Injectable Hydrogel-Encapsulating Pickering Emulsion for Overcoming Lenvatinib-Resistant Hepatocellular Carcinoma via Cuproptosis Induction and Stemness Inhibition
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials, Cell Lines, and Mice
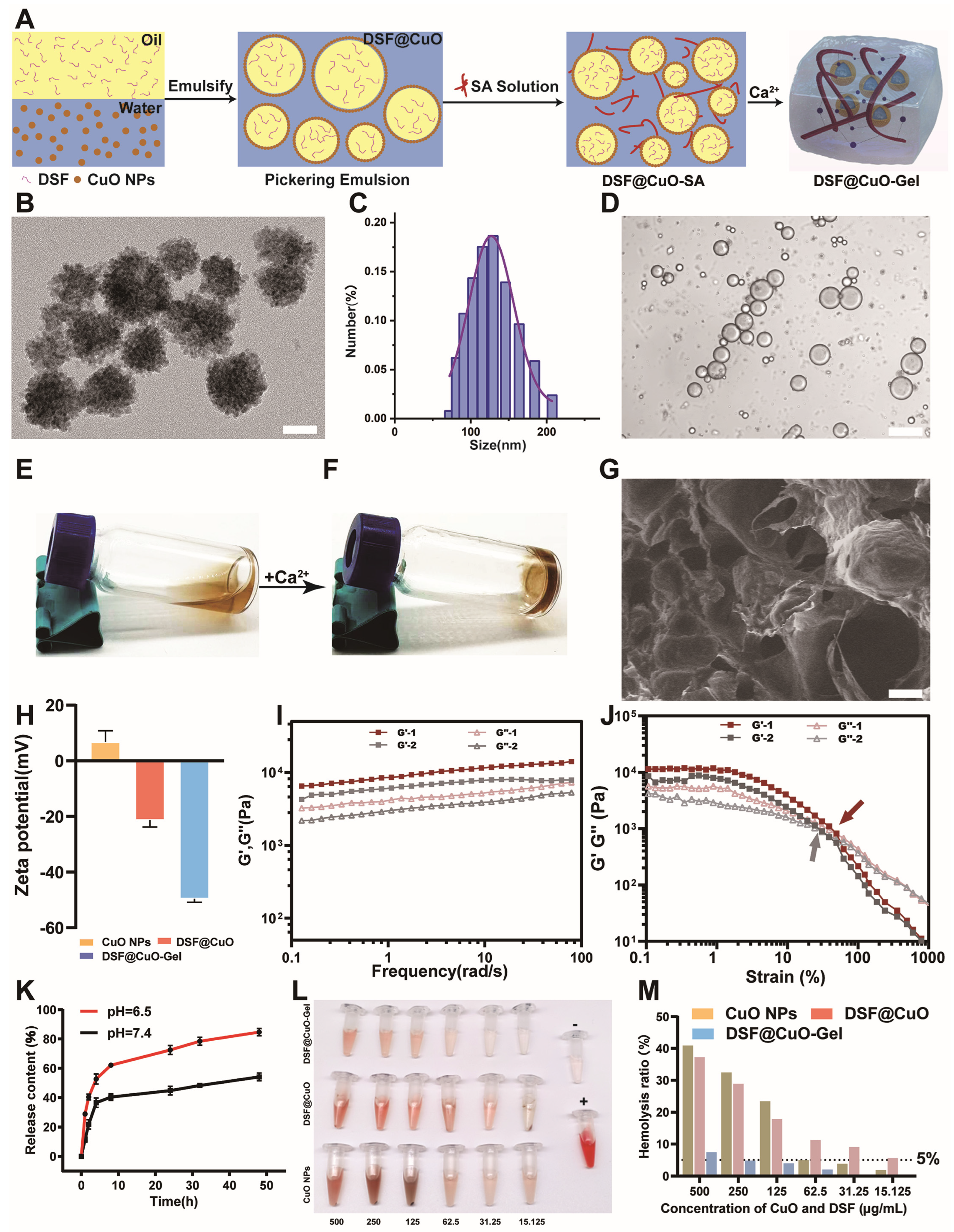
2.2. Preparation and Characterization of CuO NPs
2.3. Preparation and Characterization of DSF@CuO
2.4. Preparation and Characterization of DSF@CuO Gel
2.5. Rheological Characterization
2.6. Hemolysis Assay
2.7. Differential Gene Selection and Gene Enrichment Analysis
2.8. GSEA and GSVA Analysis
2.9. Cytotoxicity Evaluation and IC50
2.10. Inhibition of LenR HCC Cell Spheroid Formation Experiment
2.11. Cell Cloning Experiment
2.12. Transwell Experiment
2.13. Western Blot for Cell Protein Expression
2.14. In Vivo Treatment of LenR BALB/c Nude Mice Model
2.15. In Vivo Drug Retention Experiment
2.16. Evaluation of In Situ HCC Model Treatment Efficacy
2.17. Immunofluorescence Staining
2.18. In Vivo Biosafety Assessment in Mice
2.19. Statistical Analysis
3. Results and Discussion
3.1. Preparation and Characterization of Injectable Hydrogel
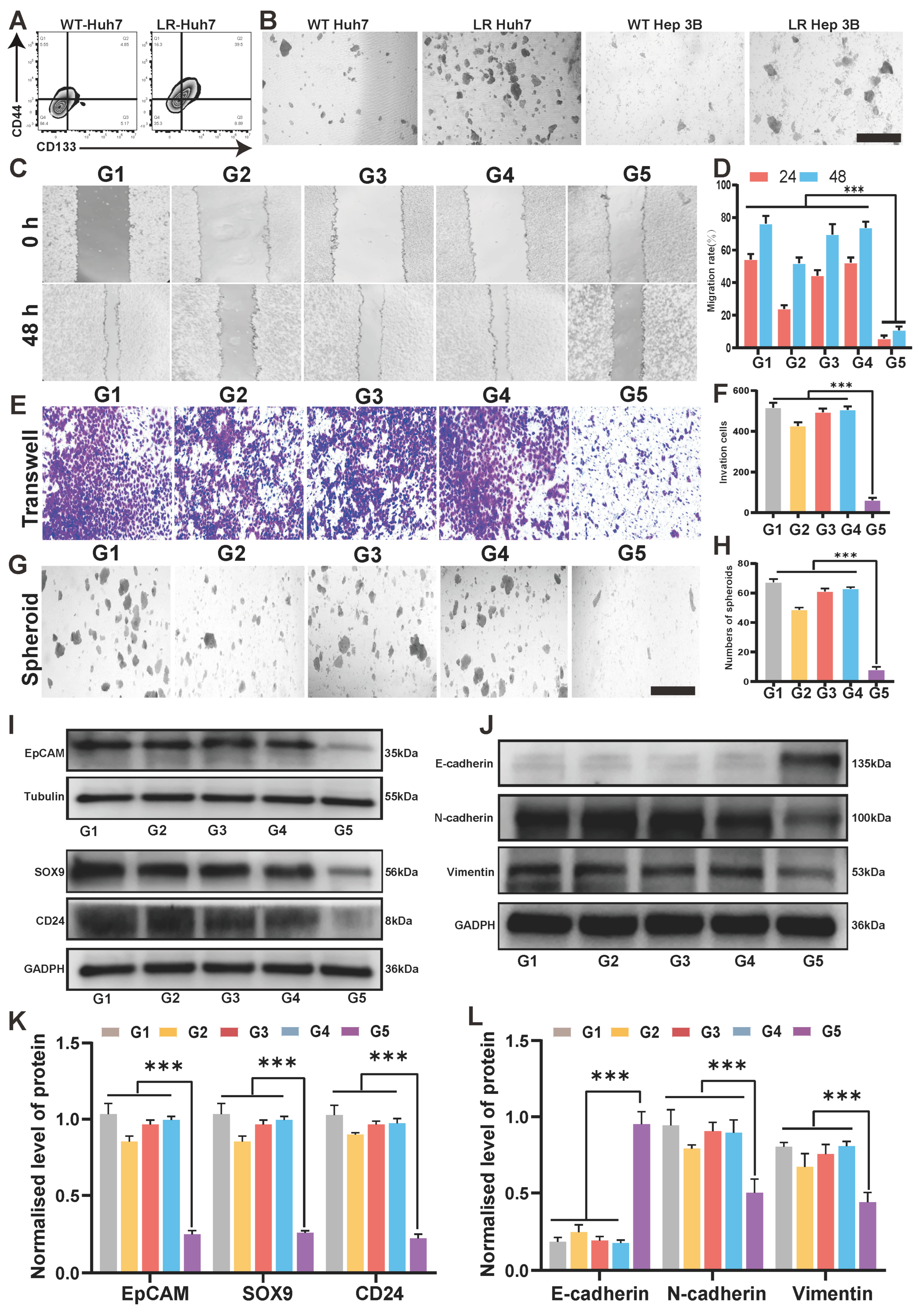
3.2. Cytotoxicity of DSF@CuO in LenR HCC
3.3. DSF@CuO Can Effectively Induce LenR HCC Cuproptosis
3.4. DSF@CuO Could Inhibit the Cell Stemness of LenR HCC
3.5. The Mechanism of DSF@CuO in Overcoming LenR HCC
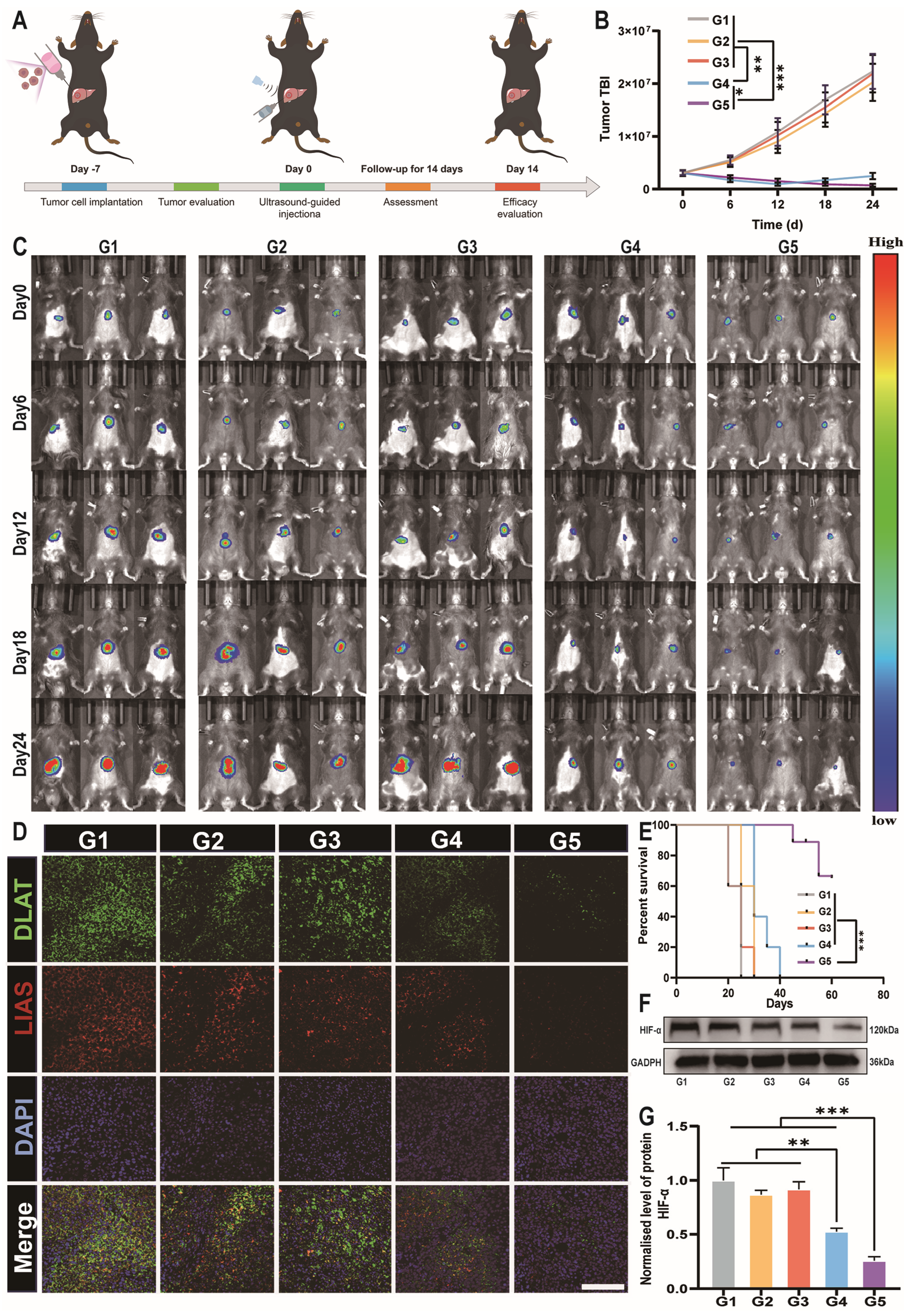
3.6. DSF@CuO Gel Could Overcome LenR In Vivo
3.7. DSF@CuO Gel Could Overcome In Situ HCC Model
3.8. Biosafety Evaluation of DSF@CuO Gel
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer statistics, 2024. CA Cancer J. Clin. 2024, 74, 12–49. [Google Scholar] [CrossRef] [PubMed]
- Mo, D.C.; Jia, R.R.; Zhong, J.H. Letter to the Editor: Hepatic Resection Compared to Chemoembolization in Intermediate- to Advanced-Stage Hepatocellular Carcinoma: A Comment For Moving Forward. Hepatology 2019, 70, 446–447. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.S.; Ikeda, M.; Zhu, A.X.; Sung, M.W.; Baron, A.D.; Kudo, M.; Okusaka, T.; Kobayashi, M.; Kumada, H.; Kaneko, S.; et al. Phase Ib Study of Lenvatinib Plus Pembrolizumab in Patients with Unresectable Hepatocellular Carcinoma. J. Clin. Oncol. 2020, 38, 2960–2970. [Google Scholar] [CrossRef] [PubMed]
- Kudo, M.; Finn, R.S.; Qin, S.; Han, K.H.; Ikeda, K.; Piscaglia, F.; Baron, A.; Park, J.W.; Han, G.; Jassem, J.; et al. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: A randomised phase 3 non-inferiority trial. Lancet 2018, 391, 1163–1173. [Google Scholar] [CrossRef]
- Jin, H.; Shi, Y.; Lv, Y.; Yuan, S.; Ramirez, C.F.A.; Lieftink, C.; Wang, L.; Wang, S.; Wang, C.; Dias, M.H.; et al. EGFR activation limits the response of liver cancer to lenvatinib. Nature 2021, 595, 730–734. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Tian, H.; Li, B.; Li, L.; Jiang, H.; Gao, Y.; Zheng, L.; Huang, C.; Zhou, Y.; Du, Z.; et al. An Ellagic Acid Coordinated Copper-Based Nanoplatform for Efficiently Overcoming Cancer Chemoresistance by Cuproptosis and Synergistic Inhibition of Cancer Cell Stemness. Small 2024, 20, e2309215. [Google Scholar] [CrossRef]
- Xiang, D.M.; Sun, W.; Zhou, T.; Zhang, C.; Cheng, Z.; Li, S.C.; Jiang, W.; Wang, R.; Fu, G.; Cui, X.; et al. Oncofetal HLF transactivates c-Jun to promote hepatocellular carcinoma development and sorafenib resistance. Gut 2019, 68, 1858–1871. [Google Scholar] [CrossRef]
- Wang, J.; Yu, H.; Dong, W.; Zhang, C.; Hu, M.; Ma, W.; Jiang, X.; Li, H.; Yang, P.; Xiang, D. N6-Methyladenosine-Mediated Up-Regulation of FZD10 Regulates Liver Cancer Stem Cells’ Properties and Lenvatinib Resistance Through WNT/β-Catenin and Hippo Signaling Pathways. Gastroenterology 2023, 164, 990–1005. [Google Scholar] [CrossRef]
- Ladd, A.D.; Duarte, S.; Sahin, I.; Zarrinpar, A. Mechanisms of drug resistance in HCC. Hepatology 2024, 79, 926–940. [Google Scholar] [CrossRef]
- Lee, T.K.; Guan, X.Y.; Ma, S. Cancer stem cells in hepatocellular carcinoma—From origin to clinical implications. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 26–44. [Google Scholar] [CrossRef]
- Zheng, P.; Wu, Y.; Wang, Y.; Hu, F. Disulfiram suppresses epithelial-mesenchymal transition (EMT), migration and invasion in cervical cancer through the HSP90A/NDRG1 pathway. Cell Signal. 2023, 109, 110771. [Google Scholar] [CrossRef] [PubMed]
- Pastushenko, I.; Mauri, F.; Song, Y.; de Cock, F.; Meeusen, B.; Swedlund, B.; Impens, F.; Van Haver, D.; Opitz, M.; Thery, M.; et al. Fat1 deletion promotes hybrid EMT state, tumour stemness and metastasis. Nature 2021, 589, 448–455. [Google Scholar] [CrossRef] [PubMed]
- Skrott, Z.; Mistrik, M.; Andersen, K.K.; Friis, S.; Majera, D.; Gursky, J.; Ozdian, T.; Bartkova, J.; Turi, Z.; Moudry, P.; et al. Alcohol-abuse drug disulfiram targets cancer via p97 segregase adaptor NPL4. Nature 2017, 552, 194–199. [Google Scholar] [CrossRef]
- Lu, Y.; Pan, Q.; Gao, W.; Pu, Y.; Luo, K.; He, B.; Gu, Z. Leveraging disulfiram to treat cancer: Mechanisms of action, delivery strategies, and treatment regimens. Biomaterials 2022, 281, 121335. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Zhang, H.; Huang, L.; Sun, C.; Yue, Y.; Cao, X.; Jia, H.; Wang, C.; Gao, Y. Disulfiram with Cu2+ alleviates dextran sulfate sodium-induced ulcerative colitis in mice. Theranostics 2023, 13, 2879–2895. [Google Scholar] [CrossRef]
- Tsvetkov, P.; Coy, S.; Petrova, B.; Dreishpoon, M.; Verma, A.; Abdusamad, M.; Rossen, J.; Joesch-Cohen, L.; Humeidi, R.; Spangler, R.D.; et al. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science 2022, 375, 1254–1261. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, Y.; Zhang, J.; Yang, Y.; Fleishman, J.S.; Wang, Y.; Wang, J.; Chen, J.; Li, Y.; Wang, H. Cuproptosis: A novel therapeutic target for overcoming cancer drug resistance. Drug Resist. Updates 2024, 72, 101018. [Google Scholar] [CrossRef]
- Oliveri, V. Selective Targeting of Cancer Cells by Copper Ionophores: An Overview. Front. Mol. Biosci. 2022, 9, 841814. [Google Scholar] [CrossRef]
- Vyas, A.; Harbison, R.A.; Faden, D.L.; Kubik, M.; Palmer, D.; Zhang, Q.; Osmanbeyoglu, H.U.; Kiselyov, K.; Méndez, E.; Duvvuri, U. Recurrent Human Papillomavirus-Related Head and Neck Cancer Undergoes Metabolic Reprogramming and Is Driven by Oxidative Phosphorylation. Clin. Cancer Res. 2021, 27, 6250–6264. [Google Scholar] [CrossRef]
- Yang, G.G.; Zhou, D.J.; Pan, Z.Y.; Yang, J.; Zhang, D.Y.; Cao, Q.; Ji, L.N.; Mao, Z.W. Multifunctional low-temperature photothermal nanodrug with in vivo clearance, ROS-Scavenging and anti-inflammatory abilities. Biomaterials 2019, 216, 119280. [Google Scholar] [CrossRef]
- Majumder, S.; Dutta, P.; Mookerjee, A.; Choudhuri, S.K. The role of a novel copper complex in overcoming doxorubicin resistance in Ehrlich ascites carcinoma cells in vivo. Chem.-Biol. Interact. 2006, 159, 90–103. [Google Scholar] [CrossRef] [PubMed]
- Kucinska, M.; Pospieszna, J.; Tang, J.; Lisiak, N.; Toton, E.; Rubis, B.; Murias, M. The combination therapy using tyrosine kinase receptors inhibitors and repurposed drugs to target patient-derived glioblastoma stem cells. Biomed. Pharmacother. 2024, 176, 116892. [Google Scholar] [CrossRef] [PubMed]
- Johansson, B. A review of the pharmacokinetics and pharmacodynamics of disulfiram and its metabolites. Acta Psychiatr. Scand. Suppl. 1992, 369, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Kannappan, V.; Ali, M.; Small, B.; Rajendran, G.; Elzhenni, S.; Taj, H.; Wang, W.; Dou, Q.P. Recent Advances in Repurposing Disulfiram and Disulfiram Derivatives as Copper-Dependent Anticancer Agents. Front. Mol. Biosci. 2021, 8, 741316. [Google Scholar] [CrossRef]
- Butcher, K.; Kannappan, V.; Kilari, R.S.; Morris, M.R.; McConville, C.; Armesilla, A.L.; Wang, W. Investigation of the key chemical structures involved in the anticancer activity of disulfiram in A549 non-small cell lung cancer cell line. BMC Cancer 2018, 18, 753. [Google Scholar] [CrossRef] [PubMed]
- Uriu-Adams, J.Y.; Keen, C.L. Copper, oxidative stress, and human health. Mol. Asp. Med. 2005, 26, 268–298. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Bao, X.; Yao, P. Protamine and BSA-dextran complex emulsion improves oral bioavailability and anti-tumor efficacy of paclitaxel. Drug Deliv. 2020, 27, 1360–1368. [Google Scholar] [CrossRef]
- Pan, J.; Chen, J.; Wang, X.; Wang, Y.; Fan, J.-B. Pickering emulsion: From controllable fabrication to biomedical application. Interdiscip. Med. 2023, 1, e20230014. [Google Scholar] [CrossRef]
- Jeong, J.; Kim, S.H.; Lee, S.; Lee, D.K.; Han, Y.; Jeon, S.; Cho, W.S. Differential Contribution of Constituent Metal Ions to the Cytotoxic Effects of Fast-Dissolving Metal-Oxide Nanoparticles. Front. Pharmacol. 2018, 9, 15. [Google Scholar] [CrossRef]
- Al-Musawi, M.M.S.; Al-Shmgani, H.; Al-Bairuty, G.A. Histopathological and Biochemical Comparative Study of Copper Oxide Nanoparticles and Copper Sulphate Toxicity in Male Albino Mice Reproductive System. Int. J. Biomater. 2022, 2022, 4877637. [Google Scholar] [CrossRef]
- Chakraborty, A.; Alexander, S.; Luo, W.; Al-Salam, N.; Van Oirschot, M.; Ranganath, S.H.; Chakrabarti, S.; Paul, A. Engineering multifunctional adhesive hydrogel patches for biomedical applications. Interdiscip. Med. 2023, 1, e20230008. [Google Scholar] [CrossRef]
- Zhang, S.; Hong, B.; Fan, Z.; Lu, J.; Xu, Y.; Pera-Titus, M. Aquivion-Carbon Composites with Tunable Amphiphilicity for Pickering Interfacial Catalysis. ACS Appl. Mater. Interfaces 2018, 10, 26795–26804. [Google Scholar] [CrossRef]
- Wang, L.; Yu, Y.; Wei, D.; Zhang, L.; Zhang, X.; Zhang, G.; Ding, D.; Xiao, H.; Zhang, D. A Systematic Strategy of Combinational Blow for Overcoming Cascade Drug Resistance via NIR-Light-Triggered Hyperthermia. Adv. Mater. 2021, 33, e2100599. [Google Scholar] [CrossRef]
- Huang, L.; Zhu, J.; Xiong, W.; Feng, J.; Yang, J.; Lu, X.; Lu, Y.; Zhang, Q.; Yi, P.; Feng, Y.; et al. Tumor-Generated Reactive Oxygen Species Storm for High-Performance Ferroptosis Therapy. ACS Nano 2023, 17, 11492–11506. [Google Scholar] [CrossRef] [PubMed]
- Shimada, K.; Reznik, E.; Stokes, M.E.; Krishnamoorthy, L.; Bos, P.H.; Song, Y.; Quartararo, C.E.; Pagano, N.C.; Carpizo, D.R.; deCarvalho, A.C.; et al. Copper-Binding Small Molecule Induces Oxidative Stress and Cell-Cycle Arrest in Glioblastoma-Patient-Derived Cells. Cell Chem. Biol. 2018, 25, 585–594.e7. [Google Scholar] [CrossRef]
- Yip, N.C.; Fombon, I.S.; Liu, P.; Brown, S.; Kannappan, V.; Armesilla, A.L.; Xu, B.; Cassidy, J.; Darling, J.L.; Wang, W. Disulfiram modulated ROS-MAPK and NFκB pathways and targeted breast cancer cells with cancer stem cell-like properties. Br. J. Cancer 2011, 104, 1564–1574. [Google Scholar] [CrossRef]
- Wang, X.; Shi, Y.; Shi, H.; Liu, X.; Liao, A.; Liu, Z.; Orlowski, R.Z.; Zhang, R.; Wang, H. MUC20 regulated by extrachromosomal circular DNA attenuates proteasome inhibitor resistance of multiple myeloma by modulating cuproptosis. J. Exp. Clin. Cancer Res. 2024, 43, 68. [Google Scholar] [CrossRef] [PubMed]
- Mok, E.H.K.; Leung, C.O.N.; Zhou, L.; Lei, M.M.L.; Leung, H.W.; Tong, M.; Wong, T.L.; Lau, E.Y.T.; Ng, I.O.L.; Ding, J.; et al. Caspase-3-Induced Activation of SREBP2 Drives Drug Resistance via Promotion of Cholesterol Biosynthesis in Hepatocellular Carcinoma. Cancer Res. 2022, 82, 3102–3115. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.L.; Hu, B.; Tang, W.G.; Xie, S.H.; Ren, N.; Guo, L.; Lu, R.Q. CD73 sustained cancer-stem-cell traits by promoting SOX9 expression and stability in hepatocellular carcinoma. J. Hematol. Oncol. 2020, 13, 11. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, X.; Wang, Y.; Lai, S.; Wang, Z.; Yang, Y.; Liu, W.; Wang, H.; Tang, B. The m(6)A demethylase ALKBH5-mediated upregulation of DDIT4-AS1 maintains pancreatic cancer stemness and suppresses chemosensitivity by activating the mTOR pathway. Mol. Cancer 2022, 21, 174. [Google Scholar] [CrossRef]
- Xiong, Y.X.; Zhang, X.C.; Zhu, J.H.; Zhang, Y.X.; Pan, Y.L.; Wu, Y.; Zhao, J.P.; Liu, J.J.; Lu, Y.X.; Liang, H.F.; et al. Collagen I-DDR1 signaling promotes hepatocellular carcinoma cell stemness via Hippo signaling repression. Cell Death Differ. 2023, 30, 1648–1665. [Google Scholar] [CrossRef] [PubMed]
- Nevi, L.; Di Matteo, S.; Carpino, G.; Zizzari, I.G.; Samira, S.; Ambrosino, V.; Costantini, D.; Overi, D.; Giancotti, A.; Monti, M.; et al. DCLK1, a Putative Stem Cell Marker in Human Cholangiocarcinoma. Hepatology 2021, 73, 144–159. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.; Chen, J.; Di, Z.; Yuan, W.; Zhou, Z.; Liu, Z.; Han, S.; Liu, Y.; Ying, G.; Shu, X.; et al. TM4SF1 promotes EMT and cancer stemness via the Wnt/β-catenin/SOX2 pathway in colorectal cancer. J. Exp. Clin. Cancer Res. 2020, 39, 232. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Liu, L.; Chen, X.; Cheng, J.; Zhang, H.; Shen, J.; Shan, J.; Xu, Y.; Yang, Z.; Lai, M.; et al. Sox9 regulates self-renewal and tumorigenicity by promoting symmetrical cell division of cancer stem cells in hepatocellular carcinoma. Hepatology 2016, 64, 117–129. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Li, H.; Zhang, B.; Xiong, Z.; Jin, Z.; Chen, J.; Zheng, Y.; Zhu, X.; Zhang, S. ABI2-mediated MEOX2/KLF4-NANOG axis promotes liver cancer stem cell and drives tumour recurrence. Liver Int. 2022, 42, 2562–2576. [Google Scholar] [CrossRef]
- Kudaravalli, S.; den Hollander, P.; Mani, S.A. Role of p38 MAP kinase in cancer stem cells and metastasis. Oncogene 2022, 41, 3177–3185. [Google Scholar] [CrossRef]
- Bharti, R.; Dey, G.; Lin, F.; Lathia, J.; Reizes, O. CD55 in cancer: Complementing functions in a non-canonical manner. Cancer Lett. 2022, 551, 215935. [Google Scholar] [CrossRef]
- de Castro, L.R.; de Oliveira, L.D.; Milan, T.M.; Eskenazi, A.P.E.; Bighetti-Trevisan, R.L.; de Almeida, O.G.G.; Amorim, M.L.M.; Squarize, C.H.; Castilho, R.M.; de Almeida, L.O. Up-regulation of TNF-alpha/NFkB/SIRT1 axis drives aggressiveness and cancer stem cells accumulation in chemoresistant oral squamous cell carcinoma. J. Cell. Physiol. 2024, 239, e31164. [Google Scholar] [CrossRef]
- Méndez-Blanco, C.; Fondevila, F.; García-Palomo, A.; González-Gallego, J.; Mauriz, J.L. Sorafenib resistance in hepatocarcinoma: Role of hypoxia-inducible factors. Exp. Mol. Med. 2018, 50, 1–9. [Google Scholar] [CrossRef]
- Gong, M.; Yang, H.; Zhang, S.; Yang, Y.; Zhang, D.; Qi, Y.; Zou, L. Superparamagnetic core/shell GoldMag nanoparticles: Size-, concentration- and time-dependent cellular nanotoxicity on human umbilical vein endothelial cells and the suitable conditions for magnetic resonance imaging. J. Nanobiotechnol. 2015, 13, 24. [Google Scholar] [CrossRef]
- Zhao, Z.; Li, Q.; Qin, X.; Zhang, M.; Du, Q.; Luan, Y. An Injectable Hydrogel Reshaping Adenosinergic Axis for Cancer Therapy. Adv. Funct. Mater. 2022, 32, 2200801. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Tang, C.; Ye, H.; Fang, C. Injectable Hydrogel-Encapsulating Pickering Emulsion for Overcoming Lenvatinib-Resistant Hepatocellular Carcinoma via Cuproptosis Induction and Stemness Inhibition. Polymers 2024, 16, 2418. https://doi.org/10.3390/polym16172418
Li X, Tang C, Ye H, Fang C. Injectable Hydrogel-Encapsulating Pickering Emulsion for Overcoming Lenvatinib-Resistant Hepatocellular Carcinoma via Cuproptosis Induction and Stemness Inhibition. Polymers. 2024; 16(17):2418. https://doi.org/10.3390/polym16172418
Chicago/Turabian StyleLi, Xin, Chuanyu Tang, Hanjie Ye, and Chihua Fang. 2024. "Injectable Hydrogel-Encapsulating Pickering Emulsion for Overcoming Lenvatinib-Resistant Hepatocellular Carcinoma via Cuproptosis Induction and Stemness Inhibition" Polymers 16, no. 17: 2418. https://doi.org/10.3390/polym16172418
APA StyleLi, X., Tang, C., Ye, H., & Fang, C. (2024). Injectable Hydrogel-Encapsulating Pickering Emulsion for Overcoming Lenvatinib-Resistant Hepatocellular Carcinoma via Cuproptosis Induction and Stemness Inhibition. Polymers, 16(17), 2418. https://doi.org/10.3390/polym16172418






