed. 1996, 115, 310–316. [Google Scholar] [CrossRef]
- Blanco, A.; Bellomo, M.P.; Cenci, A.; De Giovanni, C.; D’Ovidio, R.; Iacono, E.; Laddomada, B.; Pagnotta, M.A.; Porceddu, E.; Sciancalepore, A.; et al. A genetic linkage map of durum wheat. Theor. Appl. Genet. 1998, 97, 721–728. [Google Scholar] [CrossRef]
- Pasha, I.; Anjum, F.M.; Butt, M.S.; Sultan, J.I. Gluten quality prediction and correlation studies in spring wheats. J. Food Qual. 2007, 30, 438–449. [Google Scholar] [CrossRef]
- Shewry, P.R.; Tatham, A.S. Wheat. In The Royal Society of Chemistry; Elsevier Science B.V.: Amsterdam, The Netherlands, 2000; pp. 335–339. [Google Scholar]
- Oelofse, R.M.; Labuschagne, M.T.; van Deventer, C.S. Influencing factors of sodium dodecyl sulfate sedimentation in bread wheat. J. Cereal Sci. 2010, 52, 96–99. [Google Scholar] [CrossRef]
- Cubadda, R.E.; Carcea, M.; Marconi, E.; Trivisonno, M.C. Influence of protein content on durum wheat gluten strength determined by the SDS sedimentation test and by other methods. Cereal Foods World 2007, 52, 273–277. [Google Scholar] [CrossRef][Green Version]
- Carter, B.P.; Morris, C.F.; Anderson, J.A. Optimizing the SDS sedimentation test for end-use quality selection in a soft white and club wheat-breeding program. Cereal Chem. 1999, 76, 907–911. [Google Scholar] [CrossRef]
- De Villiers, O.T.; Laubscher, E.W. Use of the SDSS test to predict the protein content and bread volume of wheat cultivars. S. Afr. J. Plant Soil 1995, 12, 140–142. [Google Scholar] [CrossRef]
- Pogna, N.E.; Autran, J.C.; Mellini, F.; Lafiandra, D.; Feillet, P. Chromosome 1B-encoded gliadins and glutenins subunits. J. Cereal Sci. 1990, 11, 15–34. [Google Scholar] [CrossRef]
- Reif, J.C.; Gowda, M.; Maurer, H.P.; Longin, C.F.; Korzun, V.; Ebmeyer, E.; Bothe, R.; Pietsch, C.; Wurschum, T. Association mapping for quality traits in soft winter wheat. Theor. Appl. Genet. 2010, 122, 961–970. [Google Scholar] [CrossRef]
- Fiedler, J.D.; Salsman, E.; Liu, Y.; Michalak de Jimenez, M.; Hegstad, J.B.; Chen, B.; Manthey, F.A.; Chao, S.; Xu, S.; Elias, E.M.; et al. Genome-Wide Association and Prediction of Grain and Semolina Quality Traits in Durum Wheat Breeding Populations. Plant Genome 2017, 10. [Google Scholar] [CrossRef]
- Sykes, A.G. Plastocyanin and the Blue Copper Proteins. In Long-Range Electron Transfer in Biology. Structure and Bonding; Springer: Berlin/Heidelberg, Germany, 1990; pp. 175–224. [Google Scholar] [CrossRef]
- Feng, H.; Zhang, Q.; Wang, Q.; Wang, X.; Liu, J.; Li, M.; Huang, L.; Kang, Z. Target of tae-miR408, a chemocyanin-like protein gene (TaCLP1), plays positive roles in wheat response to high-salinity, heavy cupric stress and stripe rust. Plant Mol. Biol. 2013, 83, 433–443. [Google Scholar] [CrossRef]
- Yao, Z.J.; Lin, R.M.; Xu, S.C.; Li, Z.F.; Wan, A.M.; Ma, Z.Y. The molecular tagging of the yellow rust resistance gene Yr7 in wheat transferred from differential host Lee using microsatellite markers. Sci. Agric. Sin. 2006, 39, 1146–1152. [Google Scholar]
- Zhao, X.Y.; Hong, P.; Wu, J.Y.; Chen, X.B.; Ye, X.G.; Pan, Y.Y.; Wang, J.; Zhang, X.S. The tae-miR408-Mediated Control of TaTOC1 Genes Transcription Is Required for the Regulation of Heading Time in Wheat. Plant Physiol. 2016, 170, 1578–1594. [Google Scholar] [CrossRef]
- Liu, G.; Zhao, Y.; Gowda, M.; Longin, C.F.H.; Reif, J.C.; Mette, M.F. Predicting hybrid performances for quality traits through genomic-assisted approaches in central European wheat. PLoS ONE 2016, 11, e0158635. [Google Scholar] [CrossRef] [PubMed]
- Maccaferri, M.; Zhang, J.; Bulli, P.; Abate, Z.; Chao, S.; Cantu, D.; Bossolini, E.; Chen, X.; Pumphrey, M.; Dubcovsky, J. A genome-wide association study of resistance to stripe rust (Puccinia striiformis f. sp. tritici) in a worldwide collection of hexaploid spring wheat (Triticum aestivum L.). G3 Genes Genomes Genet. 2015, 5, 449–465. [Google Scholar] [CrossRef]
- Liu, W.; Maccaferri, M.; Rynearson, S.; Letta, T.; Zegeye, H.; Tuberosa, R.; Chen, X.; Pumphrey, M. Novel Sources of Stripe Rust Resistance Identified by Genome-Wide Association Mapping in Ethiopian Durum Wheat (Triticum turgidum ssp. durum). Front. Plant Sci. 2017, 8, 774. [Google Scholar] [CrossRef] [PubMed]
- Godoy, J.G.; Rynearson, S.; Chen, X.; Pumphrey, M. Genome-Wide Association Mapping of Loci for Resistance to Stripe Rust in North American Elite Spring Wheat Germplasm. Phytopathology 2018, 108, 234–245. [Google Scholar] [CrossRef]
- Liu, W.; Naruoka, Y.; Miller, K.; Garland-Campbell, K.A.; Carter, A.H. Characterizing and Validating Stripe Rust Resistance Loci in US Pacific Northwest Winter Wheat Accessions (Triticum aestivum L.) by Genome-wide Association and Linkage Mapping. Plant Genome 2018, 11. [Google Scholar] [CrossRef]
- Lemoine, R. Sucrose transporters in plants: Update on function and structure. Biochim. Biophys. Acta 1999, 1465, 246–262. [Google Scholar] [CrossRef]
- Koch, K.E. Carbohydrate-modulated gene expression in plants. Annu. Rev. Plant Physiol. Plant Mole Biol. 1996, 47, 509–540. [Google Scholar] [CrossRef]
- Smeekens, S.; Rook, F. Sugar sensing and sugar-mediated signal transduction in plants. Plant Physiol 1997, 115, 7–13. [Google Scholar] [CrossRef]
- Sun, X.-Y.; Wu, K.; Zhao, Y.; Kong, F.-M.; Han, G.-Z.; Jiang, H.-M.; Huang, X.-J.; Li, R.-J.; Wang, H.-G.; Li, S.-S. QTL analysis of kernel shape and weight using recombinant inbred lines in wheat. Euphytica 2008, 165, 615–624. [Google Scholar] [CrossRef]
- Patil, R.M.; Tamhankar, S.A.; Oak, M.D.; Raut, A.L.; Honrao, B.K.; Rao, V.S.; Misra, S.C. Mapping of QTL for agronomic traits and kernel characters in durum wheat (Triticum durum Desf.). Euphytica 2013, 190, 117–129. [Google Scholar] [CrossRef]
- Cui, F.; Fan, X.; Chen, M.; Zhang, N.; Zhao, C.; Zhang, W.; Han, J.; Ji, J.; Zhao, X.; Yang, L.; et al. QTL detection for wheat kernel size and quality and the responses of these traits to low nitrogen stress. Theor. Appl. Genet. 2016, 129, 469–484. [Google Scholar] [CrossRef] [PubMed]
- Juliana, P.; Poland, J.A.; Huerta-Espino, J.; Shrestha, S.; Crossa, J.; Crespo-Herrera, L.; Henrique Toledo, F.; Govidan, V.; Mondal, S.; Kumar, U.; et al. Improving grain yield, stress resilience and quality of bread wheat using large-scale genomics. Nat. Genet. 2019. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.H.; Wei, X.Y.; Yuan, B.; Yao, L.B.; Ma, T.T.; Zhang, P.P.; Wang, X.; Wang, P.Q.; Liu, W.T.; Li, W.Q.; et al. Genome-Wide Identification and Functional Analysis of NADPH Oxidase Family Genes in Wheat During Development and Environmental Stress Responses. Front. Plant Sci. 2018, 9, 906. [Google Scholar] [CrossRef] [PubMed]
- Kaya, H.; Nakajima, R.; Iwano, M.; Kanaoka, M.M.; Kimura, S.; Takeda, S.; Kawarazaki, T.; Senzaki, E.; Hamamura, Y.; Higashiyama, T.; et al. Ca2+-activated reactive oxygen species production by Arabidopsis RbohH and RbohJ is essential for proper pollen tube tip growth. Plant Cell 2014, 26, 1069–1080. [Google Scholar] [CrossRef]
- Gupta, D.K.; Pena, L.B.; Romero-Puertas, M.C.; Hernández, A.; Inouhe, M.; Sandalio, L.M. NADPH oxidases differentially regulate ROS metabolism and nutrient uptake under cadmium toxicity. Plant Cell Environ. 2017, 40, 509–526. [Google Scholar] [CrossRef]
- Fu, W.; Shen, Y.; Hao, J.; Wu, J.; Ke, L.; Wu, C.; Huang, K.; Luo, B.; Xu, M.; Cheng, X.; et al. Acyl-CoA N-acyltransferase influences fertility by regulating lipid metabolism and jasmonic acid biogenesis in cotton. Sci. Rep. 2015, 5, 11790. [Google Scholar] [CrossRef]
- Sui, N.; Li, M.; Meng, Q.-W.; Tian, J.-C.; Zhao, S.-J. Photosynthetic Characteristics of a Super High Yield Cultivar of Winter Wheat During Late Growth Period. Agric. Sci. China 2010, 9, 346–354. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).